Battery-Powered Portable Rotary Real-Time Fluorescent qPCR with Low Energy Consumption, Low Cost, and High Throughput
Abstract
1. Introduction
2. Materials and Methods
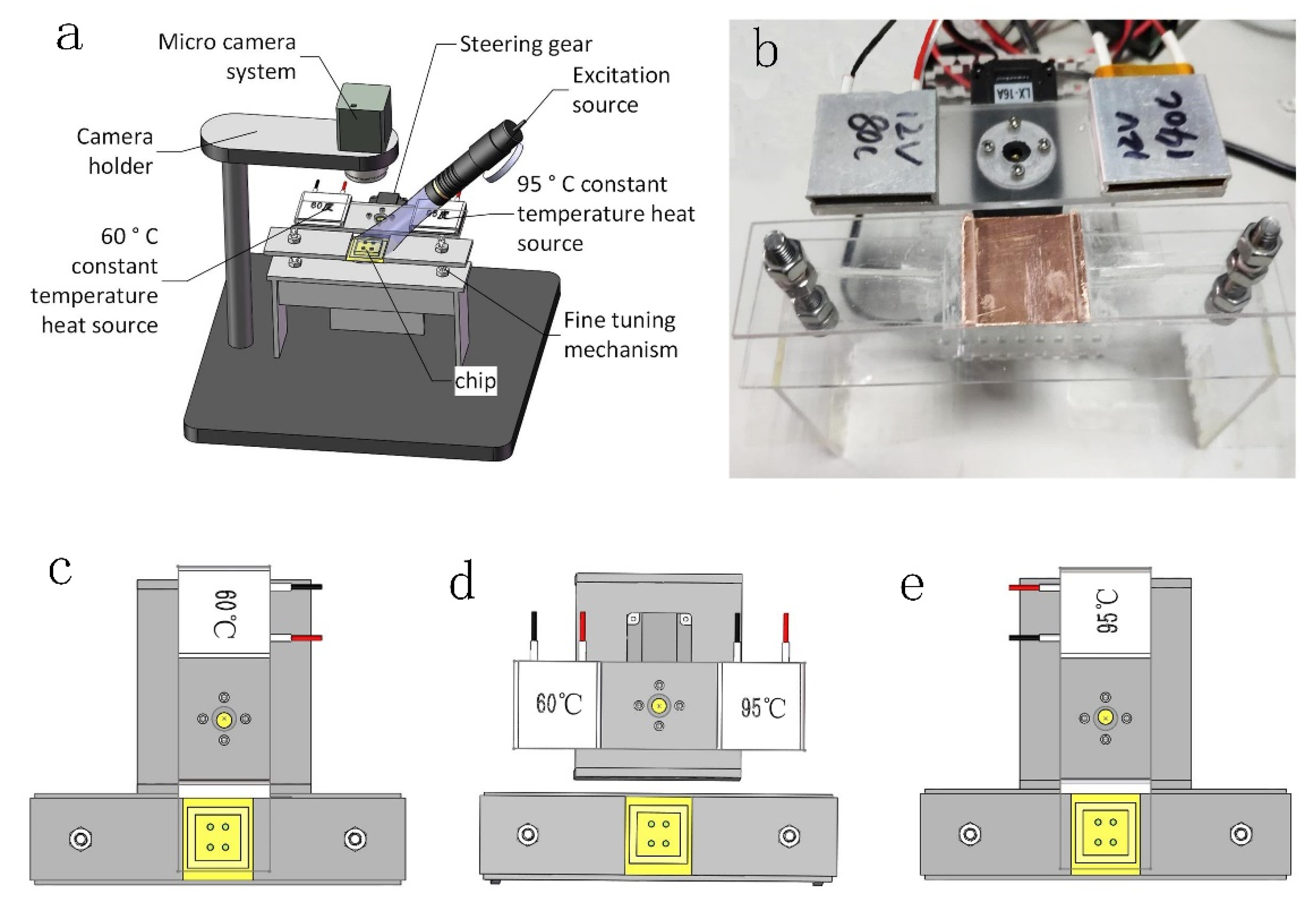
2.1. Thermal Cycling System
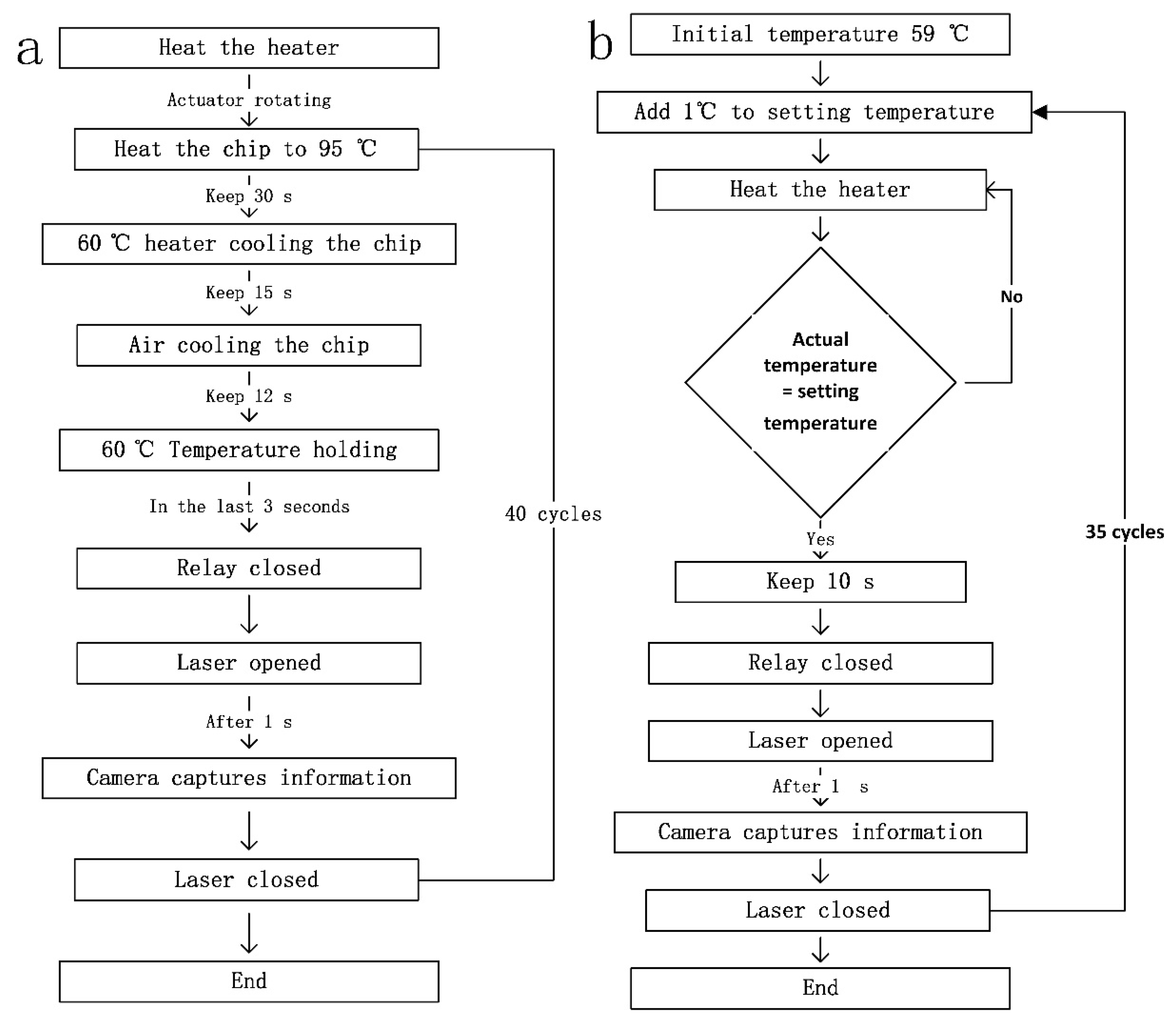
2.1.1. Amplified Heat Cycle System
2.1.2. Melting Heat Cycling System
2.2. Optical Detection System
2.3. Microfluidic Chip
2.4. Reagents
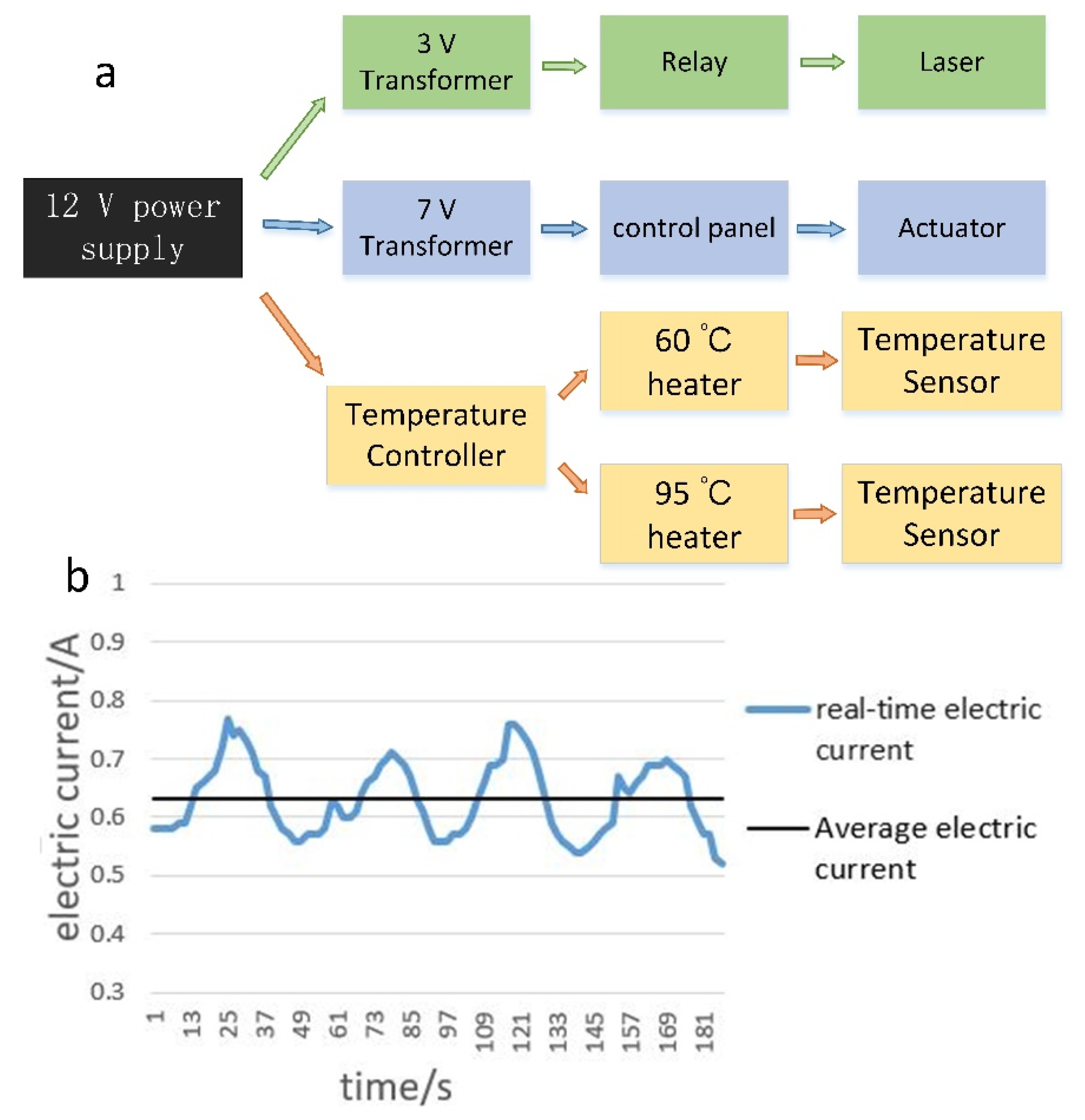
2.5. Power Management
3. Results and Discussion
3.1. Analysis of the Temperature Curve
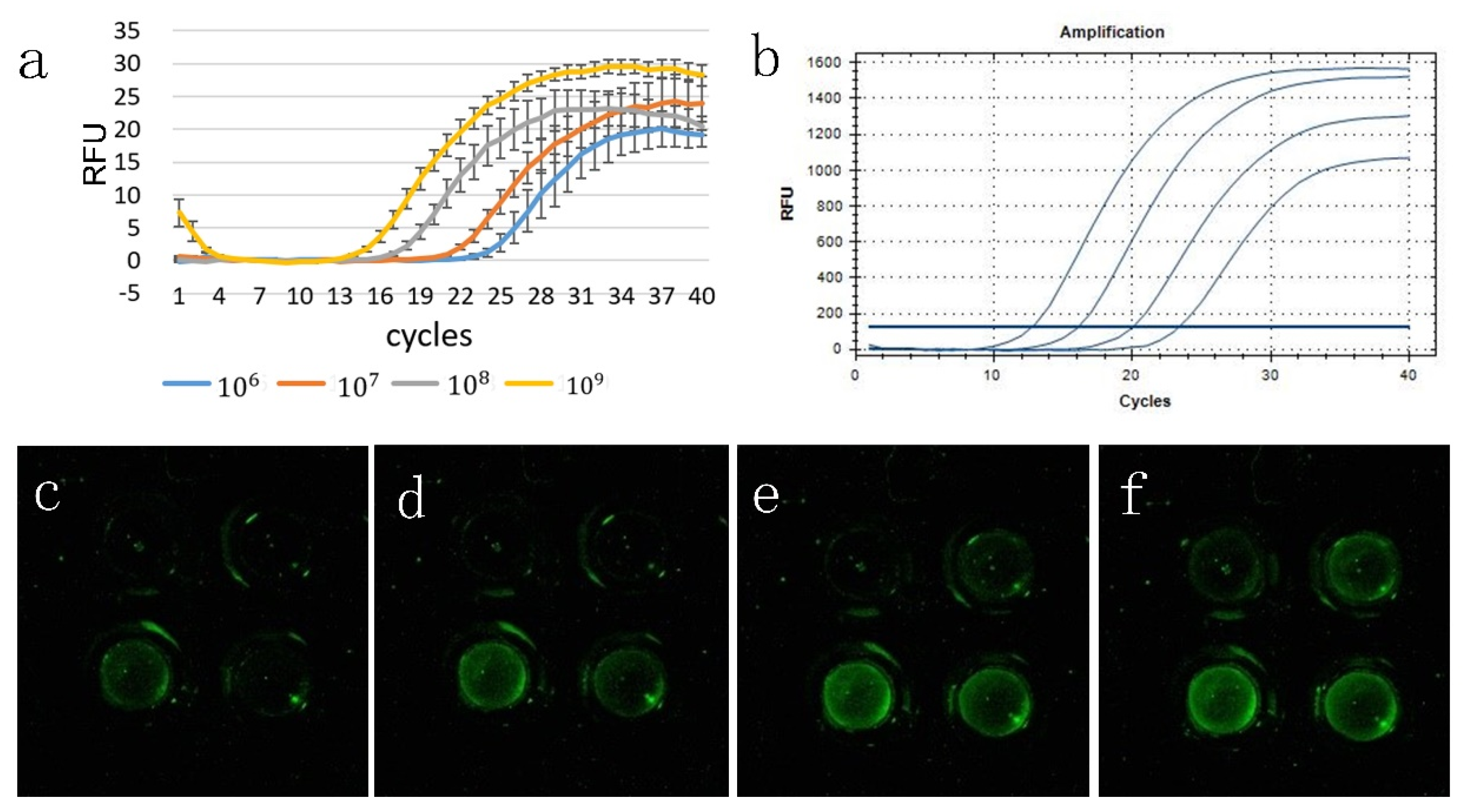
3.2. Analysis of the Gradient Template Amplification Curve
3.3. Melting Curve Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Northrup, M.A.; Gonzalez, C.; Hadley, D.; Hills, R.; Landre, P.; Lehew, S.; Saw, R.; Sninsky, J.; Watson, R. A MEMS-based miniature DNA analysis system. In Proceedings of the International Solid-State Sensors and Actuators Conference-TRANSDUCERS’95, Stockholm, Sweden, 25–29 June 1995; pp. 764–767. [Google Scholar]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Shengqi, W.; Xiaohong, W.; Suhong, C.; Wei, G. A new fluorescent quantitative polymerase chain reaction technique. Anal. Biochem. 2002, 309, 206–2113. [Google Scholar] [CrossRef]
- Arya, M.; Shergill, I.S.; Williamson, M.; Gommersall, L.; Arya, N.; Patel, H.R. Basic principles of real-time quantitative PCR. Expert Rev. Mol. Diagn. 2005, 5, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Mueller, R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. 2005, 109, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Dawson, E.D.; Moore, C.L.; Dankbar, D.M.; Mehlmann, M.; Townsend, M.B.; Smagala, J.A.; Smith, C.B.; Cox, N.J.; Kuchta, R.D.; Rowlen, K.L. Identification of A/H5N1 influenza viruses using a single gene diagnostic microarray. Anal. Chem. 2007, 79, 378–384. [Google Scholar] [CrossRef]
- Ho, S.K.; Yam, W.-C.; Leung, E.T.; Wong, L.-P.; Leung, J.K.; Lai, K.-N.; Chan, T.-M. Rapid quantification of hepatitis B virus DNA by real-time PCR using fluorescent hybridization probes. J. Med. Microbiol. 2003, 52, 397–402. [Google Scholar] [CrossRef]
- Mauk, M.; Song, J.; Bau, H.H.; Gross, R.; Bushman, F.D.; Collman, R.G.; Liu, C. Miniaturized devices for point of care molecular detection of HIV. Lab Chip 2017, 17, 382–394. [Google Scholar] [CrossRef]
- Van Elden, L.; Nijhuis, M.; Schipper, P.; Schuurman, R.; Van Loon, A. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 2001, 39, 196–200. [Google Scholar] [CrossRef]
- Templeton, K.E.; Scheltinga, S.A.; Beersma, M.F.; Kroes, A.C.; Claas, E.C. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 2004, 42, 1564–1569. [Google Scholar] [CrossRef]
- Ellis, J.; Iturriza, M.; Allan, R.; Bermingham, A.; Brown, K.; Gray, J.; Brown, D. Evaluation of four real-time PCR assays for detection of influenza A (H1N1) v viruses. Eurosurveillance 2009, 14, 19230. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.; Stat, M.; Evans, R.D.; Kennington, W.J. A fluorescence-based quantitative real-time PCR assay for accurate Pocillopora damicornis species identification. Coral Reefs 2016, 35, 895–899. [Google Scholar] [CrossRef]
- Lee, S.-H.; Ahn, M.-J.; Hong, J.-S.; Lee, J.-H. Diversity and community analysis of fermenting bacteria isolated from eight major Korean fermented foods using arbitrary-primed PCR and 16S rRNA gene sequencing. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 453–461. [Google Scholar] [CrossRef]
- Kirkizlar, E.; Zimmermann, B.; Constantin, T.; Swenerton, R.; Hoang, B.; Wayham, N.; Babiarz, J.E.; Demko, Z.; Pelham, R.J.; Kareht, S. Detection of clonal and subclonal copy-number variants in cell-free DNA from patients with breast cancer using a massively multiplexed PCR methodology. Transl. Oncol. 2015, 8, 407–416. [Google Scholar] [CrossRef]
- Todd, A.V.; Fuery, C.J.; Impey, H.L.; Applegate, T.L.; Haughton, M.A. DzyNA-PCR: Use of DNAzymes to detect and quantify nucleic acid sequences in a real-time fluorescent format. Clin. Chem. 2000, 46, 625–630. [Google Scholar] [CrossRef]
- Salman, A.; Carney, H.; Bateson, S.; Ali, Z. Shunting microfluidic PCR device for rapid bacterial detection. Talanta 2020, 207, 120303. [Google Scholar] [CrossRef]
- Neuzil, P.; Novak, L.; Pipper, J.; Lee, S.; Ng, L.F.; Zhang, C. Rapid detection of viral RNA by a pocket-size real-time PCR system. Lab Chip 2010, 10, 2632–2634. [Google Scholar] [CrossRef]
- Lagally, E.; Medintz, I.; Mathies, R. Single-molecule DNA amplification and analysis in an integrated microfluidic device. Anal. Chem. 2001, 73, 565–570. [Google Scholar] [CrossRef]
- Ahrberg, C.D.; Ilic, B.R.; Manz, A.; Neužil, P. Handheld real-time PCR device. Lab Chip 2016, 16, 586–592. [Google Scholar] [CrossRef]
- Lee, T.M.-H.; Carles, M.C.; Hsing, I.-M. Microfabricated PCR-electrochemical device for simultaneous DNA amplification and detection. Lab Chip 2003, 3, 100–105. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, B.; Wu, W. Application of automatic feedback photographing by portable smartphone in PCR. Sens. Actuators B Chem. 2019, 298, 126782. [Google Scholar] [CrossRef]
- Bialek, H.; Dawes, J.; Heer, D.; Johnston, M.L. Portable real-time PCR system using tablet-based fluorescence imaging. In Proceedings of the 2016 IEEE EMBS International Student Conference (ISC), Ottawa, ON, Canada, 29–31 May 2016; pp. 1–4. [Google Scholar]
- Zou, Q.; Miao, Y.; Chen, Y.; Sridhar, U.; Chong, C.S.; Chai, T.; Tie, Y.; Teh, C.H.L.; Lim, T.M.; Heng, C. Micro-assembled multi-chamber thermal cycler for low-cost reaction chip thermal multiplexing. Sens. Actuators A Phys. 2002, 102, 114–121. [Google Scholar] [CrossRef]
- Wu, D.; Wu, W. Battery Powered Portable Thermal Cycler for Continuous-Flow Polymerase Chain Reaction Diagnosis by Single Thermostatic Thermoelectric Cooler and Open-Loop Controller. Sensors 2019, 19, 1609. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Choi, S.J.; Park, B.H.; Choi, Y.K.; Seo, T.S. Ultrafast rotary PCR system for multiple influenza viral RNA detection. Lab Chip 2012, 12, 1598–1600. [Google Scholar] [CrossRef]
- Zhang, Y.; Ozdemir, P. Microfluidic DNA amplification—A review. Anal. Chim. Acta 2009, 638, 115–125. [Google Scholar] [CrossRef]


| Reference | Journal/Years | Temperature Control Mode | Energy Consumption | Gradient Amplification Curve | Melting Curve |
|---|---|---|---|---|---|
| [23] | IEEE Explore 2016 | TEC | 6 ah (30 cycles) | provided | provided |
| [25] | Sensors 2019 | Constant temperature heater | 6 W | Not provided | Not provided |
| [17] | Talanta 2020 | Translational motion | Not provided | Not provided | Not provided |
| [26] | Lab chip 2012 | rotation | Not provided | Not provided | Not provided |
| This study | 2020 | rotation | 7.6 W | provided | provided |
| Cooling Mode | Overall Cooling Time | Overall Cooling Rate | 95–68 °C Cooling Rate | 68–60 °C Cooling Rate |
|---|---|---|---|---|
| mode 1 | 45 s | 0.77 °C/s | 0.9 °C/s | 0.53 °C/s1 |
| mode 2 | 47 s | 0.74 °C/s | 2.25 °C/s | 0.23 °C/s |
| mode 3 | 28 s | 1.25 °C/s | 2.25 °C/s | 0.53 °C/s |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Sang, B.; Wu, W. Battery-Powered Portable Rotary Real-Time Fluorescent qPCR with Low Energy Consumption, Low Cost, and High Throughput. Biosensors 2020, 10, 49. https://doi.org/10.3390/bios10050049
He L, Sang B, Wu W. Battery-Powered Portable Rotary Real-Time Fluorescent qPCR with Low Energy Consumption, Low Cost, and High Throughput. Biosensors. 2020; 10(5):49. https://doi.org/10.3390/bios10050049
Chicago/Turabian StyleHe, Limin, Benliang Sang, and Wenming Wu. 2020. "Battery-Powered Portable Rotary Real-Time Fluorescent qPCR with Low Energy Consumption, Low Cost, and High Throughput" Biosensors 10, no. 5: 49. https://doi.org/10.3390/bios10050049
APA StyleHe, L., Sang, B., & Wu, W. (2020). Battery-Powered Portable Rotary Real-Time Fluorescent qPCR with Low Energy Consumption, Low Cost, and High Throughput. Biosensors, 10(5), 49. https://doi.org/10.3390/bios10050049






