Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems
Abstract
1. Introduction
2. Biosensors for Measuring OoC’ Metabolic Activity
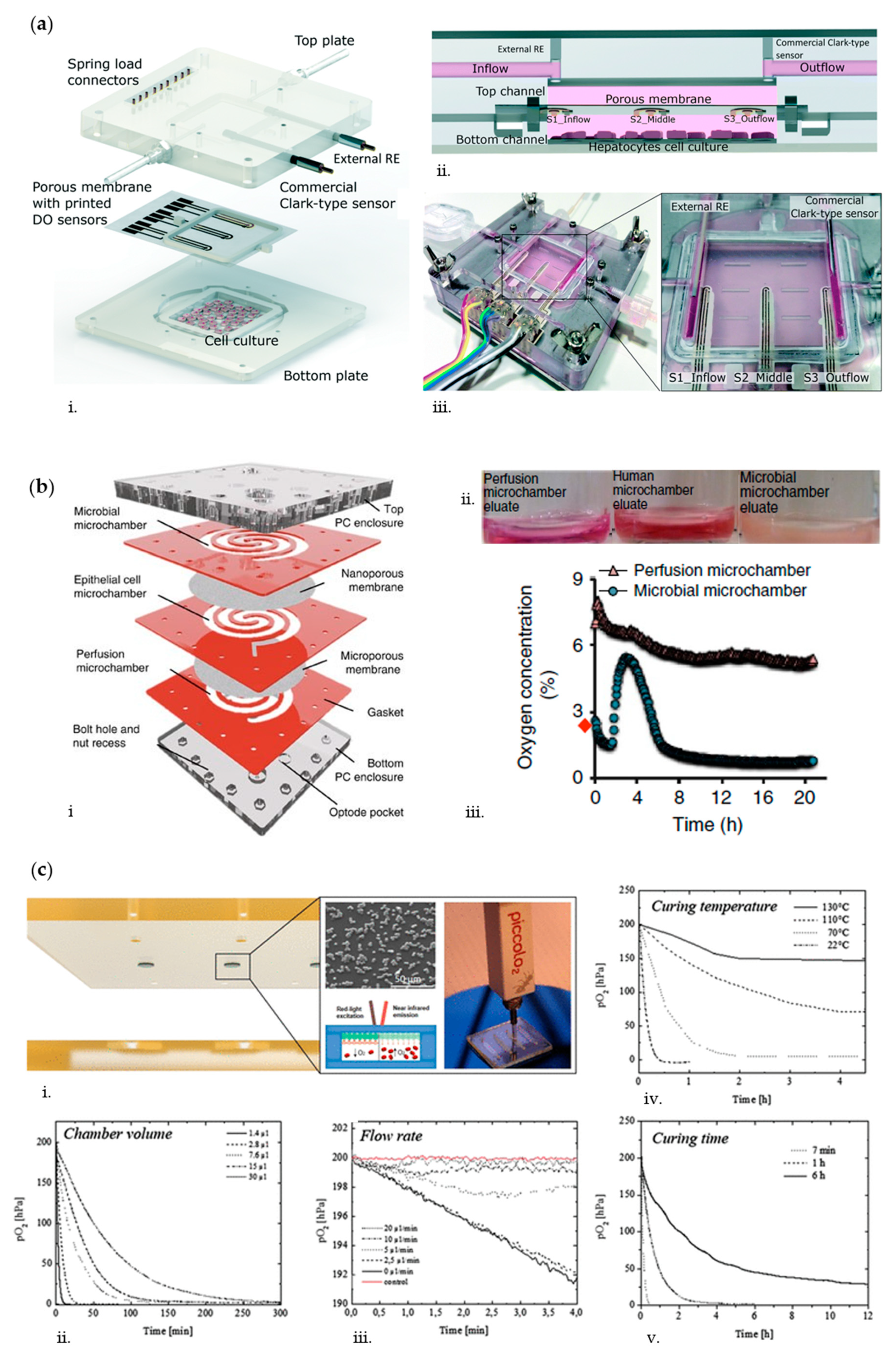
2.1. Oxygen
2.2. Glucose and Lactate
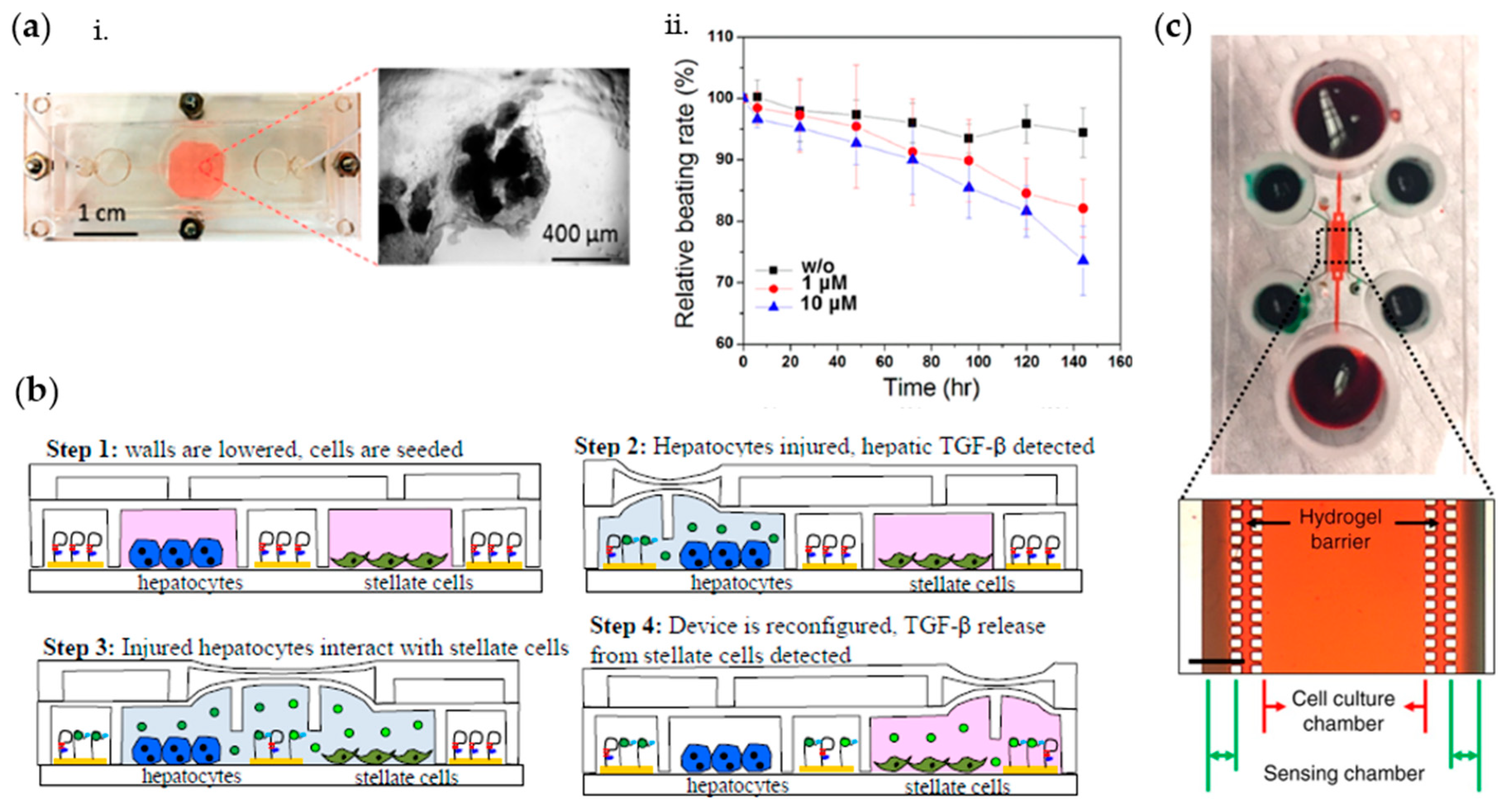
2.3. Cytokines and Other Metabolites
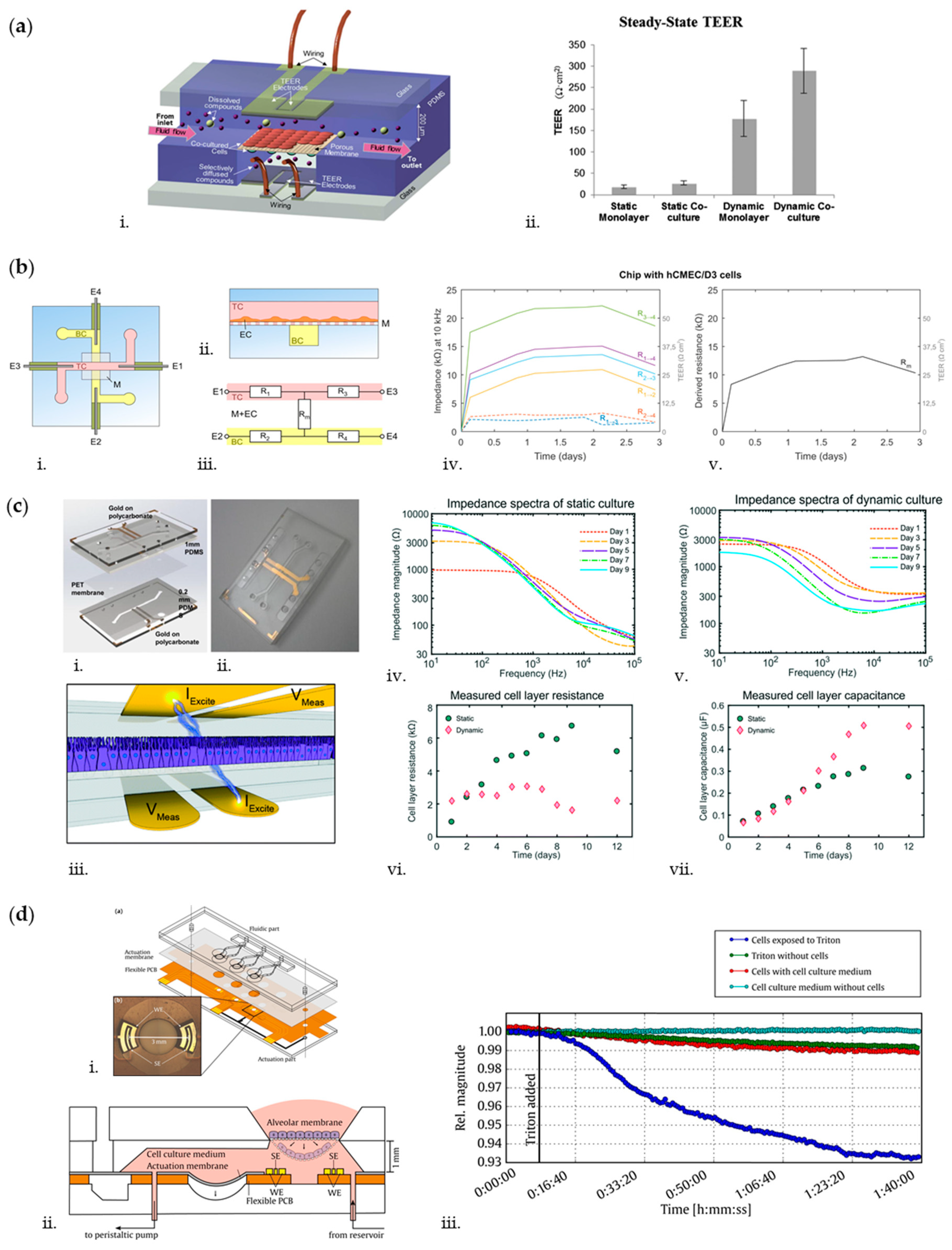
3. Biosensors for Measuring OoC’ Endothelial and Epithelial Barrier-Related Features
4. Biosensors for Measuring OoC’ Electrical Activity
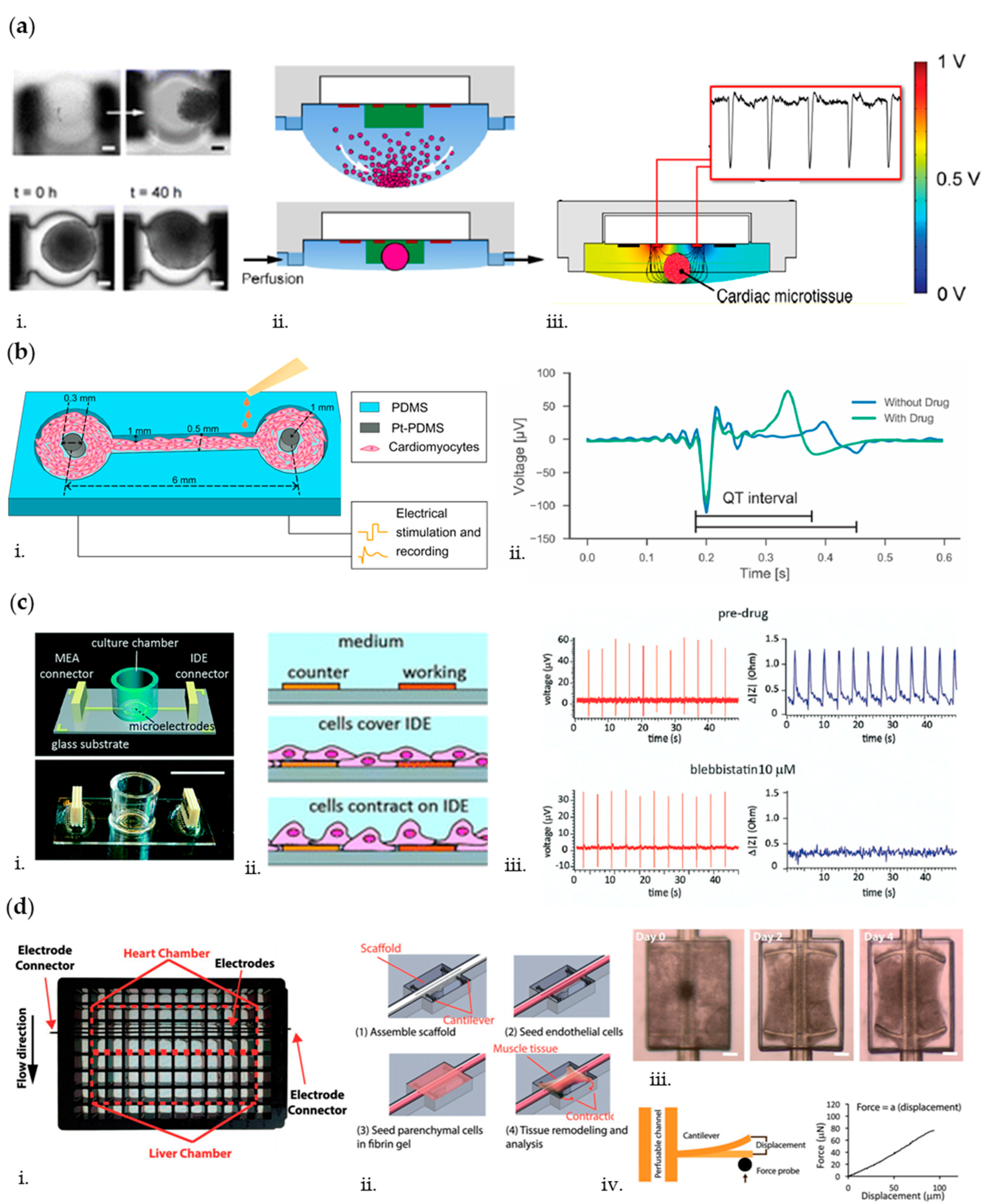
5. Biosensors for Measuring OoC’ Mechanical Activity
6. Biosensors for Measuring OoC’ Electromechanical Activity
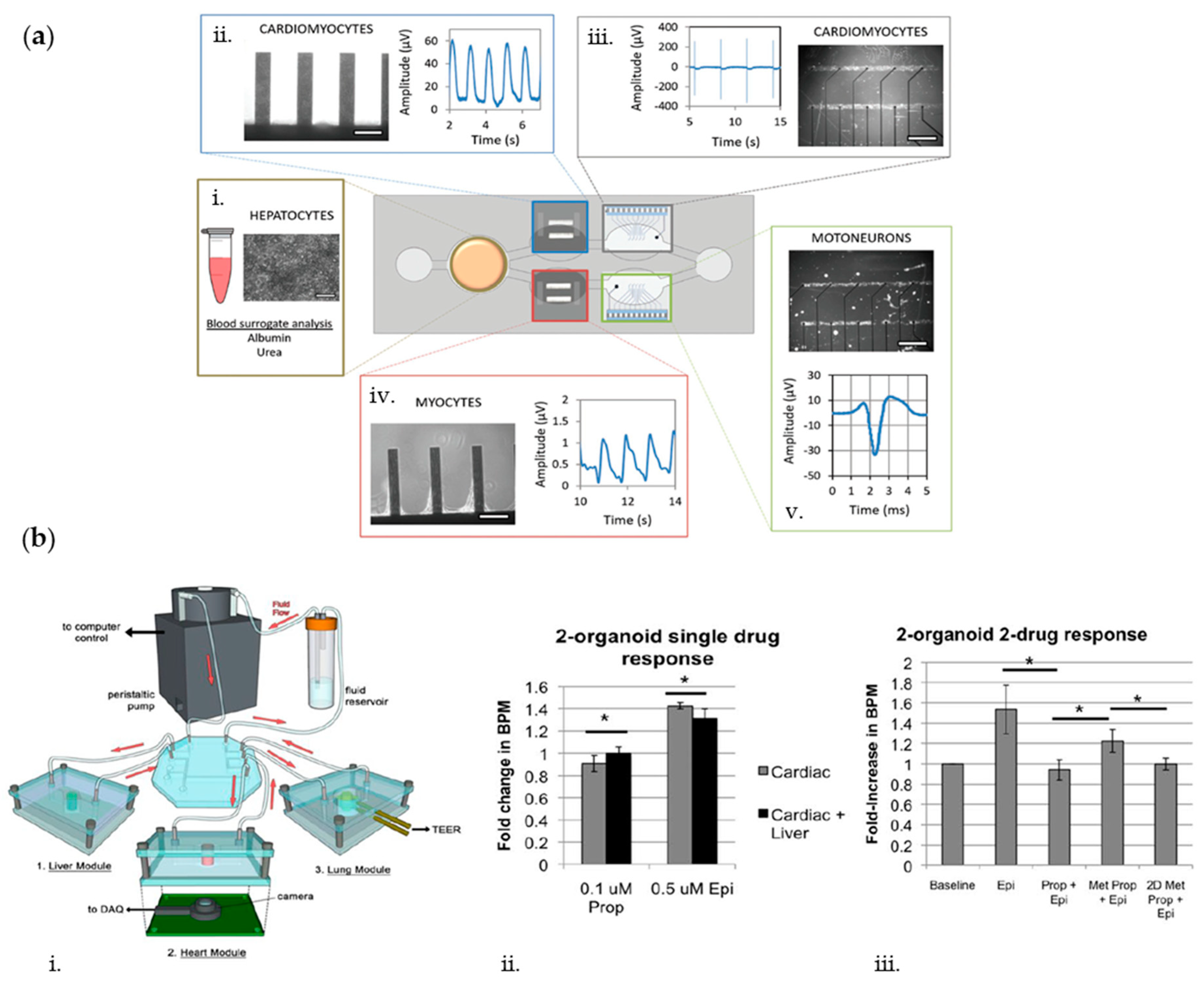
7. Multisensors for Analysis of Multiorgans-on-Chip
8. Outlook and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Caplin, J.D.; Granados, N.G.; James, M.R.; Montazami, R.; Hashemi, N. Microfluidic Organ-on-a-Chip Technology for Advancement of Drug Development and Toxicology. Adv. Healthc. Mater. 2015, 4, 1426–1450. [Google Scholar] [CrossRef] [PubMed]
- Esch, E.W.; Bahinski, A.; Huh, D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015, 14, 248–260. [Google Scholar] [CrossRef]
- Li, X.; Tian, T. Recent advances in an organ-on-a-chip: Biomarker analysis and applications. Anal. Methods 2018, 10, 3122–3130. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 6194. [Google Scholar] [CrossRef]
- Ribeiro, A.J.S.; Yang, X.; Patel, V.; Madabushi, R.; Strauss, D.G. Liver Microphysiological Systems for Predicting and Evaluating Drug Effects. Clin. Pharmacol. Ther. 2019, 106, 139–147. [Google Scholar] [CrossRef]
- Jorfi, M.; D’Avanzo, C.; Kim, D.Y.; Irimia, D. Three-Dimensional Models of the Human Brain Development and Diseases. Adv. Healthc. Mater. 2018, 7, 1–36. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 1–14. [Google Scholar] [CrossRef]
- Ugolini, G.S.; Cruz-Moreira, D.; Visone, R.; Redaelli, A.; Rasponi, M. Microfabricated physiological models for in vitro drug screening applications. Micromachines 2016, 7, 233. [Google Scholar] [CrossRef]
- Zheng, F.; Fu, F.; Cheng, Y.; Wang, C.; Zhao, Y.; Gu, Z. Organ-on-a-Chip Systems: Microengineering to Biomimic Living Systems. Small 2016, 12, 2253–2282. [Google Scholar] [CrossRef]
- Kodzius, R.; Schulze, F.; Gao, X.; Schneider, M.R. Organ-on-chip technology: Current state and future developments. Genes 2017, 8, 266. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From Three-Dimensional Cell Culture to Organs-on-Chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Hernández, J.E.; Villalba-Rodríguez, A.M.; Romero-Castillo, K.D.; Aguilar-Aguila-Isaías, M.A.; García-Reyes, I.E.; Hernández-Antonio, A.; Ahmed, I.; Sharma, A.; Parra-Saldívar, R.; Iqbal, H.M.N. Organs-on-a-Chip Module: A Review from the Development and Applications Perspective. Micromachines 2018, 9, 536. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Radisic, M. Organ-on-A-chip devices advance to market. Lab Chip 2017, 17, 2395–2420. [Google Scholar] [CrossRef] [PubMed]
- Haddrick, M.; Simpson, P.B. Organ-on-a-chip technology: Turning its potential for clinical benefit into reality. Drug Discov. Today 2019, 24, 1217–1223. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 2018, 22, 310–324. [Google Scholar] [CrossRef]
- Ishida, S. Organs-on-a-chip: Current applications and consideration points for in vitro ADME-Tox studies. Drug Metab. Pharmacokinet. 2018, 33, 49–54. [Google Scholar] [CrossRef]
- Rothbauer, M.; Rosser, J.M.; Zirath, H.; Ertl, P. Tomorrow today: Organ-on-a-chip advances towards clinically relevant pharmaceutical and medical in vitro models. Curr. Opin. Biotechnol. 2019, 55, 81–86. [Google Scholar] [CrossRef]
- Artzy-Schnirman, A.; Hobi, N.; Schneider-Daum, N.; Guenat, O.T.; Lehr, C.M.; Sznitman, J. Advanced in vitro lung-on-chip platforms for inhalation assays: From prospect to pipeline. Eur. J. Pharm. Biopharm. 2019, 144, 11–17. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Yuan Hsin, H.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating heart on a chip: A novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016, 16, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Schneider, O.; Zeifang, L.; Fuchs, S.; Sailer, C.; Loskill, P. User-Friendly and Parallelized Generation of Human Induced Pluripotent Stem Cell-Derived Microtissues in a Centrifugal Heart-on-a-Chip. Tissue Eng. Part A 2019, 25, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Chen, Z.; Zhang, X.; Luo, Y.; Wu, Z.; Lu, Y.; Liu, T.; Zhao, W.; Lin, B. A liver-chip-based alcoholic liver disease model featuring multi-non-parenchymal cells. Biomed. Microdevices 2019, 21, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Otieno, M.A.; Ronxhi, J.; Lim, H.-K.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019, 11, eaax5516. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- Beaurivage, C.; Naumovska, E.; Chang, Y.; Elstak, E.; Nicolas, A.; Wouters, H.; van Moolenbroek, G.; Lanz, H.; Trietsch, S.; Joore, J.; et al. Development of a Gut-on-a-Chip Model for High Throughput Disease Modeling and Drug Discovery. Int. J. Mol. Sci. 2019, 20, 5661. [Google Scholar] [CrossRef]
- Schutgens, F.; Rookmaaker, M.B.; Margaritis, T.; Rios, A.; Ammerlaan, C.; Jansen, J.; Gijzen, L.; Vormann, M.; Vonk, A.; Viveen, M.; et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 2019, 37, 303–313. [Google Scholar] [CrossRef]
- Jang, K.J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef]
- Osaki, T.; Uzel, S.G.M.; Kamm, R.D. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 2018, 4, 1–16. [Google Scholar] [CrossRef]
- Agrawal, G.; Aung, A.; Varghese, S. Skeletal muscle-on-a-chip: An in vitro model to evaluate tissue formation and injury. Lab Chip 2017, 17, 3447–3461. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.-P.; Deng, X.; Chen, D.-Y.; Zhu, D.; Tong, J.-L.; Zhao, T.; Ma, J.-H.; Liu, Y.-Q. A microfluidic chip-based co-culture of fibroblast-like synoviocytes with osteoblasts and osteoclasts to test bone erosion and drug evaluation. R. Soc. Open Sci. 2018, 5, 180528. [Google Scholar] [CrossRef] [PubMed]
- Sheyn, D.; Cohn-Yakubovich, D.; Ben-David, S.; De Mel, S.; Chan, V.; Hinojosa, C.; Wen, N.; Hamilton, G.A.; Gazit, D.; Gazit, Z. Bone-chip system to monitor osteogenic differentiation using optical imaging. Microfluid. Nanofluidics 2019, 23, 99. [Google Scholar] [CrossRef] [PubMed]
- Occhetta, P.; Mainardi, A.; Votta, E.; Vallmajo-Martin, Q.; Ehrbar, M.; Martin, I.; Barbero, A.; Rasponi, M. Hyperphysiological compression of articular cartilage induces an osteoarthritic phenotype in a cartilage-on-a-chip model. Nat. Biomed. Eng. 2019, 3, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, Z.; Li, E.N.; Li, X.; Del Duke, C.J.; Shen, H.; Hao, T.; O’Donnell, B.; Bunnell, B.A.; Goodman, S.B.; et al. Osteochondral Tissue Chip Derived From iPSCs: Modeling OA Pathologies and Testing Drugs. Front. Bioeng. Biotechnol. 2019, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Adriani, G.; Ma, D.; Pavesi, A.; Kamm, R.D.; Goh, E.L.K. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood-brain barrier. Lab Chip 2017, 17, 448–459. [Google Scholar] [CrossRef]
- Ugolini, G.S.; Occhetta, P.; Saccani, A.; Re, F.; Krol, S.; Rasponi, M.; Redaelli, A. Design and validation of a microfluidic device for blood–brain barrier monitoring and transport studies. J. Micromech. Microeng. 2018, 28, 044001. [Google Scholar] [CrossRef]
- Iannielli, A.; Ugolini, G.S.; Cordiglieri, C.; Bido, S.; Rubio, A.; Colasante, G.; Valtorta, M.; Cabassi, T.; Rasponi, M.; Broccoli, V. Reconstitution of the Human Nigro-striatal Pathway on-a-Chip Reveals OPA1-Dependent Mitochondrial Defects and Loss of Dopaminergic Synapses. Cell Rep. 2019, 29, 4646–4656. [Google Scholar] [CrossRef]
- Taylor, A.M.; Dieterich, D.C.; Ito, H.T.; Kim, S.A.; Schuman, E.M. Microfluidic Local Perfusion Chambers for the Visualization and Manipulation of Synapses. Neuron 2010, 66, 57–68. [Google Scholar] [CrossRef]
- Visone, R.; Talò, G.; Occhetta, P.; Cruz-Moreira, D.; Lopa, S.; Pappalardo, O.A.; Redaelli, A.; Moretti, M.; Rasponi, M. A microscale biomimetic platform for generation and electro-mechanical stimulation of 3D cardiac microtissues. APL Bioeng. 2018, 2, 046102. [Google Scholar] [CrossRef]
- Occhetta, P.; Isu, G.; Lemme, M.; Conficconi, C.; Oertle, P.; Räz, C.; Visone, R.; Cerino, G.; Plodinec, M.; Rasponi, M.; et al. A three-dimensional: In vitro dynamic micro-tissue model of cardiac scar formation. Integr. Biol. 2018, 10, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Kilic, T.; Navaee, F.; Stradolini, F.; Renaud, P.; Carrara, S. Organs-on-chip monitoring: Sensors and other strategies. Microphysiol. Syst. 2018, 1, 1. [Google Scholar] [CrossRef]
- Modena, M.M.; Chawla, K.; Misun, P.M.; Hierlemann, A. Smart Cell Culture Systems: Integration of Sensors and Actuators into Microphysiological Systems. ACS Chem. Biol. 2018, 13, 1767–1784. [Google Scholar] [CrossRef] [PubMed]
- Kratz; Höll; Schuller; Ertl; Rothbauer Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems. Biosensors 2019, 9, 110. [CrossRef]
- Kieninger, J.; Weltin, A.; Flamm, H.; Urban, G.A. Microsensor systems for cell metabolism-from 2D culture to organ-on-chip. Lab Chip 2018, 18, 1274–1291. [Google Scholar] [CrossRef] [PubMed]
- Perestrelo, A.R.; Águas, A.C.P.; Rainer, A.; Forte, G. Microfluidic organ/body-on-a-chip devices at the convergence of biology and microengineering. Sensors 2015, 15, 31142–31170. [Google Scholar] [CrossRef]
- Rothbauer, M.; Wartmann, D.; Charwat, V.; Ertl, P. Recent advances and future applications of microfluidic live-cell microarrays. Biotechnol. Adv. 2015, 33, 948–961. [Google Scholar] [CrossRef]
- Pires, N.M.M.; Dong, T.; Hanke, U.; Hoivik, N. Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors 2014, 14, 15458–15479. [Google Scholar] [CrossRef]
- Gruber, P.; Marques, M.P.C.; Szita, N.; Mayr, T. Integration and application of optical chemical sensors in microbioreactors. Lab Chip 2017, 17, 2693–2712. [Google Scholar] [CrossRef]
- Wang, T.; Green, R.; Nair, R.R.; Howell, M.; Mohapatra, S.; Guldiken, R.; Mohapatra, S.S. Surface acoustic waves (SAW)-based biosensing for quantification of cell growth in 2D and 3D Cultures. Sensors 2015, 15, 32045–32055. [Google Scholar] [CrossRef]
- Maoz, B.M.; Herland, A.; Henry, O.Y.F.; Leineweber, W.D.; Yadid, M.; Doyle, J.; Mannix, R.; Kujala, V.J.; Fitzgerald, E.A.; Parker, K.K.; et al. Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017, 17, 2294–2302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Stauffer, F.; Simona, B.R.; Zhang, F.; Zhang, Z.M.; Huang, N.P.; Vörös, J. Multifunctional 3D electrode platform for real-time in situ monitoring and stimulation of cardiac tissues. Biosens. Bioelectron. 2018, 112, 149–155. [Google Scholar] [CrossRef] [PubMed]
- van de Wijdeven, R.; Ramstad, O.H.; Valderhaug, V.D.; Köllensperger, P.; Sandvig, A.; Sandvig, I.; Halaas, Ø. A novel lab-on-chip platform enabling axotomy and neuromodulation in a multi-nodal network. Biosens. Bioelectron. 2019, 140, 111329. [Google Scholar] [CrossRef]
- Caluori, G.; Pribyl, J.; Pesl, M.; Jelinkova, S.; Rotrekl, V.; Skladal, P.; Raiteri, R. Non-invasive electromechanical cell-based biosensors for improved investigation of 3D cardiac models. Biosens. Bioelectron. 2019, 124–125, 129–135. [Google Scholar] [CrossRef]
- Liu, H.; MacQueen, L.A.; Usprech, J.F.; Maleki, H.; Sider, K.L.; Doyle, M.G.; Sun, Y.; Simmons, C.A. Microdevice arrays with strain sensors for 3D mechanical stimulation and monitoring of engineered tissues. Biomaterials 2018, 172, 30–40. [Google Scholar] [CrossRef]
- Zirath, H.; Rothbauer, M.; Spitz, S.; Bachmann, B.; Jordan, C.; Müller, B.; Ehgartner, J.; Priglinger, E.; Mühleder, S.; Redl, H.; et al. Every breath you take: Non-invasive real-time oxygen biosensing in two-and three-dimensional microfluidic cell models. Front. Physiol. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Rennert, K.; Steinborn, S.; Gröger, M.; Ungerböck, B.; Jank, A.-M.; Ehgartner, J.; Nietzsche, S.; Dinger, J.; Kiehntopf, M.; Funke, H.; et al. A microfluidically perfused three dimensional human liver model. Biomaterials 2015, 71, 119–131. [Google Scholar] [CrossRef]
- Moya, A.; Ortega-Ribera, M.; Guimerà, X.; Sowade, E.; Zea, M.; Illa, X.; Ramon, E.; Villa, R.; Gracia-Sancho, J.; Gabriel, G. Online oxygen monitoring using integrated inkjet-printed sensors in a liver-on-a-chip system. Lab Chip 2018, 18, 2023–2035. [Google Scholar] [CrossRef]
- Weltin, A.; Slotwinski, K.; Kieninger, J.; Moser, I.; Jobst, G.; Wego, M.; Ehret, R.; Urban, G.A. Cell culture monitoring for drug screening and cancer research: A transparent, microfluidic, multi-sensor microsystem. Lab Chip 2014, 14, 138–146. [Google Scholar] [CrossRef]
- Misun, P.M.; Rothe, J.; Schmid, Y.R.F.; Hierlemann, A.; Frey, O. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst. Nanoeng. 2016, 2, 16022. [Google Scholar] [CrossRef]
- Ortega, M.A.; Fernández-Garibay, X.; Castaño, A.G.; De Chiara, F.; Hernández-Albors, A.; Balaguer-Trias, J.; Ramón-Azcón, J. Muscle-on-a-chip with an on-site multiplexed biosensing system for: In situ monitoring of secreted IL-6 and TNF-α. Lab Chip 2019, 19, 2568–2580. [Google Scholar] [CrossRef] [PubMed]
- Young, A.T.; Rivera, K.R.; Erb, P.D.; Daniele, M.A. Monitoring of Microphysiological Systems: Integrating Sensors and Real-Time Data Analysis toward Autonomous Decision-Making. ACS Sens. 2019, 4, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Carreau, A.; Hafny-Rahbi, B.E.; Matejuk, A.; Grillon, C.; Kieda, C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Klimant, I.; Wolfbeis, O.S. Oxygen-Sensitive Luminescent Materials Based on Silicone-Soluble Ruthenium Diimine Complexes. Anal. Chem. 1995, 67, 3160–3166. [Google Scholar] [CrossRef]
- Oomen, P.E.; Skolimowski, M.D.; Verpoorte, E. Implementing oxygen control in chip-based cell and tissue culture systems. Lab Chip 2016, 16, 3394–3414. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Alborzinia, H.; Can, S.; Holenya, P.; Scholl, C.; Lederer, E.; Kitanovic, I.; Wölfl, S. Real-Time Monitoring of Cisplatin-Induced Cell Death. PLoS ONE 2011, 6, e19714. [Google Scholar] [CrossRef]
- Solaini, G.; Baracca, A.; Lenaz, G.; Sgarbi, G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta-Bioenerg. 2010, 1797, 1171–1177. [Google Scholar] [CrossRef]
- Bavli, D.; Prill, S.; Ezra, E.; Levy, G.; Cohen, M.; Vinken, M.; Vanfleteren, J.; Jaeger, M.; Nahmias, Y. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 2016, 113, E2231–E2240. [Google Scholar] [CrossRef]
- Kietzmann, T. Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 2017, 11, 622–630. [Google Scholar] [CrossRef]
- Siddiqui, M.A.; Ahmad, J.; Farshori, N.N.; Saquib, Q.; Jahan, S.; Kashyap, M.P.; Ahamed, M.; Musarrat, J.; Al-Khedhairy, A.A. Rotenone-induced oxidative stress and apoptosis in human liver HepG2 cells. Mol. Cell. Biochem. 2013, 384, 59–69. [Google Scholar] [CrossRef]
- Julie, N.L.; Julie, I.M.; Kende, A.I.; Wilson, G.L. Mitochondrial dysfunction and delayed hepatotoxicity: Another lesson from troglitazone. Diabetologia 2008, 51, 2108–2116. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Espey, M.G. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic. Biol. Med. 2013, 55, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed]
- Mauleon, G.; Fall, C.P.; Eddington, D.T. Precise Spatial and Temporal Control of Oxygen within In Vitro Brain Slices via Microfluidic Gas Channels. PLoS ONE 2012, 7, e43309. [Google Scholar] [CrossRef]
- Bonnekoh, P.; Kuroiwa, T.; Oschlies, U.; Hossmann, K.A. Barbiturate promotes post-ischemic reaggregation of polyribosomes in gerbil hippocampus. Neurosci. Lett. 1992, 146, 75–78. [Google Scholar] [CrossRef]
- Sticker, D.; Rothbauer, M.; Ehgartner, J.; Steininger, C.; Liske, O.; Liska, R.; Neuhaus, W.; Mayr, T.; Haraldsson, T.; Kutter, J.P.; et al. Oxygen Management at the Microscale: A Functional Biochip Material with Long-Lasting and Tunable Oxygen Scavenging Properties for Cell Culture Applications. ACS Appl. Mater. Interfaces 2019, 11, 9730–9739. [Google Scholar] [CrossRef]
- Carioscia, J.A.; Stansbury, J.W.; Bowman, C.N. Evaluation and Control of Thiol-ene/Thiol-epoxy Hybrid Networks. Polymer 2007, 23, 1–7. [Google Scholar]
- Brennan, M.D.; Rexius-Hall, M.L.; Elgass, L.J.; Eddington, D.T. Oxygen control with microfluidics. Lab Chip 2014, 14, 4305–4318. [Google Scholar] [CrossRef]
- Weltin, A.; Hammer, S.; Noor, F.; Kaminski, Y.; Kieninger, J.; Urban, G.A. Accessing 3D microtissue metabolism: Lactate and oxygen monitoring in hepatocyte spheroids. Biosens. Bioelectron. 2017, 87, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yu, P.; Hao, J.; Wang, Y.; Ohsaka, T.; Mao, L. Continuous and simultaneous electrochemical measurements of glucose, lactate, and ascorbate in rat brain following brain ischemia. Anal. Chem. 2014, 86, 3895–3901. [Google Scholar] [CrossRef] [PubMed]
- Dervisevic, E.; Tuck, K.L.; Voelcker, N.H.; Cadarso, V.J. Recent Progress in Lab-On-a-Chip Systems for the Monitoring of Metabolites for Mammalian and Microbial Cell Research. Sensors 2019, 19, 5027. [Google Scholar] [CrossRef] [PubMed]
- Bange, A.; Halsall, H.B.; Heineman, W.R. Microfluidic immunosensor systems. Biosens. Bioelectron. 2005, 20, 2488–2503. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.A.; Ahmed, M.U. Electrochemical immunosensors and their recent nanomaterial-based signal amplification strategies: A review. RSC Adv. 2016, 6, 24995–25014. [Google Scholar] [CrossRef]
- Stenken, J.A.; Poschenrieder, A.J. Bioanalytical Chemistry of Cytokines-A Review. Anal. Chim. Acta 2015, 853, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Qi, M.; Hutchinson, M.R.; Yang, G.; Goldys, E.M. Recent advances in cytokine detection by immunosensing. Biosens. Bioelectron. 2016, 79, 810–821. [Google Scholar] [CrossRef]
- Li, X.; Soler, M.; Özdemir, C.I.; Belushkin, A.; Yesilköy, F.; Altug, H. Plasmonic nanohole array biosensor for label-free and real-time analysis of live cell secretion. Lab Chip 2017, 17, 2208–2217. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.R.; Kilic, T.; Zhang, Y.S.; Avci, H.; Hu, N.; Kim, D.; Branco, C.; Aleman, J.; Massa, S.; Silvestri, A.; et al. Label-Free and Regenerative Electrochemical Microfluidic Biosensors for Continual Monitoring of Cell Secretomes. Adv. Sci. 2017, 4, 1600522. [Google Scholar] [CrossRef] [PubMed]
- Baganizi, D.R.; Leroy, L.; Laplatine, L.; Fairley, S.J.; Heidmann, S.; Menad, S.; Livache, T.; Marche, P.N.; Roupioz, Y. A Simple microfluidic platform for long-term analysis and continuous dual-imaging detection of T-Cell secreted IFN-γ and IL-2 on antibody-based biochip. Biosensors 2015, 5, 750–767. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.R.; Zhang, Y.S.; Kim, D.J.; Manbohi, A.; Avci, H.; Silvestri, A.; Aleman, J.; Hu, N.; Kilic, T.; Keung, W.; et al. Aptamer-Based Microfluidic Electrochemical Biosensor for Monitoring Cell-Secreted Trace Cardiac Biomarkers. Anal. Chem. 2016, 88, 10019–10027. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Patel, D.; Kwa, T.; Haque, A.; Matharu, Z.; Stybayeva, G.; Gao, Y.; Diehl, A.M.; Revzin, A. Liver injury-on-a-chip: Microfluidic co-cultures with integrated biosensors for monitoring liver cell signaling during injury. Lab Chip 2015, 15, 4467–4478. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Y.; Matharu, Z.; Rahimian, A.; Revzin, A. Detecting multiple cell-secreted cytokines from the same aptamer-functionalized electrode. Biosens. Bioelectron. 2015, 64, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Son, K.J.; Gheibi, P.; Stybayeva, G.; Rahimian, A.; Revzin, A. Detecting cell-secreted growth factors in microfluidic devices using bead-based biosensors. Microsyst. Nanoeng. 2017, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Riahi, R.; Shaegh, S.A.M.; Ghaderi, M.; Zhang, Y.S.; Shin, S.R.; Aleman, J.; Massa, S.; Kim, D.; Dokmeci, M.R.; Khademhosseini, A. Automated microfluidic platform of bead-based electrochemical immunosensor integrated with bioreactor for continual monitoring of cell secreted biomarkers. Sci. Rep. 2016, 6, 24598. [Google Scholar] [CrossRef]
- Wong, J.F.; Simmons, C.A. Microfluidic assay for the on-chip electrochemical measurement of cell monolayer permeability. Lab Chip 2019, 19, 1060–1070. [Google Scholar] [CrossRef]
- Matter, K.; Balda, M.S. Functional analysis of tight junctions. Methods 2003, 30, 228–234. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER Measurement Techniques for In Vitro Barrier Model Systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Cai, Z.; Qiao, P.F.; Wan, C.Q.; Cai, M.; Zhou, N.K.; Li, Q. Role of Blood-Brain Barrier in Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 63, 1223–1234. [Google Scholar] [CrossRef]
- Vargas-Osorio, Z.; Da Silva-Candal, A.; Piñeiro, Y.; Iglesias-Rey, R.; Sobrino, T.; Campos, F.; Castillo, J.; Rivas, J. Multifunctional Superparamagnetic Stiff Nanoreservoirs for Blood Brain Barrier Applications. Nanomaterials 2019, 9, 449. [Google Scholar] [CrossRef]
- Abdullahi, W.; Tripathi, D.; Ronaldson, P.T. Blood-brain barrier dysfunction in ischemic stroke: Targeting tight junctions and transporters for vascular protection. Am. J. Physiol.-Cell Physiol. 2018, 315, C343–C356. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, F.; Nishihara, H.; Kanda, T. Blood–brain barrier dysfunction in immuno-mediated neurological diseases. Immunol. Med. 2018, 41, 120–128. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Merali, Z.; Huang, K.; Mikulis, D.; Silver, F.; Kassner, A. Evolution of blood-brain-barrier permeability after acute ischemic stroke. PLoS ONE 2017, 12, e0171558. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.D.; Sagare, A.P.; Zlokovic, B.V. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018, 14, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Akaishi, T.; Takahashi, T.; Nakashima, I. Oligoclonal bands and periventricular lesions in multiple sclerosis will not increase blood-brain barrier permeability. J. Neurol. Sci. 2018, 387, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, S.; Zheng, J.; Li, Y.; Huang, H. Recent progress in microfluidic models of the blood-brain barrier. Micromachines 2019, 10, 375. [Google Scholar] [CrossRef]
- Heidari, H.; Taylor, H. Review Article: Capturing the physiological complexity of the brain’s neuro-vascular unit in vitro. Biomicrofluidics 2018, 12, 051502. [Google Scholar] [CrossRef]
- Booth, R.; Kim, H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab Chip 2012, 12, 1784–1792. [Google Scholar] [CrossRef]
- Cucullo, L.; Hossain, M.; Puvenna, V.; Marchi, N.; Janigro, D. The role of shear stress in Blood-Brain Barrier endothelial physiology. BMC Neurosci. 2011, 12, 40. [Google Scholar] [CrossRef]
- Vastag, M.; Keseru, G. Current in vitro and in silico models of blood-brain barrier penetration: A practical view. Curr. Opin. Drug Discov. Dev. 2009, 12, 115–124. [Google Scholar]
- Kim, S.; Buonocore, J.; Park, J.; Welsh, C.J.; Li, J.; Han, A. A Three-Dimensional Arrayed Microfluidic Blood-Brain Barrier Model With Integrated Electrical Sensor Array. IEEE Trans. Biomed. Eng. 2018, 65, 431–439. [Google Scholar]
- Wang, Y.I.; Abaci, H.E.; Shuler, M.L. Microfluidic blood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017, 114, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Griep, L.M.; Wolbers, F.; De Wagenaar, B.; Ter Braak, P.M.; Weksler, B.B.; Romero, I.A.; Couraud, P.O.; Vermes, I.; Van Der Meer, A.D.; Van Den Berg, A. BBB on CHIP: Microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed. Microdevices 2013, 15, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.D.; Khafagy, E.S.; Khanafer, K.; Takayama, S.; Elsayed, M.E.H. Organization of Endothelial Cells, Pericytes, and Astrocytes into a 3D Microfluidic in Vitro Model of the Blood-Brain Barrier. Mol. Pharm. 2016, 13, 895–906. [Google Scholar] [CrossRef]
- van der Helm, M.W.; Odijk, M.; Frimat, J.-P.; van der Meer, A.D.; Eijkel, J.C.T.; van den Berg, A.; Segerink, L.I. Direct quantification of transendothelial electrical resistance in organs-on-chips. Biosens. Bioelectron. 2016, 85, 924–929. [Google Scholar] [CrossRef]
- van der Helm, M.W.; Odijk, M.; Frimat, J.P.; van der Meer, A.D.; Eijkel, J.C.T.; van den Berg, A.; Segerink, L.I. Fabrication and validation of an organ-on-chip system with integrated electrodes to directly quantify transendothelial electrical resistance. J. Vis. Exp. 2017, 2017, 1–8. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef]
- Henry, O.Y.F.; Villenave, R.; Cronce, M.J.; Leineweber, W.D.; Benz, M.A.; Ingber, D.E. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 2017, 17, 2264–2271. [Google Scholar] [CrossRef]
- van der Helm, M.W.; Henry, O.Y.F.; Bein, A.; Hamkins-Indik, T.; Cronce, M.J.; Leineweber, W.D.; Odijk, M.; van der Meer, A.D.; Segerink, L.I.; Ingber, D.E. Non-invasive sensing of transepithelial barrier function and tissue differentiation in organs-on-chips using impedance spectroscopy. Lab Chip 2019, 19, 452–463. [Google Scholar] [CrossRef]
- Odijk, M.; Van Der Meer, A.D.; Levner, D.; Kim, H.J.; Van Der Helm, M.W.; Segerink, L.I.; Frimat, J.P.; Hamilton, G.A.; Ingber, D.E.; Van Den Berg, A. Measuring direct current trans-epithelial electrical resistance in organ-on-a-chip microsystems. Lab Chip 2015, 15, 745–752. [Google Scholar] [CrossRef]
- Mermoud, Y.; Felder, M.; Stucki, J.D.; Stucki, A.O.; Guenat, O.T. Microimpedance tomography system to monitor cell activity and membrane movements in a breathing lung-on-chip. Sens. Actuators B Chem. 2018, 255, 3647–3653. [Google Scholar] [CrossRef]
- Wegener, J.; Seebach, J. Experimental tools to monitor the dynamics of endothelial barrier function: A survey of in vitro approaches. Cell Tissue Res. 2014, 355, 485–514. [Google Scholar] [CrossRef] [PubMed]
- Millet, L.J.; Gillette, M.U. New perspectives on neuronal development via microfluidic environments. Trends Neurosci. 2012, 35, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Vahidi, B.; Taylor, A.M.; Rhee, S.W.; Jeon, N.L. Microfluidic culture platform for neuroscience research. Nat. Protoc. 2006, 1, 2128–2136. [Google Scholar] [CrossRef]
- Jadhav, D.A.; Wei, L.; Shi, P. Compartmentalized Platforms for Neuro-Pharmacological Research. Curr. Neuropharmacol. 2015, 14, 72–86. [Google Scholar] [CrossRef]
- Taylor, A.M.; Rhee, S.W.; Tu, C.H.; Cribbs, D.H.; Cotman, C.W.; Jeon, N.L. Microfluidic multicompartment device for neuroscience research. Langmuir 2003, 19, 1551–1556. [Google Scholar] [CrossRef]
- Taylor, A.M.; Cotman, C.W.; Cribbs, D.H.; Blurton-Jones, M.; Jeon, N.L.; Rhee, S.W. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods 2005, 2, 599–605. [Google Scholar] [CrossRef]
- Park, J.; Kim, S.; Park, S.I.; Choe, Y.; Li, J.; Han, A. A microchip for quantitative analysis of CNS axon growth under localized biomolecular treatments. J. Neurosci. Methods 2014, 221, 166–174. [Google Scholar] [CrossRef]
- Kilinc, D.; Peyrin, J.M.; Soubeyre, V.; Magnifico, S.; Saias, L.; Viovy, J.L.; Brugg, B. Wallerian-like degeneration of central neurons after synchronized and geometrically registered mass axotomy in a three-compartmental microfluidic chip. Neurotox. Res. 2011, 19, 149–161. [Google Scholar] [CrossRef]
- Hosmane, S.; Fournier, A.; Wright, R.; Rajbhandari, L.; Siddique, R.; Yang, I.H.; Ramesh, K.T.; Venkatesan, A.; Thakor, N. Valve-based microfluidic compression platform: Single axon injury and regrowth. Lab Chip 2011, 11, 3888–3895. [Google Scholar] [CrossRef]
- Lu, X.; Kim-Han, J.S.; O’Malley, K.L.; Sakiyama-Elbert, S.E. A microdevice platform for visualizing mitochondrial transport in aligned dopaminergic axons. J. Neurosci. Methods 2012, 209, 35–39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Virlogeux, A.; Moutaux, E.; Christaller, W.; Genoux, A.; Bruyère, J.; Fino, E.; Charlot, B.; Cazorla, M.; Saudou, F. Reconstituting Corticostriatal Network on-a-Chip Reveals the Contribution of the Presynaptic Compartment to Huntington’s Disease. Cell Rep. 2018, 22, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Biffi, E.; Piraino, F.; Pedrocchi, A.; Fiore, G.B.; Ferrigno, G.; Redaelli, A.; Menegon, A.; Rasponi, M. A microfluidic platform for controlled biochemical stimulation of twin neuronal networks. Biomicrofluidics 2012, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Habibey, R.; Latifi, S.; Mousavi, H.; Pesce, M.; Arab-Tehrany, E.; Blau, A. A multielectrode array microchannel platform reveals both transient and slow changes in axonal conduction velocity. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gladkov, A.; Pigareva, Y.; Kutyina, D.; Kolpakov, V.; Bukatin, A.; Mukhina, I.; Kazantsev, V.; Pimashkin, A. Design of Cultured Neuron Networks in vitro with Predefined Connectivity Using Asymmetric Microfluidic Channels. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Moutaux, E.; Charlot, B.; Genoux, A.; Saudou, F.; Cazorla, M. An integrated microfluidic/microelectrode array for the study of activity-dependent intracellular dynamics in neuronal networks. Lab Chip 2018, 18, 3425–3435. [Google Scholar] [CrossRef]
- Shen, X.; Wu, J.; Wang, Z.; Chen, T. Characterization of in vitro neural functional connectivity on a neurofluidic device. Electrophoresis 2019, 40, 2996–3004. [Google Scholar] [CrossRef]
- Morin, F.; Nishimura, N.; Griscom, L.; LePioufle, B.; Fujita, H.; Takamura, Y.; Tamiya, E. Constraining the connectivity of neuronal networks cultured on microelectrode arrays with microfluidic techniques: A step towards neuron-based functional chips. Biosens. Bioelectron. 2006, 21, 1093–1100. [Google Scholar] [CrossRef]
- Pine, J. Recording action potentials from cultured neurons with extracellular microcircuit electrodes. J. Neurosci. Methods 1980, 2, 19–31. [Google Scholar] [CrossRef]
- Gross, G.W. Simultaneous Single Unit Recording in vitro with a Photoetched Laser Deinsulated Gold Multimicroelectrode Surface. IEEE Trans. Biomed. Eng. 1979, BME-26, 273–279. [Google Scholar] [CrossRef]
- Perelman, Y.; Ginosar, R. An integrated system for multichannel neuronal recording with spike/LFP separation, integrated A/D conversion and threshold detection. IEEE Trans. Biomed. Eng. 2007, 54, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Shimba, K.; Sakai, K.; Isomura, T.; Kotani, K.; Jimbo, Y. Axonal conduction slowing induced by spontaneous bursting activity in cortical neurons cultured in a microtunnel device. Integr. Biol. 2015, 7, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Riss, M.; Buitrago, J.O.; Claverol-Tinturé, E. Biophysics of microchannel-enabled neuron–electrode interfaces. J. Neural Eng. 2012, 9, 026010. [Google Scholar] [CrossRef] [PubMed]
- Hong, N.; Joo, S.; Nam, Y. Characterization of axonal spikes in cultured neuronal networks using microelectrode arrays and microchannel devices. IEEE Trans. Biomed. Eng. 2017, 64, 492–498. [Google Scholar] [CrossRef]
- Pedraza, E.; Karajić, A.; Raoux, M.; Perrier, R.; Pirog, A.; Lebreton, F.; Arbault, S.; Gaitan, J.; Renaud, S.; Kuhn, A.; et al. Guiding pancreatic beta cells to target electrodes in a whole-cell biosensor for diabetes. Lab Chip 2015, 15, 3880–3890. [Google Scholar] [CrossRef] [PubMed]
- Rorsman, P.; Braun, M. Regulation of Insulin Secretion in Human Pancreatic Islets. Annu. Rev. Physiol. 2013, 75, 155–179. [Google Scholar] [CrossRef]
- Koutsouras, D.A.; Perrier, R.; Villarroel Marquez, A.; Pirog, A.; Pedraza, E.; Cloutet, E.; Renaud, S.; Raoux, M.; Malliaras, G.G.; Lang, J. Simultaneous monitoring of single cell and of micro-organ activity by PEDOT:PSS covered multi-electrode arrays. Mater. Sci. Eng. C 2017, 81, 84–89. [Google Scholar] [CrossRef]
- Lebreton, F.; Pirog, A.; Belouah, I.; Bosco, D.; Berney, T.; Meda, P.; Bornat, Y.; Catargi, B.; Renaud, S.; Raoux, M.; et al. Slow potentials encode intercellular coupling and insulin demand in pancreatic beta cells. Diabetologia 2015, 58, 1291–1299. [Google Scholar] [CrossRef]
- Raoux, M.; Bontorin, G.; Bornat, Y.; Lang, J.; Renaud, S. Bioelectronic Sensing of Insulin Demand. In Biohybrid Systems; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2011; pp. 191–202. [Google Scholar]
- Raoux, M.; Bornat, Y.; Quotb, A.; Catargi, B.; Renaud, S.; Lang, J. Non-invasive long-term and real-time analysis of endocrine cells on micro-electrode arrays. J. Physiol. 2012, 590, 1085–1091. [Google Scholar] [CrossRef]
- Johnston, N.R.; Mitchell, R.K.; Haythorne, E.; Trauner, D.; Rutter, G.A.; Hodson, D.J.; Johnston, N.R.; Mitchell, R.K.; Haythorne, E.; Pessoa, M.P.; et al. Beta Cell Hubs Dictate Pancreatic Islet Responses to Glucose Article Beta Cell Hubs Dictate Pancreatic Islet Responses to Glucose. Cell Metab. 2016, 24, 389–401. [Google Scholar] [CrossRef]
- Renaud, S.; Catargi, B.; Lang, J. Biosensors in Diabetes: How to get the most out of evolution and transpose it into a signal. IEEE Pulse 2014, 5, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Perrier, R.; Pirog, A.; Jaffredo, M.; Gaitan, J.; Catargi, B.; Renaud, S.; Raoux, M.; Lang, J. Bioelectronic organ-based sensor for microfluidic real-time analysis of the demand in insulin. Biosens. Bioelectron. 2018, 117, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Chal, J.; Pourquié, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, R.; Sherratt, M.J.; Cruickshank, J.K.; Derby, B. Characterizing the elastic properties of tissues. Mater Today 2012, 14, 96–105. [Google Scholar] [CrossRef]
- Palchesko, R.N.; Zhang, L.; Sun, Y.; Feinberg, A.W. Development of Polydimethylsiloxane Substrates with Tunable Elastic Modulus to Study Cell Mechanobiology in Muscle and Nerve. PLoS ONE 2012, 7, e51499. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Singh, S.P.; Schwartz, M.P.; Tokuda, E.Y.; Luo, Y.; Rogers, R.E.; Fujita, M.; Ahn, N.G.; Anseth, K.S. A synthetic modular approach for modeling the role of the 3D microenvironment in tumor progression. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef]
- Lin, C.H.; Wang, C.K.; Chen, Y.A.; Peng, C.C.; Liao, W.H.; Tung, Y.C. Measurement of in-plane elasticity of live cell layers using a pressure sensor embedded microfluidic device. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Tandon, N.; Cannizzaro, C.; Chao, P.P.-H.G.; Maidhof, R.; Marsano, A.; Au, H.T.H.; Radisic, M.; Vunjak-Novakovic, G. Electrical stimulation systems for cardiac tissue engineering. Nat. Protoc. 2009, 4, 155–173. [Google Scholar] [CrossRef]
- Feinberg, A.W.; Alford, P.W.; Jin, H.; Ripplinger, C.M.; Werdich, A.A.; Sheehy, S.P.; Grosberg, A.; Parker, K.K. Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture. Biomaterials 2012, 23, 5732–5741. [Google Scholar] [CrossRef]
- Wang, Y.I.; Oleaga, C.; Long, C.J.; Esch, M.B.; McAleer, C.W.; Miller, P.G.; Hickman, J.J.; Shuler, M.L. Self-contained, low-cost Body-on-a-Chip systems for drug development. Exp. Biol. Med. 2017, 242, 1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Schmid, Y.R.F.; Bürgel, S.C.; Misun, P.M.; Hierlemann, A.; Frey, O. Electrical Impedance Spectroscopy for Microtissue Spheroid Analysis in Hanging-Drop Networks. ACS Sens. 2016, 1, 1028–1035. [Google Scholar] [CrossRef]
- Bürgel, S.C.; Diener, L.; Frey, O.; Kim, J.Y.; Hierlemann, A. Automated, multiplexed electrical impedance spectroscopy platform for continuous monitoring of microtissue spheroids. Anal. Chem. 2016, 88, 10876–10883. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, P.; Moritz, W.; Kelm, J.M.; Ullrich, N.D.; Agarkova, I.; Anson, B.D.; Suter, T.M.; Zuppinger, C. Development and Characterization of a Scaffold-Free 3D Spheroid Model of Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes. Tissue Eng.-Part C Methods 2015, 21, 852–861. [Google Scholar] [CrossRef]
- Gómez, A.M.; Valdivia, H.H.; Cheng, H.; Lederer, M.R.; Santana, L.F.; Mccune, S.A.; Altschuld, R.A.; Lederer, W.J.; Gomez, A.M.; Valdivia, H.H.; et al. Defective Excitation-Contraction Coupling in Experimental Cardiac Hypertrophy and Heart Failure. Science 1997, 276, 800–806. [Google Scholar] [CrossRef]
- Farman, G.P.; Tachampa, K.; Mateja, R.; Cazorla, O.; Lacampagne, A.; De Tombe, P.P. Blebbistatin: Use as inhibitor of muscle contraction. Pflug. Arch. Eur. J. Physiol. 2008, 455, 995–1005. [Google Scholar] [CrossRef]
- Oudit, G.Y.; Sun, H.; Trivieri, M.G.; Koch, S.E.; Dawood, F.; Ackerley, C.; Yazdanpanah, M.; Wilson, G.J.; Schwartz, A.; Liu, P.P.; et al. L-type Ca2+ channels provide a major pathway for iron entry into cardiomyocytes in iron-overload cardiomyopathy. Nat. Med. 2003, 9, 1187–1194. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, T.; Wang, P.; Hu, N. High-Throughput Assessment of Drug Cardiac Safety Using a High-Speed Impedance Detection Technology-Based Heart-on-a-Chip. Micromachines 2016, 7, 122. [Google Scholar] [CrossRef]
- Radisic, M.; Park, H.; Gerecht, S.; Cannizzaro, C.; Langer, R.; Vunjak-Novakovic, G. Biomimetic approach to cardiac tissue engineering. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1357–1368. [Google Scholar] [CrossRef]
- Qian, F.; Huang, C.; Lin, Y.D.; Ivanovskaya, A.N.; O’Hara, T.J.; Booth, R.H.; Creek, C.J.; Enright, H.A.; Soscia, D.A.; Belle, A.M.; et al. Simultaneous electrical recording of cardiac electrophysiology and contraction on chip. Lab Chip 2017, 17, 1732–1739. [Google Scholar] [CrossRef]
- Wang, T.; Hu, N.; Cao, J.; Wu, J.; Su, K.; Wang, P. A cardiomyocyte-based biosensor for antiarrhythmic drug evaluation by simultaneously monitoring cell growth and beating. Biosens. Bioelectron. 2013, 49, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Nguemo, F.; Šarić, T.; Pfannkuche, K.; Watzele, M.; Reppel, M.; Hescheler, J. In vitro model for assessing arrhythmogenic properties of drugs based on high-resolution impedance measurements. Cell. Physiol. Biochem. 2012, 29, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Munch, P.A.; Thoren, P.N.; Brown, A.M. Dual effects of norepinephrine and mechanisms of baroreceptor stimulation. Circ. Res. 1987, 61, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.F.L.; Huyer, L.D.; Lu, R.X.Z.; Drecun, S.; Radisic, M.; Zhang, B. InVADE: Integrated Vasculature for Assessing Dynamic Events. Adv. Funct. Mater. 2017, 27, 1–11. [Google Scholar] [CrossRef]
- Oleaga, C.; Lavado, A.; Riu, A.; Rothemund, S.; Carmona-Moran, C.A.; Persaud, K.; Yurko, A.; Lear, J.; Narasimhan, N.S.; Long, C.J.; et al. Long-Term Electrical and Mechanical Function Monitoring of a Human-on-a-Chip System. Adv. Funct. Mater. 2019, 29, 1–12. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Shaegh, S.A.M.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef]
- Skardal, A.; Murphy, S.V.; Devarasetty, M.; Mead, I.; Kang, H.W.; Seol, Y.J.; Zhang, Y.S.; Shin, S.R.; Zhao, L.; Aleman, J.; et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrari, E.; Palma, C.; Vesentini, S.; Occhetta, P.; Rasponi, M. Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems. Biosensors 2020, 10, 110. https://doi.org/10.3390/bios10090110
Ferrari E, Palma C, Vesentini S, Occhetta P, Rasponi M. Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems. Biosensors. 2020; 10(9):110. https://doi.org/10.3390/bios10090110
Chicago/Turabian StyleFerrari, Erika, Cecilia Palma, Simone Vesentini, Paola Occhetta, and Marco Rasponi. 2020. "Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems" Biosensors 10, no. 9: 110. https://doi.org/10.3390/bios10090110
APA StyleFerrari, E., Palma, C., Vesentini, S., Occhetta, P., & Rasponi, M. (2020). Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems. Biosensors, 10(9), 110. https://doi.org/10.3390/bios10090110







