Structural and Functional Stability of DNA Nanopores in Biological Media
Abstract
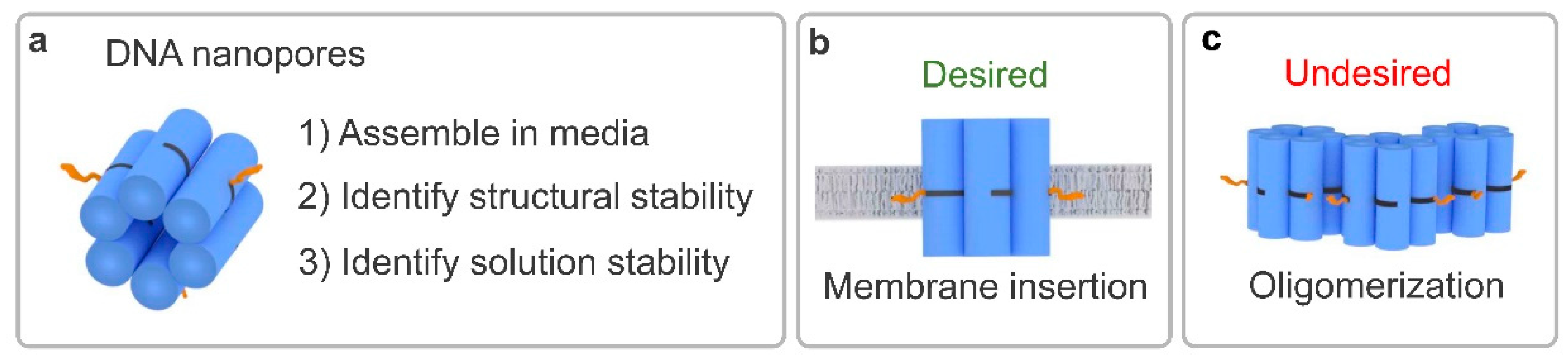
1. Introduction
2. Materials and Methods
3. Results
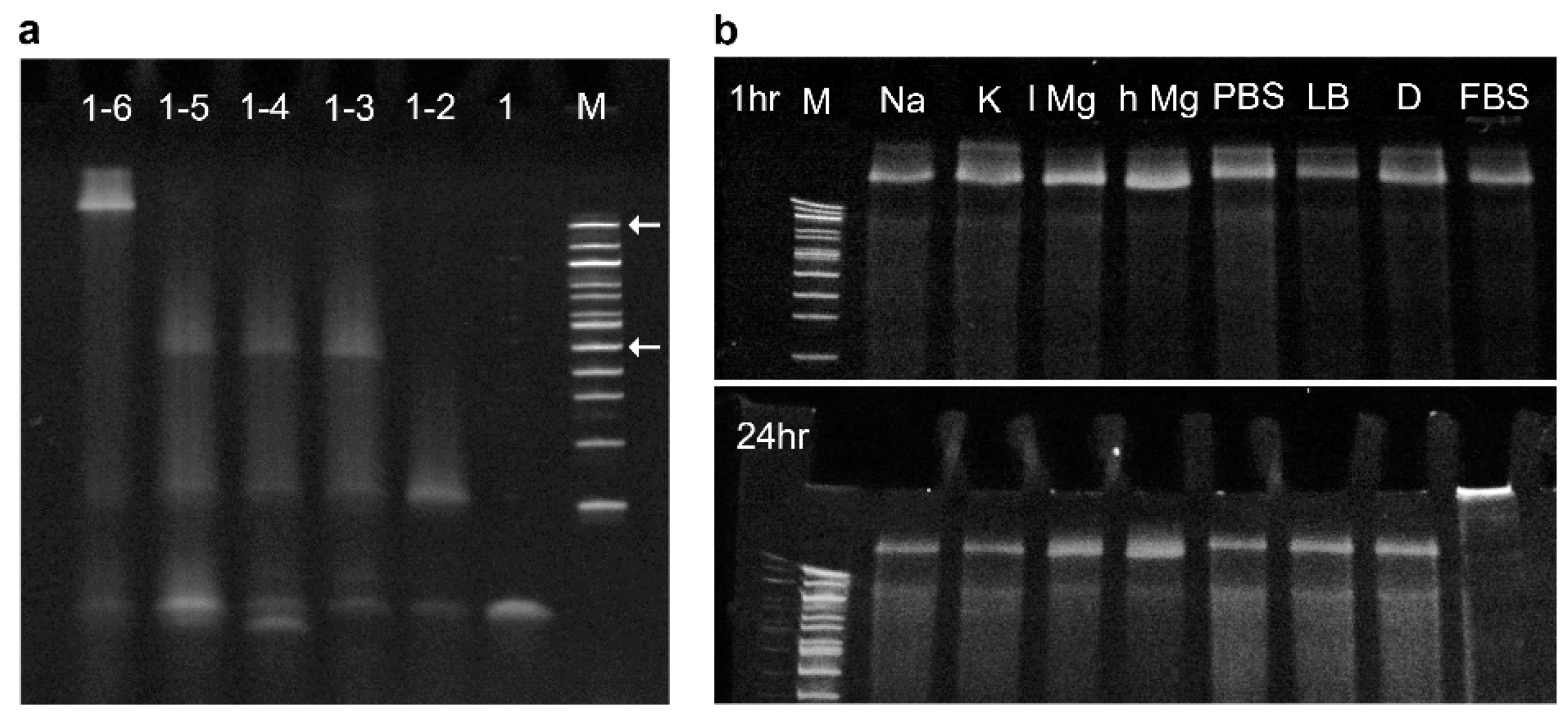
3.1. Determining Nanopore Formation in Media
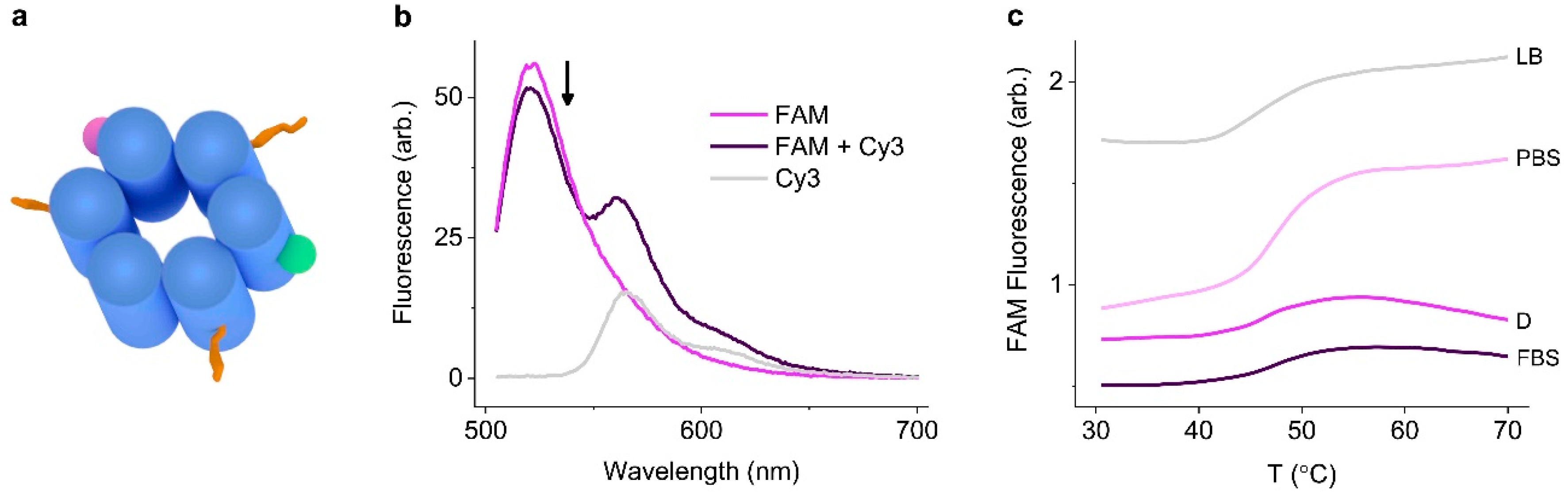
3.2. Identifying Nanopore Melting Temperatures in Biological Media
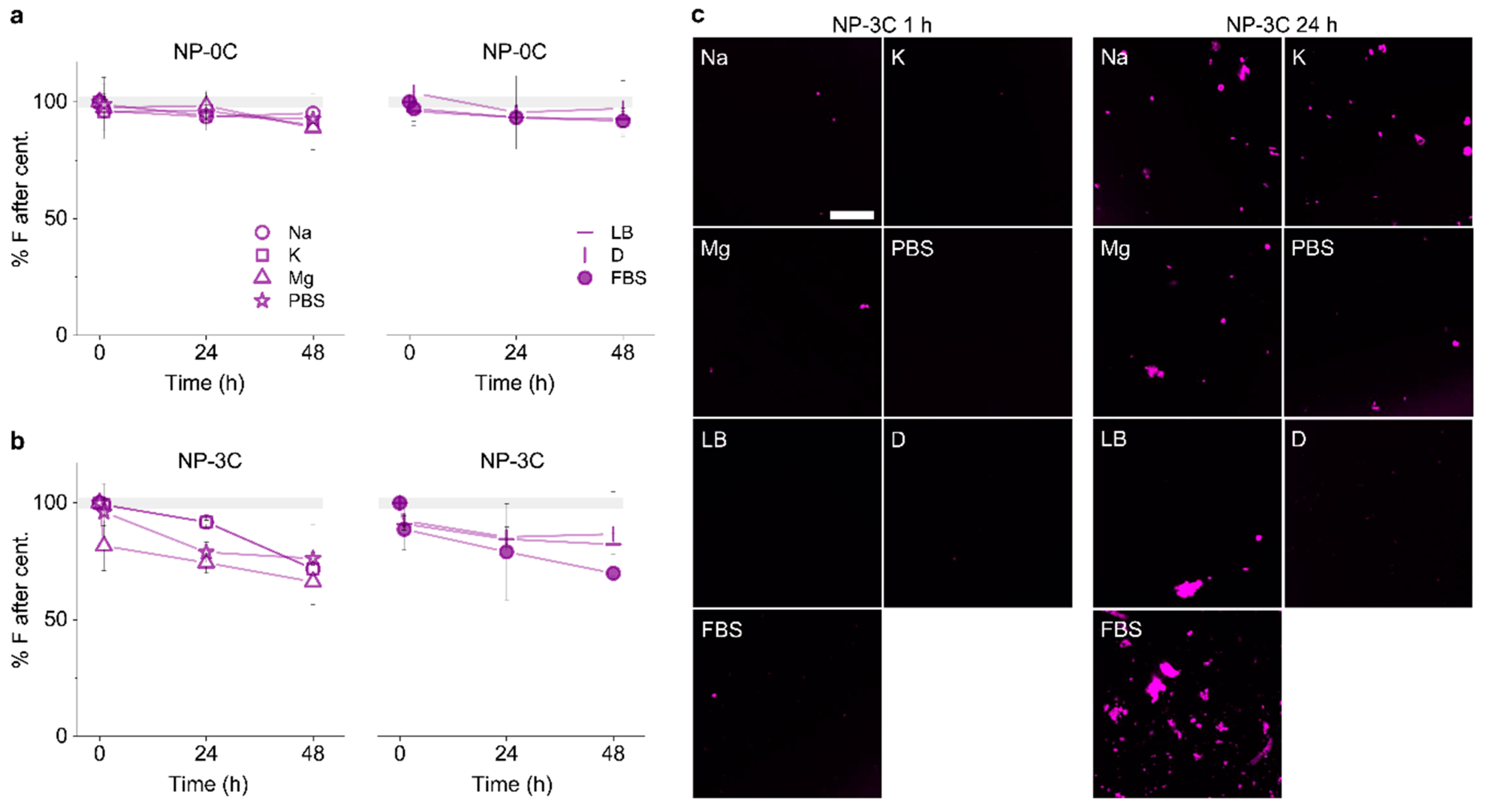
3.3. Identifying Time-Dependent Nanopore Water-Solubility
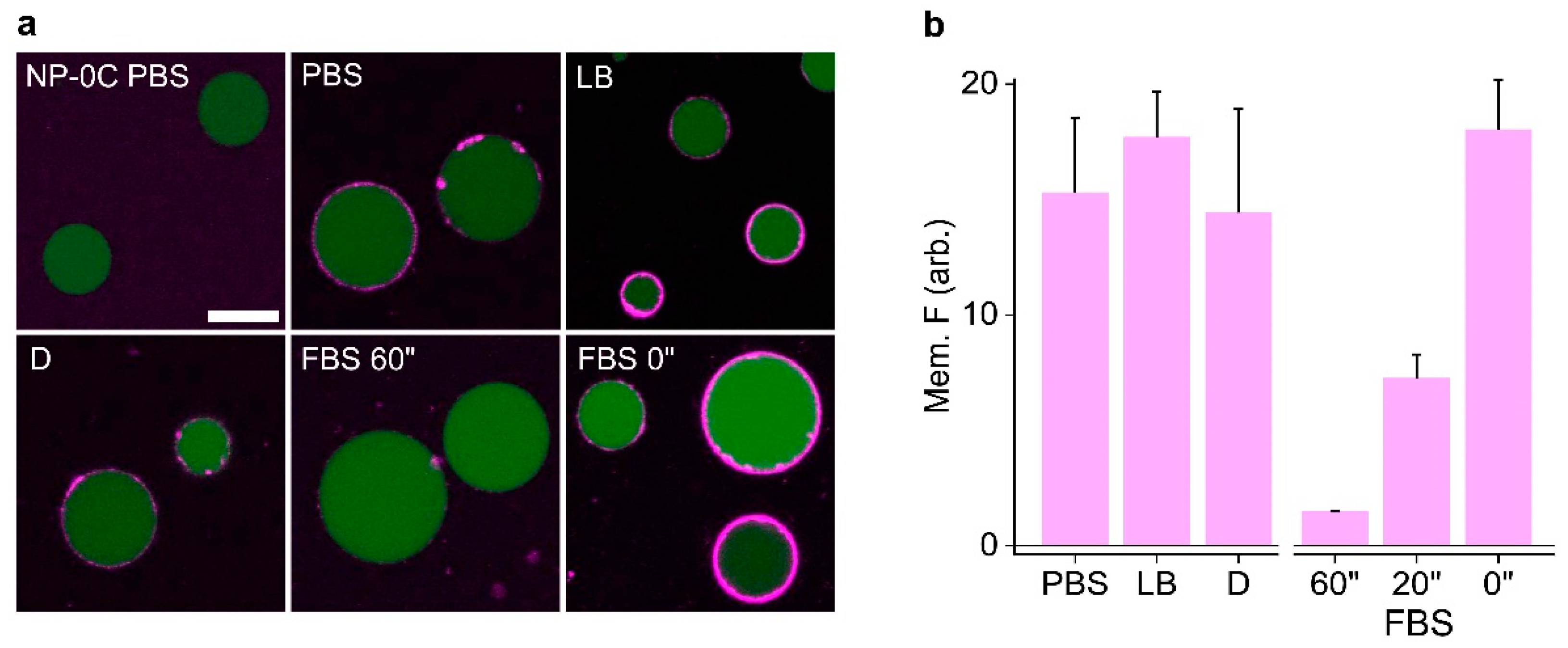
3.4. Identifying Nanopore Membrane Binding Activity in Media
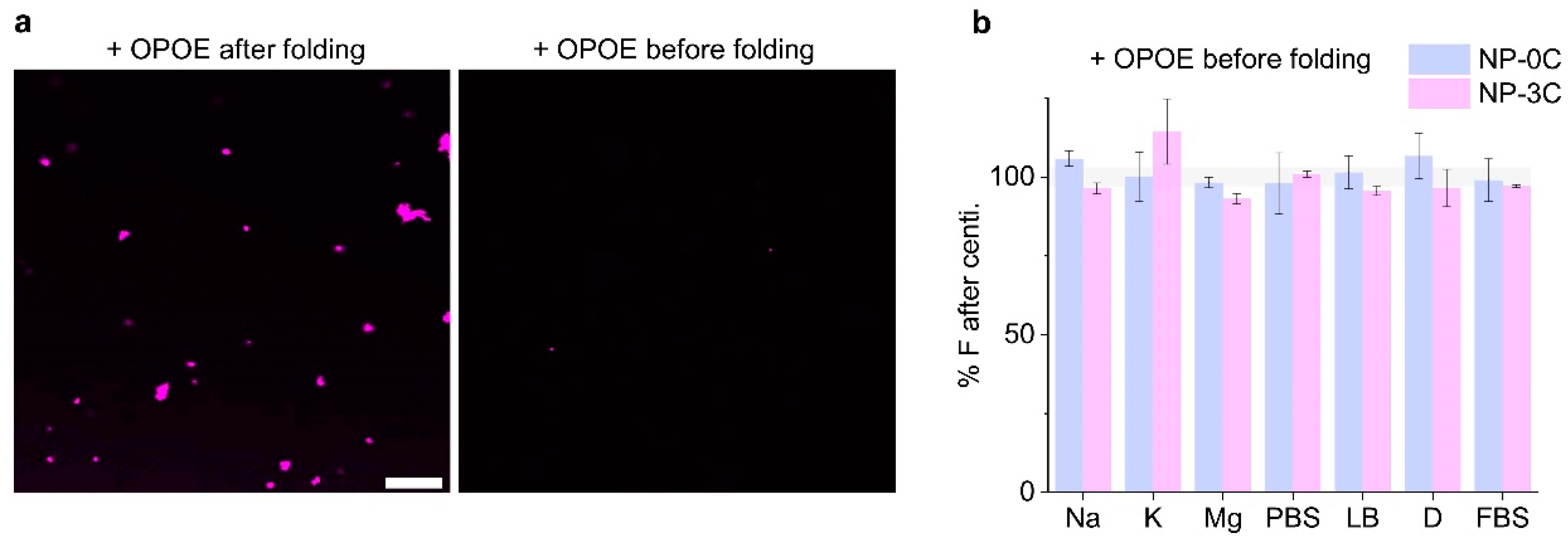
3.5. Adding Detergent Prevents Nanopore Aggregation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into twisted and curved nanoscale shapes. Science 2009, 325, 725–730. [Google Scholar] [CrossRef]
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. [Google Scholar] [CrossRef]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Hogberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Eckstein, F. Modified oligonucleotides: Synthesis and strategy for users. Ann. Rev. Biochem. 1998, 67, 99–134. [Google Scholar] [CrossRef] [PubMed]
- Edwardson, T.G.W.; Carneiro, K.M.M.; Serpell, C.J.; Sleiman, H.F. An efficient and modular route to sequence-defined polymers appended to DNA. Angew. Chem. Int. Ed. 2014, 53, 4567–4571. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef] [PubMed]
- Halley, P.D.; Lucas, C.R.; McWilliams, E.M.; Webber, M.J.; Patton, R.A.; Kural, C.; Lucas, D.M.; Byrd, J.C.; Castro, C.E. Daunorubicin-loaded DNA origami nanostructures circumvent drug-resistance mechanisms in a leukemia model. Small 2016, 12, 308–320. [Google Scholar] [CrossRef]
- Mikkila, J.; Eskelinen, A.P.; Niemela, E.H.; Linko, V.; Frilander, M.J.; Torma, P.; Kostiainen, M.A. Virus-encapsulated DNA origami nanostructures for cellular delivery. Nano Lett. 2014, 14, 2196–2200. [Google Scholar] [CrossRef]
- Kiviaho, J.K.; Linko, V.; Ora, A.; Tiainen, T.; Järvihaavisto, E.; Mikkilä, J.; Tenhu, H.; Nonappa; Kostiainen, M.A. Cationic polymers for DNA origami coating—Examining their binding efficiency and tuning the enzymatic reaction rates. Nanoscale 2016, 8, 11674–11680. [Google Scholar] [CrossRef] [PubMed]
- Strauss, M.T.; Schueder, F.; Haas, D.; Nickels, P.C.; Jungmann, R. Quantifying absolute addressability in DNA origami with molecular resolution. Nat. Commun. 2018, 9, 1600. [Google Scholar] [CrossRef] [PubMed]
- Voigt, N.V.; Torring, T.; Rotaru, A.; Jacobsen, M.F.; Ravnsbaek, J.B.; Subramani, R.; Mamdouh, W.; Kjems, J.; Mokhir, A.; Besenbacher, F.; et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 2010, 5, 200–203. [Google Scholar] [CrossRef]
- Woo, S.; Rothemund, P.W. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 2011, 3, 620–627. [Google Scholar] [CrossRef]
- Li, J.; Green, A.A.; Yan, H.; Fan, C. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem. 2017, 9, 1056–1067. [Google Scholar] [CrossRef]
- Gerling, T.; Wagenbauer, K.F.; Neuner, A.M.; Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 2015, 347, 1446–1452. [Google Scholar] [CrossRef]
- Chen, Y.J.; Groves, B.; Muscat, R.A.; Seelig, G. DNA nanotechnology from the test tube to the cell. Nat. Nanotechnol. 2015, 10, 748–760. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Halman, J.R.; Shah, A.B.; Khisamutdinov, E.F.; Dobrovolskaia, M.A.; Afonin, K.A. Structure and composition define immunorecognition of nucleic acid nanoparticles. Nano Lett. 2018, 18, 4309–4321. [Google Scholar] [CrossRef]
- Ke, W.; Hong, E.; Saito, R.F.; Rangel, M.C.; Wang, J.; Viard, M.; Richardson, M.; Khisamutdinov, E.F.; Panigaj, M.; Dokholyan, N.V.; et al. RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef]
- Saha, S.; Prakash, V.; Halder, S.; Chakraborty, K.; Krishnan, Y. A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat. Nanotechnol. 2015, 10, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Hartman, M.R.; Derrien, T.L.; Hamada, S.; An, D.; Yancey, K.G.; Cheng, R.; Ma, M.; Luo, D. DNA materials: Bridging nanotechnology and biotechnology. Acc. Chem. Res. 2014, 47, 1902–1911. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Grabow, W.W.; Walker, F.M.; Bindewald, E.; Dobrovolskaia, M.A.; Shapiro, B.A.; Jaeger, L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 2011, 6, 2022–2034. [Google Scholar] [CrossRef]
- Jiang, Q.; Song, C.; Nangreave, J.; Liu, X.; Lin, L.; Qiu, D.; Wang, Z.-G.; Zou, G.; Liang, X.; Yan, H.; et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403. [Google Scholar] [CrossRef] [PubMed]
- Veetil, A.T.; Chakraborty, K.; Xiao, K.; Minter, M.R.; Sisodia, S.S.; Krishnan, Y. Cell-targetable DNA nanocapsules for spatiotemporal release of caged bioactive small molecules. Nat. Nanotechnol. 2017, 12, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Mo, F.; Wu, J.; Huang, Y.; Zhou, H.; Ding, S.; Chen, W. A multifunctional DNA nano-scorpion for highly efficient targeted delivery of mRNA therapeutics. Sci. Rep. 2018, 8, 10196. [Google Scholar] [CrossRef]
- Zhao, Z.; Fu, J.; Dhakal, S.; Johnson-Buck, A.; Liu, M.; Zhang, T.; Woodbury, N.W.; Liu, Y.; Walter, N.G.; Yan, H. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 2016, 7, 10619. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Lamarre, B.; Pyne, A.L.B.; Noble, J.E.; Ryadnov, M.G. DNA origami inside-out viruses. ACS Synth. Biol. 2018, 7, 767–773. [Google Scholar] [CrossRef]
- Bastings, M.M.C.; Anastassacos, F.M.; Ponnuswamy, N.; Leifer, F.G.; Cuneo, G.; Lin, C.; Ingber, D.E.; Ryu, J.H.; Shih, W.M. Modulation of the cellular uptake of DNA origami through control over mass and shape. Nano Lett. 2018, 18, 3557–3564. [Google Scholar] [CrossRef]
- Wang, P.; Rahman, M.A.; Zhao, Z.; Weiss, K.; Zhang, C.; Chen, Z.; Hurwitz, S.J.; Chen, Z.G.; Shin, D.M.; Ke, Y. Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells. J. Am. Chem. Soc. 2018, 140, 2478–2484. [Google Scholar] [CrossRef]
- Ko, S.; Liu, H.; Chen, Y.; Mao, C. DNA nanotubes as combinatorial vehicles for cellular delivery. Biomacromolecules 2008, 9, 3039–3043. [Google Scholar] [CrossRef]
- Ponnuswamy, N.; Bastings, M.M.C.; Nathwani, B.; Ryu, J.H.; Chou, L.Y.T.; Vinther, M.; Li, W.A.; Anastassacos, F.M.; Mooney, D.J.; Shih, W.M. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Ijas, H.; Linko, V.; Keller, A. Structural stability of DNA origami nanostructures under application-specific conditions. Comput. Struct. Biotechnol. J. 2018, 16, 342–349. [Google Scholar] [CrossRef]
- Jiang, D.; Ge, Z.; Im, H.J.; England, C.G.; Ni, D.; Hou, J.; Zhang, L.; Kutyreff, C.J.; Yan, Y.; Liu, Y.; et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat. Biomed. Eng. 2018, 2, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.; Wickham, S.F.; Shih, W.M.; Perrault, S.D. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef] [PubMed]
- Kocabey, S.; Meinl, H.; MacPherson, S.I.; Cassinelli, V.; Manetto, A.; Rothenfusser, S.; Liedl, T.; Lichtenegger, S.F. Cellular uptake of tile-assembled DNA nanotubes. Nanomaterials 2015, 5, 47–60. [Google Scholar] [CrossRef]
- Kielar, C.; Xin, Y.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Linko, V.; Keller, A. On the stability of DNA origami nanostructures in low-magnesium buffers. Angew. Chem. Int. Ed. 2018, 57, 9470–9474. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, P.; Rothemund, P.W.K.; Kumar, A.; Fygenson, D.K. Sturdier DNA nanotubes via ligation. Nano Lett. 2006, 6, 1379–1383. [Google Scholar] [CrossRef]
- Michelotti, N.; Johnson-Buck, A.; Manzo, A.J.; Walter, N.G. Beyond DNA origami: The unfolding prospects of nucleic acid nanotechnology. Wiley Interdiscip. Rev. Nanomed. 2012, 4, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012, 338, 932–936. [Google Scholar] [CrossRef]
- Burns, J.R.; Stulz, E.; Howorka, S. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 2013, 13, 2351–2356. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Seifert, A.; Fertig, N.; Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016, 11, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Gopfrich, K.; Li, C.Y.; Ricci, M.; Bhamidimarri, S.P.; Yoo, J.; Gyenes, B.; Ohmann, A.; Winterhalter, M.; Aksimentiev, A.; Keyser, U.F. Large-conductance transmembrane porin made from DNA origami. ACS Nano 2016, 10, 8207–8214. [Google Scholar] [CrossRef]
- Birkholz, O.; Burns, J.R.; Richter, C.P.; Psathaki, O.E.; Howorka, S.; Piehler, J. Multi-functional DNA nanostructures that puncture and remodel lipid membranes into hybrid materials. Nat. Commun. 2018, 9, 1521. [Google Scholar] [CrossRef] [PubMed]
- Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 2017, 12, 619–630. [Google Scholar] [CrossRef]
- Pugh, G.C.; Burns, J.R.; Howorka, S. Comparing proteins and nucleic acids for next-generation biomolecular engineering. Nat. Rev. Chem. 2018, 2, 113–130. [Google Scholar] [CrossRef]
- Ohmann, A.; Li, C.-Y.; Maffeo, C.; Al Nahas, K.; Baumann, K.N.; Göpfrich, K.; Yoo, J.; Keyser, U.F.; Aksimentiev, A. A synthetic enzyme built from DNA flips 107 lipids per second in biological membranes. Nat. Commun. 2018, 9, 2426. [Google Scholar] [CrossRef]
- Burns, J.R.; Howorka, S. Defined bilayer interactions of DNA nanopores revealed with a nuclease-based nanoprobe strategy. ACS Nano 2018, 12, 3263–3271. [Google Scholar] [CrossRef] [PubMed]
- List, J.; Weber, M.; Simmel, F.C. Hydrophobic actuation of a DNA origami bilayer structure. Angew. Chem. Int. Ed. 2014, 53, 4236–4239. [Google Scholar] [CrossRef]
- Edwardson, T.G.W.; Carneiro, K.M.M.; McLaughlin, C.K.; Serpell, C.J.; Sleiman, H.F. Site-specific positioning of dendritic alkyl chains on DNA cages enables their geometry-dependent self-assembly. Nat. Chem. 2013, 5, 868. [Google Scholar] [CrossRef]
- Arora, M. Cell Culture Media: A Review. Mater. Methods 2013, 3. [Google Scholar] [CrossRef]
- Moore, T.L.; Rodriguez-Lorenzo, L.; Hirsch, V.; Balog, S.; Urban, D.; Jud, C.; Rothen-Rutishauser, B.; Lattuada, M.; Petri-Fink, A. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chem. Soc. Rev. 2015, 44, 6287–6305. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.S.; Walter, G.L.; Walker, R.M. Clinical pathology in non-clinical toxicology testing. In Haschek and Rousseaux’s Handbook of Toxicologic Pathology; Academic Press: Cambridge, MA, USA, 2013; pp. 565–594. [Google Scholar] [CrossRef]
- Seifert, A.; Gopfrich, K.; Burns, J.R.; Fertig, N.; Keyser, U.F.; Howorka, S. Bilayer-spanning DNA nanopores with voltage-switching between open and closed state. ACS Nano 2015, 9, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Nangreave, J.; Jiang, S.; Yan, H.; Liu, Y. Mapping the thermal behavior of DNA origami nanostructures. J. Am. Chem. Soc. 2013, 135, 6165–6176. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Branzoi, I.V.; Iordoc, M.; Branzoi, F.; Vasilescu-Mirea, R.; Sbarcea, G. Influence of diamond-like carbon coating on the corrosion resistance of the NITINOL shape memory alloy. Surf. Interface Anal. 2010, 42, 502–509. [Google Scholar] [CrossRef]
- Krishnan, S.; Ziegler, D.; Arnaut, V.; Martin, T.G.; Kapsner, K.; Henneberg, K.; Bausch, A.R.; Dietz, H.; Simmel, F.C. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 2016, 7, 12787. [Google Scholar] [CrossRef] [PubMed]
- Norman, D.G.; Grainger, R.J.; Uhrín, D.; Lilley, D.M.J. Location of Cyanine-3 on Double-Stranded DNA: Importance for Fluorescence Resonance Energy Transfer Studies. Biochemistry 2000, 39, 6317–6324. [Google Scholar] [CrossRef]
- Wolfe, A.J.; Si, W.; Zhang, Z.; Blanden, A.R.; Hsueh, Y.C.; Gugel, J.F.; Pham, B.; Chen, M.; Loh, S.N.; Rozovsky, S.; et al. Quantification of membrane protein-detergent complex interactions. J. Phys. Chem. B 2017, 121, 10228–10241. [Google Scholar] [CrossRef]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, M.; Hamming, J.B.R.; Voldum, A.; Tsakiridou, G.; Larsen, M.T.; Schmokel, J.S.; Sohn, E.; Bienk, K.; Schaffert, D.; Sorensen, E.S.; et al. An albumin-oligonucleotide assembly for potential combinatorial drug delivery and half-life extension applications. Mol. Ther. Nucleic Acids 2017, 9, 284–293. [Google Scholar] [CrossRef]
- Osborn, M.F.; Coles, A.H.; Biscans, A.; Haraszti, R.A.; Roux, L.; Davis, S.; Ly, S.; Echeverria, D.; Hassler, M.R.; Godinho, B.; et al. Hydrophobicity drives the systemic distribution of lipid-conjugated siRNAs via lipid transport pathways. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, Y.; De Llano, E.; Barisic, I. (Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures. Nanoscale 2018, 10, 7494–7504. [Google Scholar] [CrossRef] [PubMed]
| Abbreviation | Na | K | Mg | PBS | LB | D | FBS |
|---|---|---|---|---|---|---|---|
| Salt/media | NaCl | KCl | MgCl2 TAE | Phosphate buffered-saline | Lysogeny broth | Dulbecco’s modified Eagle medium | D + 10% fetal bovine serum |
| Ionic strength | 0.32 M | 0.32 M | 0.11 M | 0.17 M | 0.17 M | 0.17 M | 0.19 M |
| Construct | Na | K | Mg | PBS | LB | D | FBS |
|---|---|---|---|---|---|---|---|
| NP-0C | 49.7 ± 0.3 | 50.9 ± 1.2 | 52.7 ± 0.3 | 46.4 ± 0.9 | 46.6 ± 0.3 | 45.8 ± 0.3 | 45.7 ± 0.3 |
| NP-3C | 51.3 ± 0.6 | 52.2 ± 0.8 | 53.8 ± 1.5 | 46.7 ± 0.3 | 45.0 ± 1.0 | 46.8 ± 0.3 | 47.2 ± 0.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burns, J.R.; Howorka, S. Structural and Functional Stability of DNA Nanopores in Biological Media. Nanomaterials 2019, 9, 490. https://doi.org/10.3390/nano9040490
Burns JR, Howorka S. Structural and Functional Stability of DNA Nanopores in Biological Media. Nanomaterials. 2019; 9(4):490. https://doi.org/10.3390/nano9040490
Chicago/Turabian StyleBurns, Jonathan R., and Stefan Howorka. 2019. "Structural and Functional Stability of DNA Nanopores in Biological Media" Nanomaterials 9, no. 4: 490. https://doi.org/10.3390/nano9040490
APA StyleBurns, J. R., & Howorka, S. (2019). Structural and Functional Stability of DNA Nanopores in Biological Media. Nanomaterials, 9(4), 490. https://doi.org/10.3390/nano9040490





