Towards Bacteria Counting in DI Water of Several Microliters or Growing Suspension Using Impedance Biochips
Abstract
1. Introduction
2. Materials and Methods
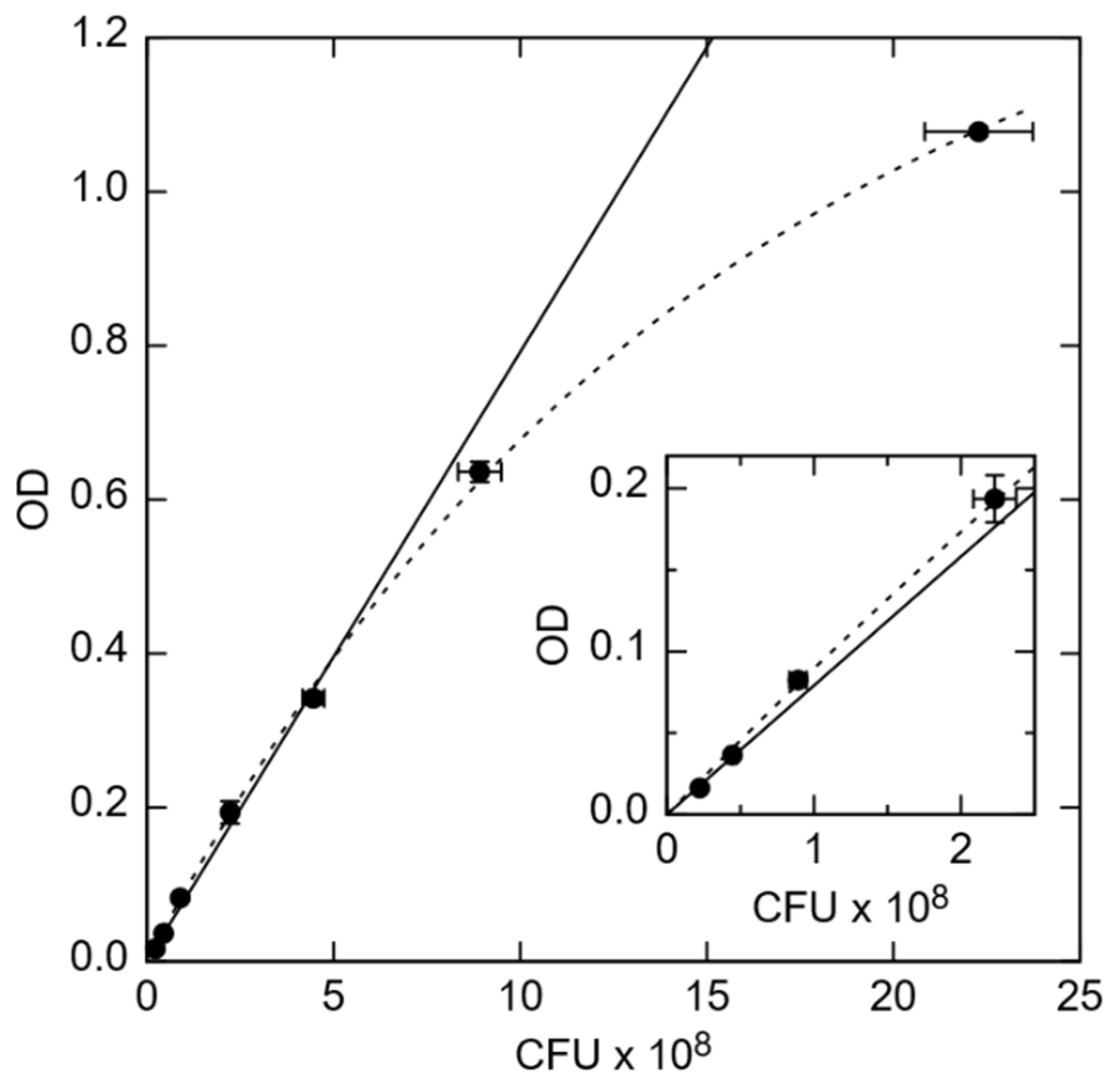
2.1. Preparation of Bacteria and OD Measurements
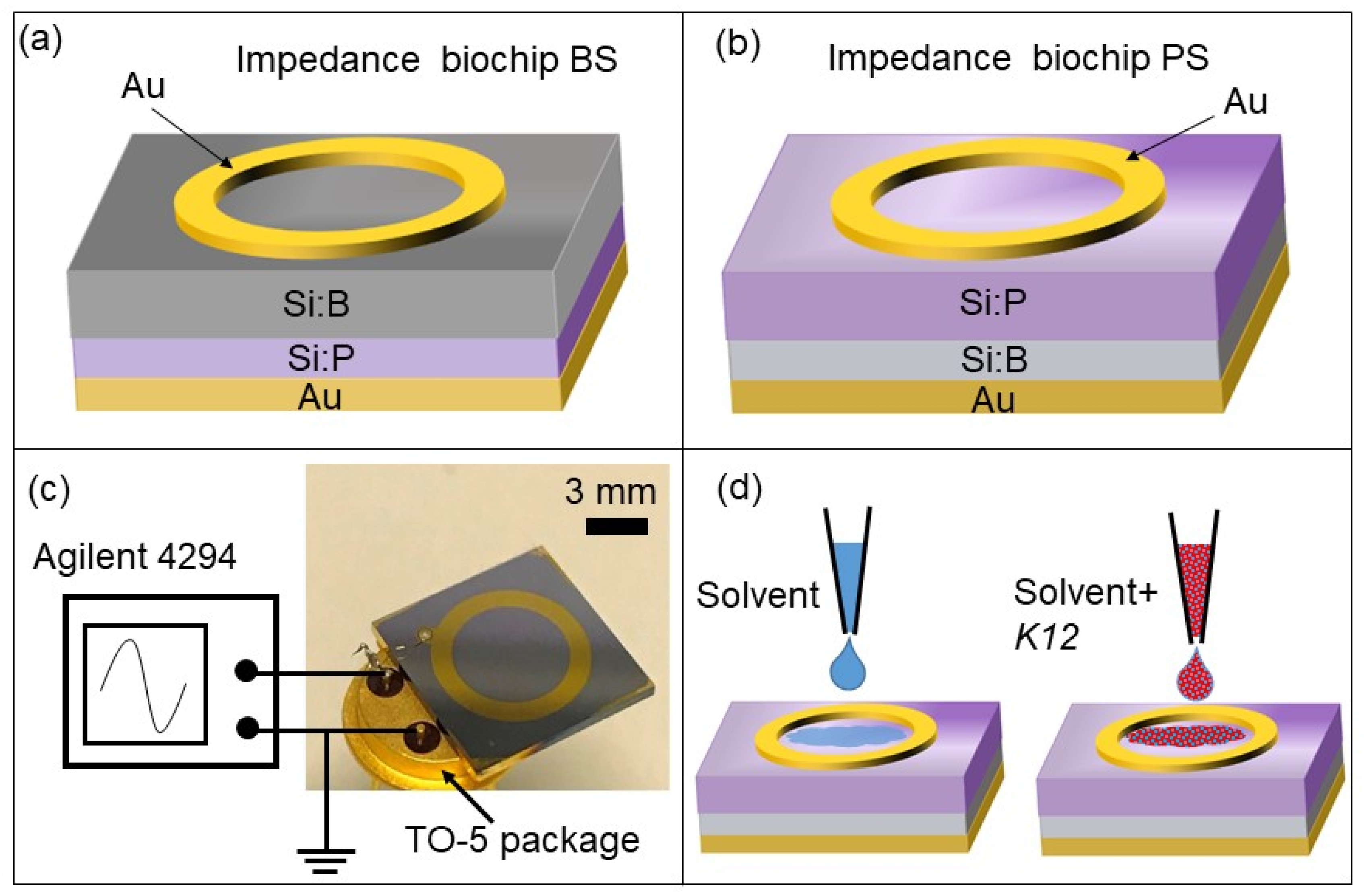
2.2. Structural Description
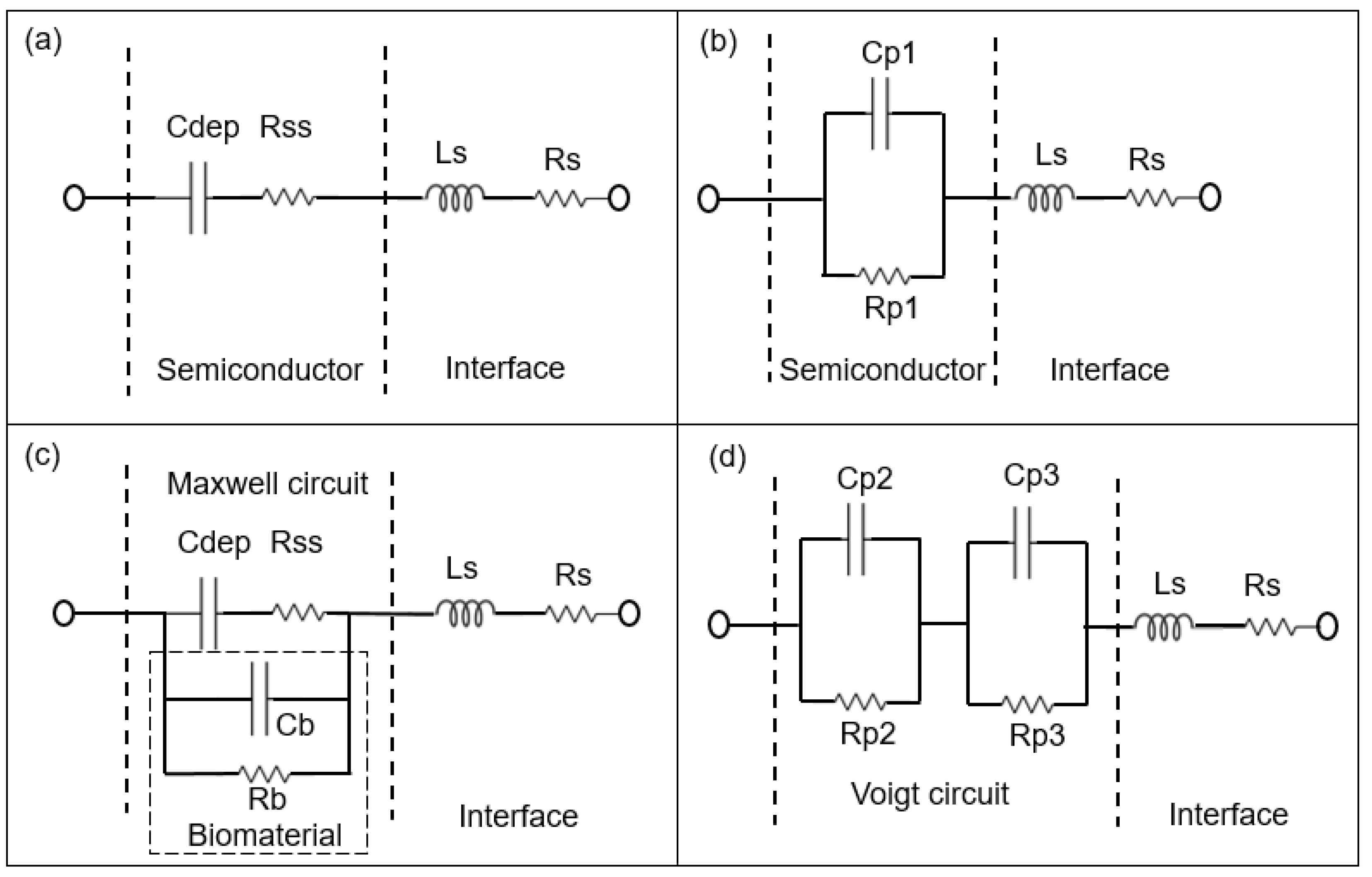
2.3. Modelling
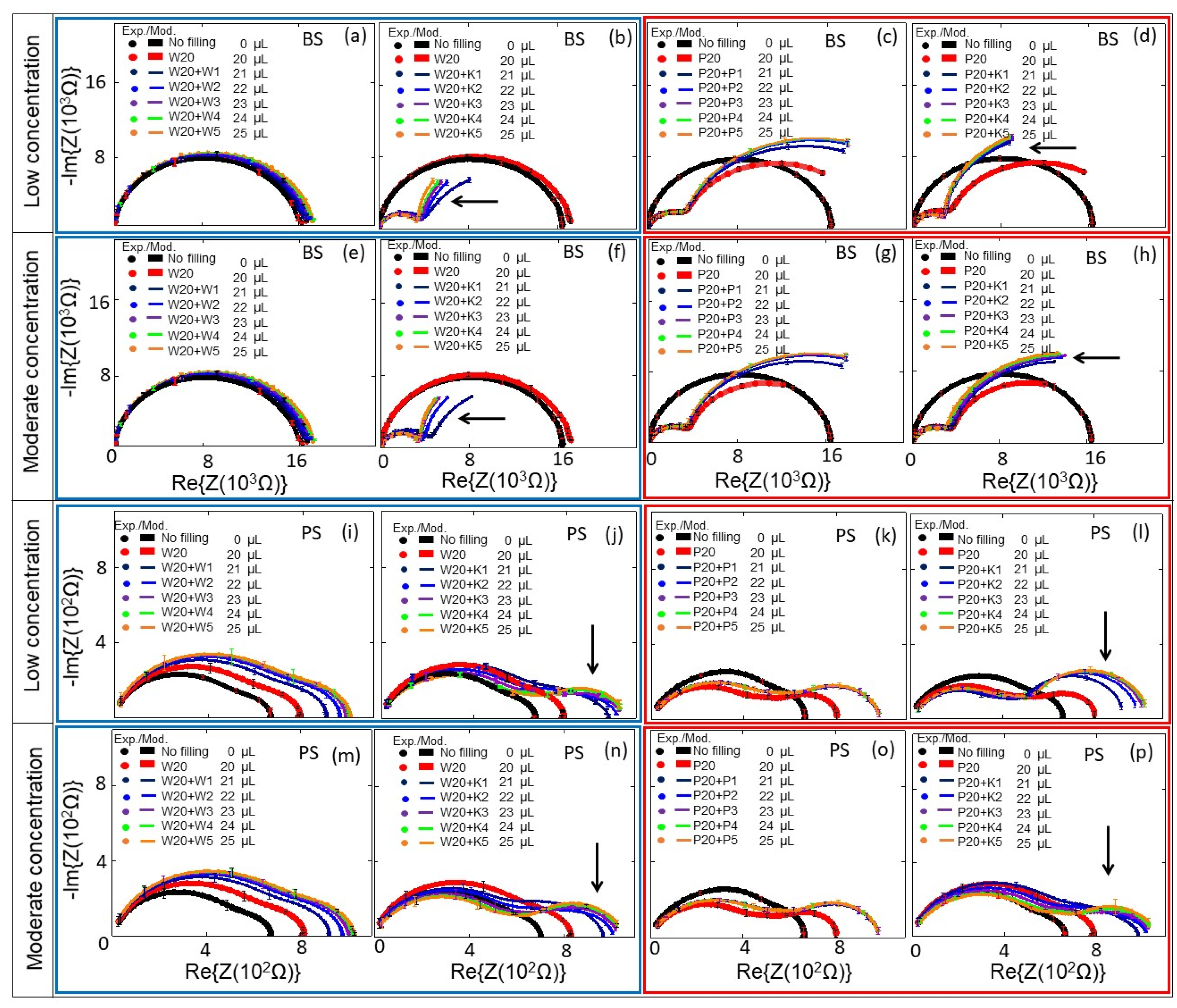
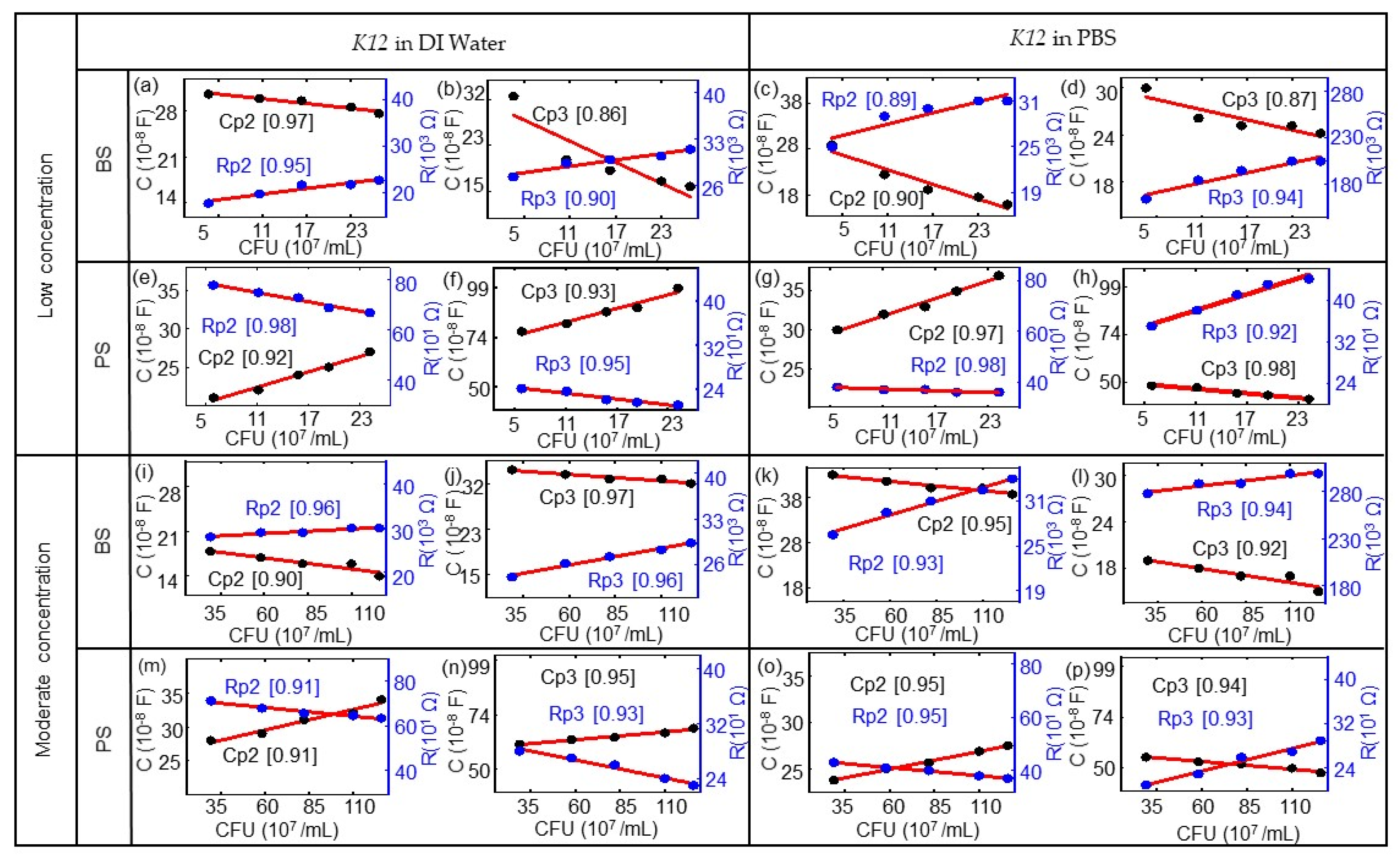
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|
| BS + W20 + W0 | 6.2 × 10−9 | 1.7 × 104 | 4.1 × 10−7 | 1.31× 104 |
| BS + W20 + W1 | 6.0 × 10−9 | 1.9 × 104 | 4.3 × 10−7 | 1.3 × 104 |
| BS + W20 + W2 | 5.9 × 10−9 | 2.0 × 104 | 4.9 × 10−7 | 1.4 × 104 |
| BS + W20 + W3 | 5.8 × 10−9 | 2.1 × 104 | 5.5 × 10−7 | 1.6 × 104 |
| BS + W20 + W4 | 5.7 × 10−9 | 2.2 × 104 | 5.8 × 10−7 | 1.8 × 104 |
| BS + W20 + W5 | 5.6 × 10−9 | 2.3 × 104 | 6.0 × 10−7 | 1.9 × 104 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|
| BS + W20 + W0 | 6.2 × 10−9 | 1.7 × 104 | 4.1 × 10−7 | 1.31× 104 |
| BS + W20 + K1 | 3.1 × 10−9 | 1.8 × 104 | 3.3 × 10−7 | 2.4 × 104 |
| BS + W20 + K2 | 3.0 × 10−9 | 2.0 × 104 | 2.1 × 10−7 | 3.0 × 104 |
| BS + W20 + K3 | 3.0 × 10−9 | 2.2 × 104 | 1.9 × 10−7 | 3.0 × 104 |
| BS + W20 + K4 | 2.9 × 10−9 | 2.2 × 104 | 1.7 × 10−7 | 3.1 × 104 |
| BS + W20 + K5 | 2.8 × 10−9 | 2.3 × 104 | 1.6 × 10−7 | 3.2 × 104 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|
| BS + W20 + W0 | 6.2 × 10−9 | 1.7 × 104 | 4.1 × 10−7 | 1.31× 104 |
| BS + W20 + K1 | 3.0 × 10−9 | 3.0 × 104 | 2.1 × 10−7 | 3.4 × 104 |
| BS + W20 + K2 | 2.9 × 10−9 | 3.2 × 104 | 2.0 × 10−7 | 3.7 × 104 |
| BS + W20 + K3 | 2.8 × 10−9 | 3.5 × 104 | 1.8 × 10−7 | 3.8 × 104 |
| BS + W20 + K4 | 2.6 × 10−9 | 4.2 × 104 | 1.7 × 10−7 | 3.9 × 104 |
| BS + W20 + K5 | 2.5 × 10−9 | 4.7 × 104 | 1.6 × 10−7 | 4.0 × 104 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cp3 (F | Rp3 (Ω) |
|---|---|---|---|---|
| BS + P20 + P0 | 2.0 × 10−9 | 3.3 × 104 | 1.9 × 10−7 | 2.4× 104 |
| BS + P20 + P1 | 2.0 × 10−9 | 3.3 × 104 | 2.2 × 10−7 | 2.6 × 104 |
| BS + P20 + P2 | 2.1 × 10−9 | 3.4 × 104 | 2.3 × 10−7 | 2.7 × 104 |
| BS + P20 + P3 | 2.2 × 10−9 | 3.4 × 104 | 2.4 × 10−7 | 2.8 × 104 |
| BS + P20 + P4 | 2.3 × 10−9 | 3.6 × 104 | 2.5 × 10−7 | 2.9 × 104 |
| BS + P20 + P5 | 2.5 × 10−9 | 3.7 × 104 | 2.7 × 10−7 | 3.0 × 104 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|
| BS + P20 + P0 | 2.0 × 10−9 | 3.3 × 104 | 1.9 × 10−7 | 2.4× 104 |
| BS + P20 + K1 | 2.5 × 10−9 | 2.4 × 104 | 3.1 × 10−7 | 1.8 × 104 |
| BS + P20 + K2 | 2.1 × 10−9 | 2.8 × 104 | 2.7 × 10−7 | 2.0 × 104 |
| BS + P20 + K3 | 1.9 × 10−9 | 2.9 × 104 | 2.6 × 10−7 | 2.1 × 104 |
| BS + P20 + K4 | 1.8 × 10−9 | 3.0 × 104 | 2.6 × 10−7 | 2.2 × 104 |
| BS + P20 + K5 | 1.7 × 10−9 | 3.0 × 104 | 2.5 × 10−7 | 2.2 × 104 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|
| BS + P20 + P0 | 2.0 × 10−9 | 3.3 × 104 | 1.9 × 10−7 | 2.4× 104 |
| BS + P20 +K1 | 1.9 × 10−9 | 2.8 × 104 | 3.8 × 10−7 | 2.5 × 104 |
| BS + P20 +K2 | 1.8 × 10−9 | 2.9 × 104 | 3.7 × 10−7 | 2.7 × 104 |
| BS + P20 +K3 | 1.7 × 10−9 | 2.9 × 104 | 3.6 × 10−7 | 2.8 × 104 |
| BS + P20 +K4 | 1.7 × 10−9 | 3.0 × 104 | 3.6 × 10−7 | 2.9 × 104 |
| BS + P20 +K5 | 1.5 × 10−9 | 3.0 × 104 | 3.5 × 10−7 | 3.0 × 104 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Csch (F) | Rsch (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|---|---|
| PS + W20 + W0 | 5.9 × 10−9 | 7.4 × 102 | 1.7 × 10−7 | 5.5 × 102 | 2.2 × 10−9 | 2.4 × 102 |
| PS + W20 + W1 | 5.7 × 10−9 | 8.2 × 102 | 1.4 × 10−7 | 5.5 × 102 | 1.9 × 10−9 | 2.8 × 102 |
| PS + W20 + W2 | 5.5 × 10−9 | 8.5 × 102 | 1.4 × 10−7 | 5.5 × 102 | 1.7 × 10−9 | 3.1 × 102 |
| PS + W20 + W3 | 5.4 × 10−9 | 8.7 × 102 | 1.3 × 10−7 | 5.4 × 102 | 1.6 × 10−9 | 3.5 × 102 |
| PS + W20 + W4 | 5.2 × 10−9 | 8.9 × 102 | 1.3 × 10−7 | 5.4 × 102 | 1.5 × 10−9 | 3.8 × 102 |
| PS + W20 + W5 | 5.0 × 10−9 | 9.1 × 102 | 1.3 × 10−7 | 5.4 × 102 | 1.2 × 10−9 | 4.1 × 102 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Csch (F) | Rsch (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|---|---|
| PS + W20 + K0 | 5.9 × 10−9 | 7.4 × 102 | 1.7 × 10−7 | 5.5 × 102 | 2.2 × 10−9 | 2.4 × 102 |
| PS + W20 + K1 | 2.1 × 10−9 | 7.8 × 102 | 1.4 × 10−7 | 5.5 × 102 | 7.8 × 10−9 | 2.4 × 102 |
| PS + W20 + K2 | 2.2 × 10−9 | 7.5 × 102 | 1.4 × 10−7 | 5.5 × 102 | 8.2 × 10−9 | 2.4 × 102 |
| PS + W20 + K3 | 2.4 × 10−9 | 7.3 × 102 | 1.3 × 10−7 | 5.4 × 102 | 8.8 × 10−9 | 2.2 × 102 |
| PS + W20 + K4 | 2.5 × 10−9 | 6.9 × 102 | 1.3 × 10−7 | 5.4 × 102 | 9.0 × 10−9 | 2.1 × 102 |
| PS + W20 + K5 | 2.7 × 10−9 | 6.7 × 102 | 1.3 × 10−7 | 5.4 × 102 | 10 × 10−9 | 2.1 × 102 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cpsch (F) | Rsch (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|---|---|
| PS + W20 + K0 | 5.9 × 10−9 | 7.4 × 102 | 1.7 × 10−7 | 5.5 × 102 | 2.2 × 10−9 | 2.4 × 102 |
| PS + W20 + K1 | 2.8 × 10−9 | 6.8 × 102 | 1.4 × 10−7 | 5.5 × 102 | 6.1 × 10−9 | 2.8 × 102 |
| PS + W20 + K2 | 2.9 × 10−9 | 6.5 × 102 | 1.4 × 10−7 | 5.5 × 102 | 6.3 × 10−9 | 2.7 × 102 |
| PS + W20 + K3 | 3.1 × 10−9 | 6.3 × 102 | 1.3 × 10−7 | 5.4 × 102 | 6.4 × 10−9 | 2.6 × 102 |
| PS + W20 + K4 | 3.2× 10−9 | 6.2 × 102 | 1.3 × 10−7 | 5.4 × 102 | 6.6 × 10−9 | 2.4 × 102 |
| PS + W20 + K5 | 3.4 × 10−9 | 6.1 × 102 | 1.3 × 10−7 | 5.4 × 102 | 6.8 × 10−9 | 2.3 × 102 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cpsch (F) | Rsch (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|---|---|
| PS + P20 + P0 | 7.8 × 10−9 | 4.7 × 102 | 2.7 × 10−8 | 3.5 × 102 | 1.4 × 10−7 | 2.8 × 102 |
| PS + P20 + P1 | 7.9 × 10−9 | 6.8 × 102 | 1.4 × 10−8 | 5.5 × 102 | 1.1 × 10−9 | 3.4 × 102 |
| PS + P20 + P2 | 7.9 × 10−9 | 6.7 × 102 | 1.4 × 10−8 | 5.4 × 102 | 1.2 × 10−9 | 3.4 × 102 |
| PS + P20 + P3 | 7.9 × 10−9 | 6.7 × 102 | 1.4 × 10−8 | 5.4 × 102 | 1.2 × 10−9 | 3.4 × 102 |
| PS + P20 + P4 | 7.9 × 10−9 | 6.7 × 102 | 1.4 × 10−8 | 5.4 × 102 | 1.2 × 10−9 | 3.4 × 102 |
| PS + P20 + P5 | 7.9 × 10−9 | 6.7 × 102 | 1.4 × 10−8 | 5.4 × 102 | 1.2 × 10−9 | 3.4 × 102 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cpsch (F) | Rsch (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|---|---|
| PS + P20 + K0 | 7.8 × 10−9 | 4.7 × 102 | 2.7 × 10−8 | 3.5 × 102 | 1.4 × 10−7 | 2.8 × 102 |
| PS + P20 + K1 | 3.0 × 10−9 | 3.8 × 102 | 1.4 × 10−7 | 5.5 × 102 | 4.8 × 10−9 | 3.5 × 102 |
| PS + P20 + K2 | 3.2 × 10−9 | 3.7 × 102 | 1.3 × 10−7 | 5.4 × 102 | 4.7 × 10−9 | 3.8 × 102 |
| PS + P20 + K3 | 3.3 × 10−9 | 3.7 × 102 | 1.3 × 10−7 | 5.4 × 102 | 4.4 × 10−9 | 4.1 × 102 |
| PS + P20 + K4 | 3.5 × 10−9 | 3.6 × 102 | 1.3 × 10−7 | 5.4 × 102 | 4.3 × 10−9 | 4.3 × 102 |
| PS + P20 + K5 | 3.7 × 10−9 | 3.6 × 102 | 1.4 × 10−7 | 5.5 × 102 | 4.1 × 10−9 | 4.4 × 102 |
| Circuit Element | Cp2 (F) | Rp2 (Ω) | Cpsch (F) | Rsch (Ω) | Cp3 (F) | Rp3 (Ω) |
|---|---|---|---|---|---|---|
| PS + P20 + K0 | 7.8 × 10−9 | 4.7 × 102 | 2.7 × 10−8 | 3.5 × 102 | 1.4 × 10−7 | 2.8 × 102 |
| PS + P20 + K1 | 1.8 × 10−9 | 4.3 × 102 | 1.4 × 10−7 | 5.5 × 102 | 5.5 × 10−9 | 2.9 × 102 |
| PS + P20 + K2 | 2.0 × 10−9 | 4.1 × 102 | 1.4 × 10−7 | 5.5 × 102 | 5.3 × 10−9 | 2.7 × 102 |
| PS + P20 + K3 | 2.1 × 10−9 | 4.0 × 102 | 1.3 × 10−7 | 5.4 × 102 | 5.2 × 10−9 | 2.6 × 102 |
| PS + P20 + K4 | 2.3 × 10−9 | 3.8 × 102 | 1.3 × 10−7 | 5.4 × 102 | 5.0 × 10−9 | 2.3 × 102 |
| PS + P20 + K5 | 2.4 × 10−9 | 3.7 × 102 | 1.3 × 10−7 | 5.4 × 102 | 4.8 × 10−9 | 2.1 × 102 |
Appendix B
| |Z|(Ω) @ 40 Hz | |Z|(Ω) @ 400 Hz | |Z|(Ω) @ 4000 Hz | |
|---|---|---|---|
| BS + W20 + K1 | 17956 | 16325 | 8732 |
| BS + W20 + K2 | 17202 | 16088 | 8425 |
| BS + W20 + K3 | 16871 | 15851 | 8321 |
| BS + W20 + K4 | 16689 | 15122 | 8258 |
| BS + W20 + K5 | 16198 | 14712 | 8104 |
| |Z|(Ω) @ 40 Hz | |Z|(Ω) @ 400 Hz | |Z|(Ω) @ 4000 Hz | |
|---|---|---|---|
| PS + W20 +K1 | 820 | 805 | 768 |
| PS + W20 +K2 | 812 | 796 | 761 |
| PS + W20 +K3 | 804 | 789 | 752 |
| PS + W20 +K4 | 796 | 783 | 748 |
| PS + W20 +K5 | 790 | 778 | 741 |
References
- Turano, A.; Pirali, F. Quantification Methods in Microbiology. In Laboratory Diagnosis of Infectious Diseases; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 1988; pp. 8–13. [Google Scholar]
- Stevenson, K.; McVey, A.F.; Clark, I.B.N.; Swain, P.S.; Pilizota, T. General calibration of microbial growth in microplate readers. Sci. Rep. 2016, 6, 38828. [Google Scholar] [CrossRef] [PubMed]
- Meredith, W.J.; Massey, J.B. Fundamental Physics of Radiology; Butterworth-Heinemann: Oxford, UK, 1977. [Google Scholar]
- Bereiter-Hahn, J.; Fox, C.H.; Thorell, B. Quantitative reflection contrast microscopy of living cells. J. Cell. Biol. 1979, 82, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Fruin, J.T.; Idll, T.M.; Clarke, J.B.; Fowler, J.L.; Guthertz, L.S. Accuracy and Speed in Counting Agar Plates1. J. Food Prot. 1977, 40, 596–599. [Google Scholar] [CrossRef] [PubMed]
- Haandbæk, N.; Bürgel, S.C.; Heer, F.; Hierlemann, A. Characterization of subcellular morphology of single yeast cells using high frequency microfluidic impedance cytometer. Lab Chip 2014, 14, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Krommenhoek, E.E.; Gardeniers, H.J.; Bomer, J.; Berg, A.V.D.; Li, X.; Ottens, M.; Van Der Wielen, L.; Van Dedem, G.; Van Leeuwen, M.; Van Gulik, W.; et al. Monitoring of yeast cell concentration using a micromachined impedance sensor. Sens. Actuators B Chem. 2006, 115, 384–389. [Google Scholar] [CrossRef]
- Turolla, A.; Di Mauro, M.; Mezzera, L.; Antonelli, M.; Carminati, M. Development of a Miniaturized and Selective Impedance Sensor for Real-Time Slime Monitoring in Pipes and Tanks. Sens. Actuators B Chem. 2019, 281, 288–295. [Google Scholar] [CrossRef]
- Suehiro, J.; Ohtsubo, A.; Hatano, T.; Hara, M. Selective detection of bacteria by a dielectrophoretic impedance measurement method using an antibody-immobilized electrode chip. Sens. Actuators B Chem. 2006, 119, 319–326. [Google Scholar] [CrossRef]
- Gawad, S.; Schild, L.; Renaud, P. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab Chip 2001, 1, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Geddes, L.A.; Baker, L.E. Principles of Applied Biotent, Medical Instrumentation, 3rd ed.; Wiley: New York, NY, USA, 1989; pp. 566–576. [Google Scholar]
- Felice, C.J.; Madrid, R.E.; Olivera, J.M.; Rotger, V.I.; Valentinuzzi, M.E. Impedance microbiology: Quantification of bacterial content in milk by means of capacitance growth curves. J. Microbiol. Methods 1999, 35, 37–42. [Google Scholar] [CrossRef]
- Cady, P. Progress in Impedance Measurements in Microlayer—Biology. In Mechanizing Microbiology; Sharpe, A.N., Clark, D.S., Eds.; Thomas: Springfield, IL, USA, 1978; pp. 199–239. [Google Scholar]
- Kiani, M.; Du, N.; Vogel, M.; Raff, J.; Hübner, U.; Skorupa, I.; Bürger, D.; Schulz, S.E.; Schmidt, O.G.; Schmidt, H. P-N Junction-Based Si Biochips with Ring Electrodes for Novel Biosensing Applications. Biosensors 2019, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Kiani, M.; Du, N.; Vogel, M.; Raff, J.; Hübner, U.; Skorupa, I.; Bürger, D.; Schulz, S.E.; Schmidt, O.G.; Blaschke, D.; et al. Disturbing-Free Determination of Yeast Concentration in DI Water and in Glucose Using Impedance Biochips. Biosensors 2020, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Jungerman, J.A. Fourth-order uniform electric field from two charged rings. Rev. Sci. Instrum. 1984, 55, 1479. [Google Scholar] [CrossRef]
- Scully, J.R.; Silverman, D.C.; Kendig, M.W. Electrochemical Impedance—Analysis and Interpretation; ASTM: Philadelphia, PA, USA, 1993; p. 54. [Google Scholar]
- Fricke, H. The theory of Electrolyte Polarization. Philos. Mag. 1953, 14, 310–318. [Google Scholar] [CrossRef]
- Kerrison, L. The capacitor. Stud. Q. J. 1933, 4, 100. [Google Scholar] [CrossRef]
- Cole, K.S.; Cole, R.H. Dispersion and Absorption in Dielectrics I. Alternating Current Characteristics. J. Chem. Phys. 1941, 9, 341. [Google Scholar] [CrossRef]
- McKubre, M.C.H.; Macdonald, D.D. Electronic Instrumentation for Electrochemical Studies. In A Comprehensive Treatise of Electrochemistry; Bockris, J.O., Conway, B.E., Yeager, E., Eds.; Plenum Press: Boston, MA, USA, 1984. [Google Scholar]
- Zandman, F.; Stein, S. A New Precision Film Resistor Exhibiting Bulk Properties. IEEE Trans. Compon. Parts 1964, 11, 107–119. [Google Scholar] [CrossRef]
- Usui, Y. Equivalent Circuit of Distributed Constant Circuit and Transmission Equation. J. Jpn. Inst. Electron. Packag. 2018, 21, 311–316. [Google Scholar] [CrossRef]
- Bauerle, J.E. Study of Solid Electrolyte Polarization by a Complex Admittance Method. J. Phys. Chem. Solids 1969, 30, 2657–2670. [Google Scholar] [CrossRef]
- Sihvola, A. Electromagnetic Mixing Formulas and Applications; The Institute of Electrical Engineers: London, UK, 1999. [Google Scholar]
- Barsoukov, E.; Ross, M.J. Impedance Spectroscopy: Theory, Experiment, and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Novoseleskii, M.; Gudina, N.N.; Fetistov, Y.I. Identical Equivalent Impedance Circuits. Sov. Electrochem. 1972, 8, 546–548. [Google Scholar]

| Biochip | Implanted Ion | Ion Energy (MeV) | Ion Fluence (cm−2) |
|---|---|---|---|
| PS | Phosphorus | 1.00 | 3 × 1013 |
| BS | Boron | 0.45 | 3 × 1013 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiani, M.; Tannert, A.; Du, N.; Hübner, U.; Skorupa, I.; Bürger, D.; Zhao, X.; Blaschke, D.; Rebohle, L.; Cherkouk, C.; et al. Towards Bacteria Counting in DI Water of Several Microliters or Growing Suspension Using Impedance Biochips. Biosensors 2020, 10, 82. https://doi.org/10.3390/bios10080082
Kiani M, Tannert A, Du N, Hübner U, Skorupa I, Bürger D, Zhao X, Blaschke D, Rebohle L, Cherkouk C, et al. Towards Bacteria Counting in DI Water of Several Microliters or Growing Suspension Using Impedance Biochips. Biosensors. 2020; 10(8):82. https://doi.org/10.3390/bios10080082
Chicago/Turabian StyleKiani, Mahdi, Astrid Tannert, Nan Du, Uwe Hübner, Ilona Skorupa, Danilo Bürger, Xianyue Zhao, Daniel Blaschke, Lars Rebohle, Charaf Cherkouk, and et al. 2020. "Towards Bacteria Counting in DI Water of Several Microliters or Growing Suspension Using Impedance Biochips" Biosensors 10, no. 8: 82. https://doi.org/10.3390/bios10080082
APA StyleKiani, M., Tannert, A., Du, N., Hübner, U., Skorupa, I., Bürger, D., Zhao, X., Blaschke, D., Rebohle, L., Cherkouk, C., Neugebauer, U., Schmidt, O. G., & Schmidt, H. (2020). Towards Bacteria Counting in DI Water of Several Microliters or Growing Suspension Using Impedance Biochips. Biosensors, 10(8), 82. https://doi.org/10.3390/bios10080082






