Out-of-Plane Aptamer Functionalization of RNA Three-Helix Tiles
Abstract
1. Introduction
2. Materials and Methods
2.1. Design of Three-Helix RNA Tiles (3H-AE)
2.2. Preparation of RNA Tiles
2.3. Characterization of RNA Tiles and Tile Assemblies
3. Results and Discussion
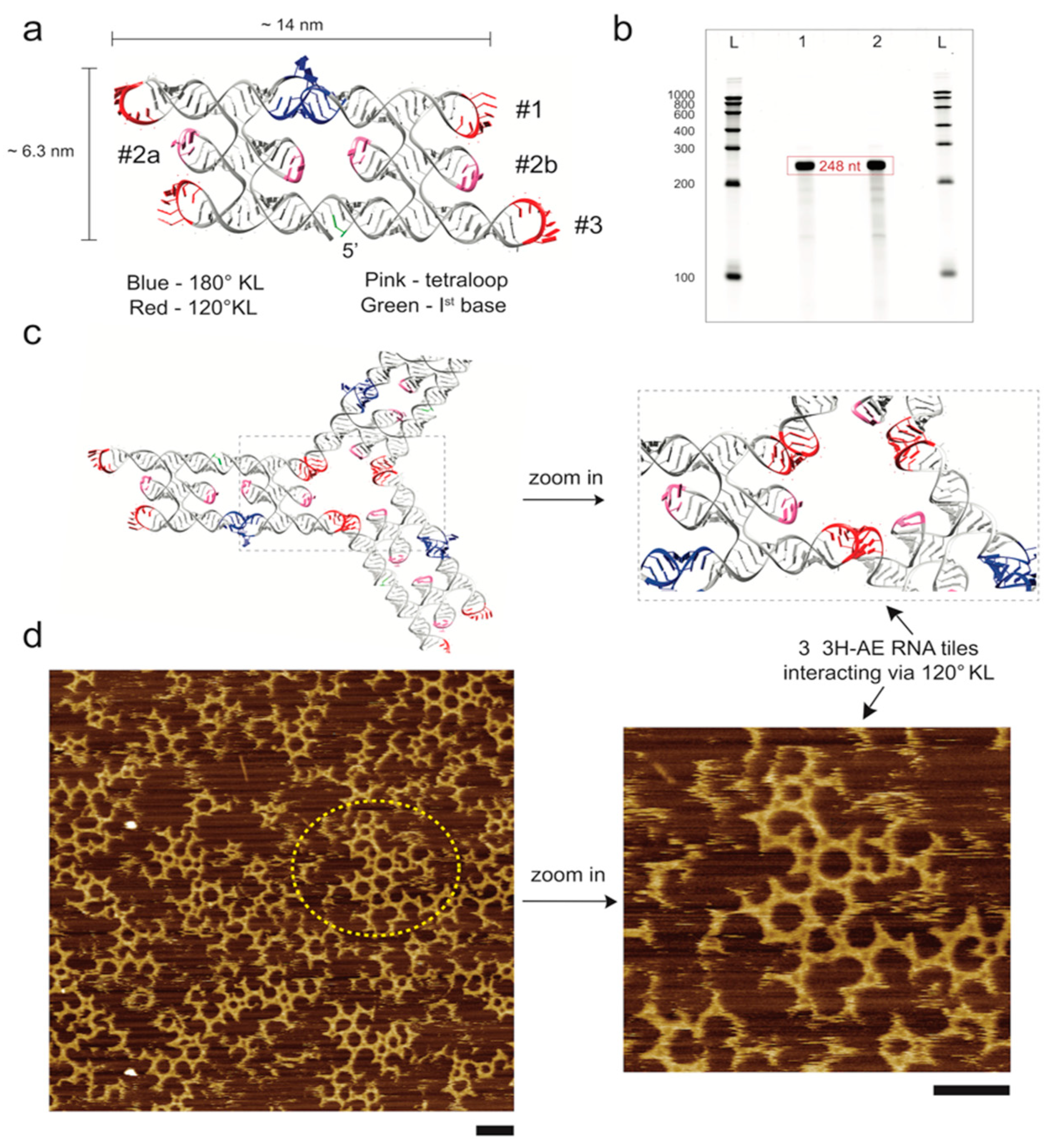
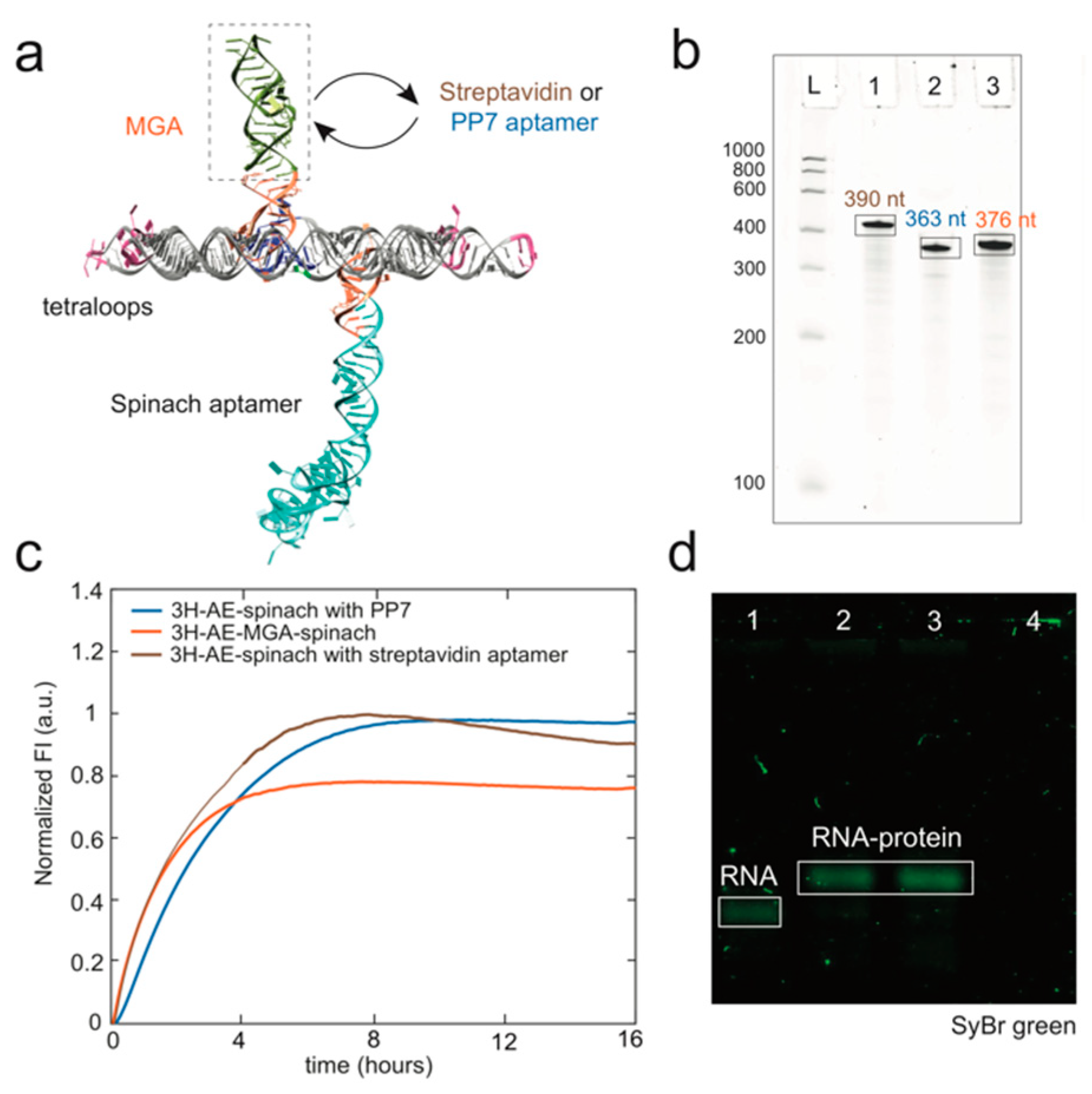
3.1. Design and Folding of the 3H-AE RNA Tile
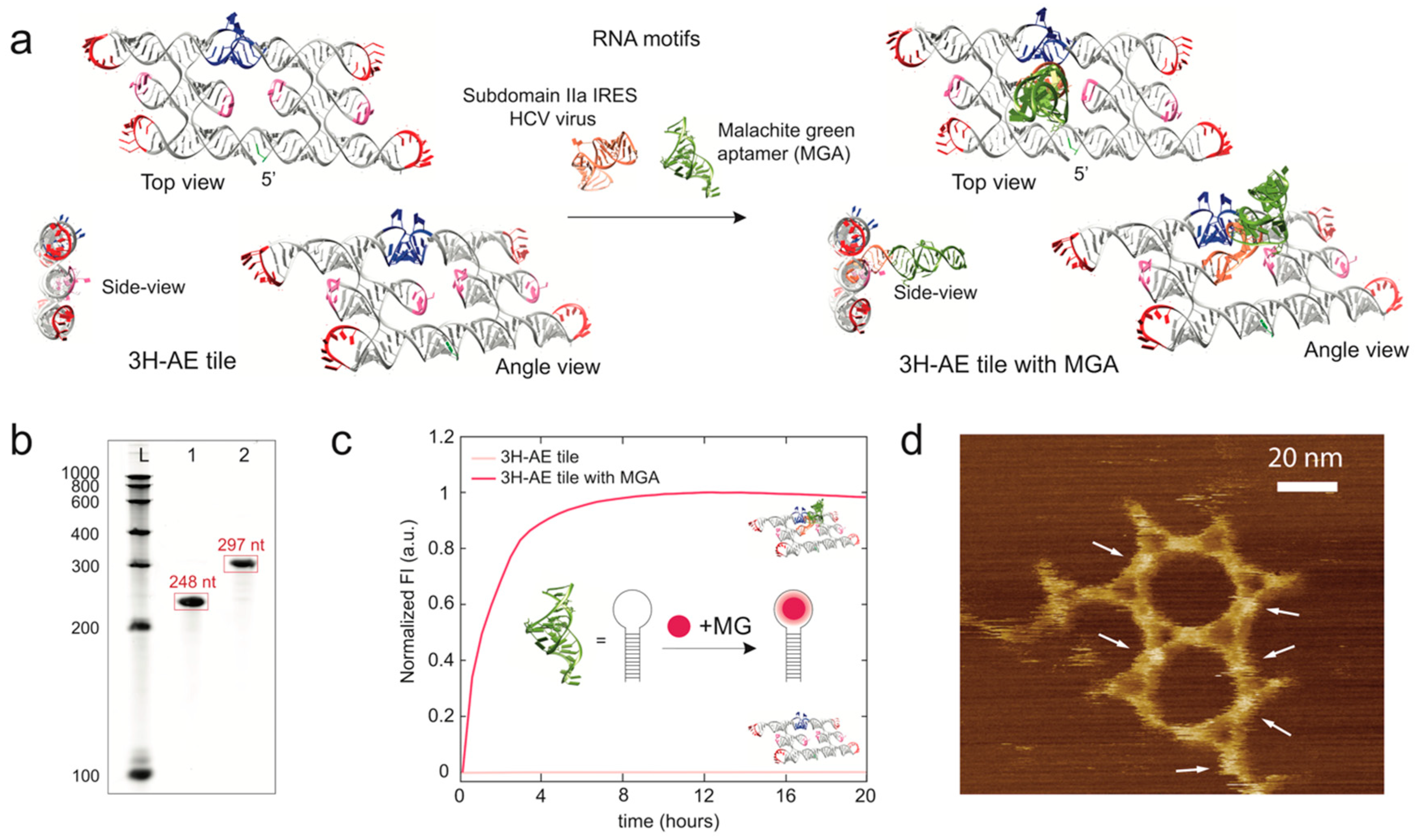
3.2. Modification of the 3H-AE RNA Tile with Aptamers
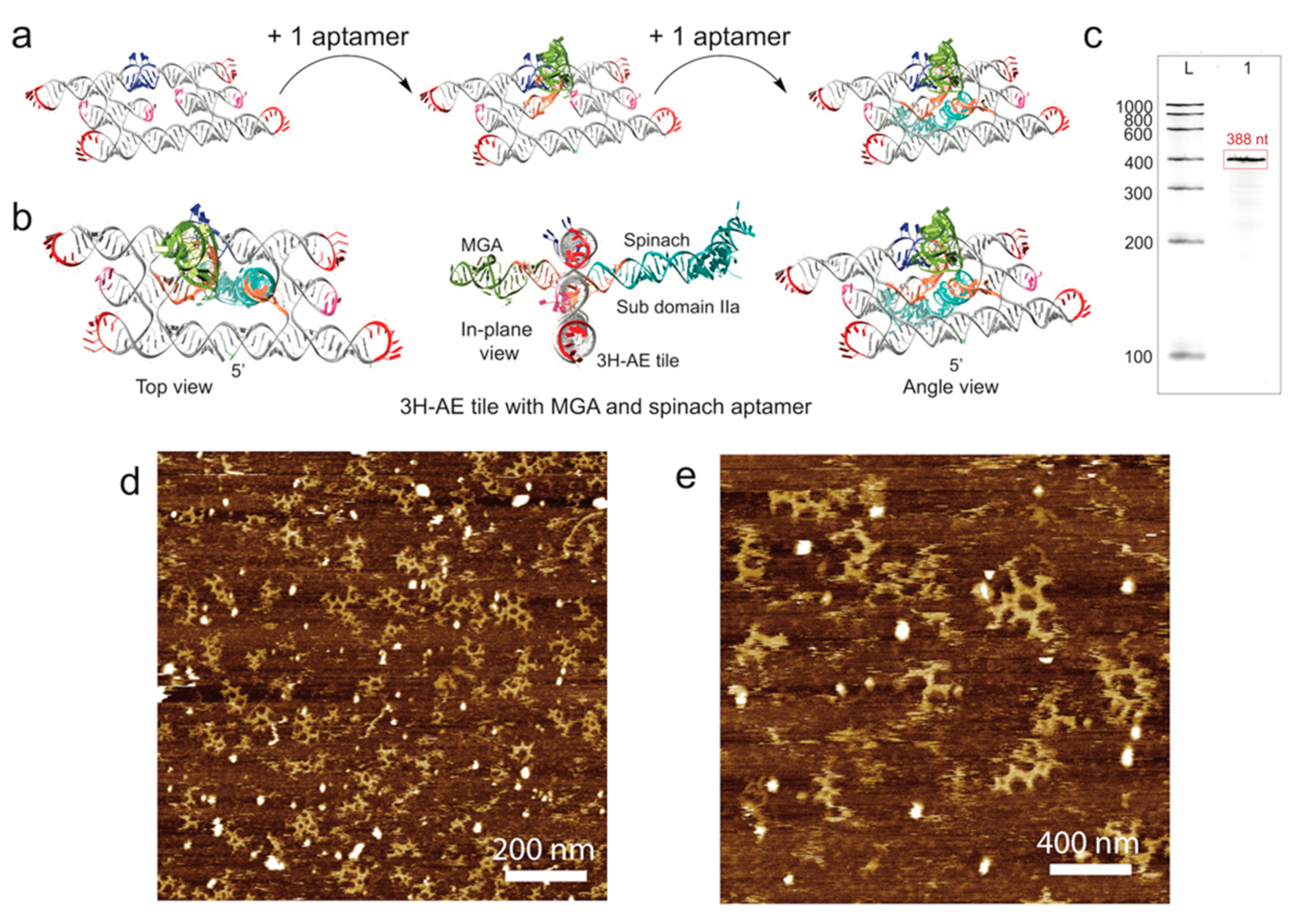
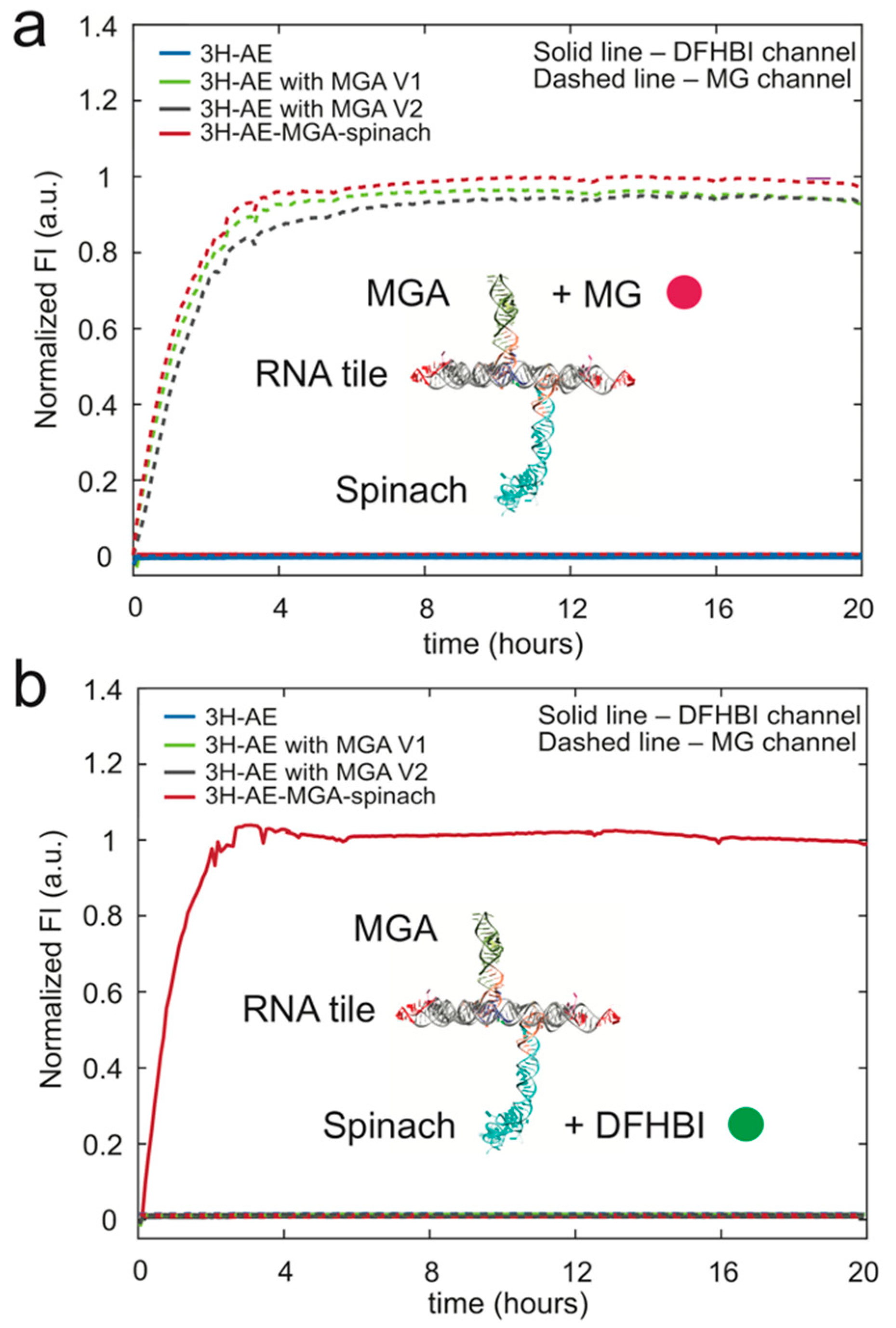
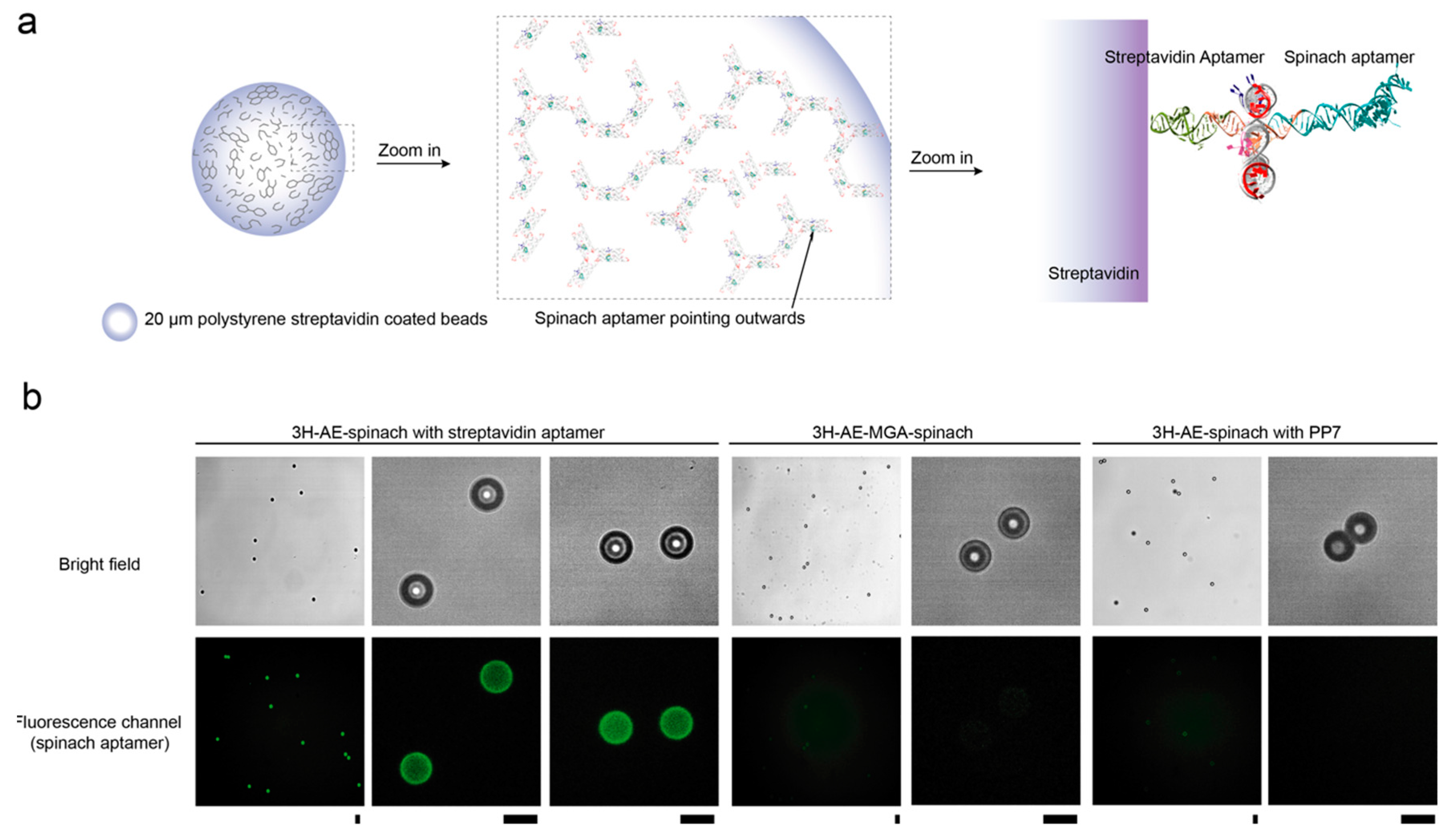
3.3. Double Functionalization of the 3H-AE RNA Tile with Two Aptamers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ribeiro, D.M.; Zanzoni, A.; Cipriano, A.; Delli Ponti, R.; Spinelli, L.; Ballarino, M.; Bozzoni, I.; Tartaglia, G.G.; Brun, C. Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res. 2017, 46, 917–928. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, D.; Haque, F.; Binzel, D.W.; Guo, P. Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Batey, R.T.; Rambo, R.P.; Doudna, J.A. Tertiary Motifs in RNA Structure and Folding. Angew. Chem. Int. Ed. Engl. 1999, 38, 2326–2343. [Google Scholar] [CrossRef]
- Leontis, N.B.; Lescoute, A.; Westhof, E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006, 16, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Bindewald, E.; Hayes, R.; Yingling, Y.G.; Kasprzak, W.; Shapiro, B.A. RNAJunction: A database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Res. 2007, 36, D392–D397. [Google Scholar] [CrossRef] [PubMed]
- Delebecque, C.J.; Lindner, A.B.; Silver, P.A.; Aldaye, F.A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 2011, 333, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, G.; Garg, A.; Godding, D.; Way, J.C.; Silver, P.A. In vivo co-localization of enzymes on RNA scaffolds increases metabolic production in a geometrically dependent manner. Nucleic Acids Res. 2014, 42, 9493–9503. [Google Scholar] [CrossRef]
- Jaeger, L.; Chworos, A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006, 16, 531–543. [Google Scholar] [CrossRef]
- Jaeger, L.; Westhof, E.; Leontis, N.B. TectoRNA: Modular assembly units. Nucleic Acids Res. 2001, 29, 455–463. [Google Scholar] [CrossRef]
- Ko, S.H.; Su, M.; Zhang, C.; Ribbe, A.E.; Jiang, W.; Mao, C. Synergistic self-assembly of RNA and DNA molecules. Nat. Chem. 2010, 2, 1050. [Google Scholar] [CrossRef]
- Afonin, K.A.; Bindewald, E.; Yaghoubian, A.J.; Voss, N.; Jacovetty, E.; Shapiro, B.A.; Jaeger, L. In vitro Assembly of Cubic RNA-Based Scaffolds Designed in silico. Nat. Nanotechnol. 2010, 5, 676–682. [Google Scholar] [CrossRef]
- Geary, C.; Rothemund, P.W.K.; Andersen, E.S. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 2014, 345, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Parlea, L.; Bindewald, E.; Sharan, R.; Bartlett, N.; Moriarty, D.; Oliver, J.; Afonin, K.A.; Shapiro, B.A. Ring Catalog: A resource for designing self-assembling RNA nanostructures. Methods 2016, 103, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Qi, X.; Myhrvold, C.; Wang, B.; Dai, M.; Jiang, S.; Bates, M.; Liu, Y.; An, B.; Zhang, F.; et al. Single-stranded DNA and RNA origami. Science 2017, 358, eaao2648. [Google Scholar] [CrossRef]
- Afonin, K.A.; Grabow, W.W.; Walker, F.M.; Bindewald, E.; Dobrovolskaia, M.A.; Shapiro, B.A.; Jaeger, L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 2011, 6, 2022. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Kireeva, M.; Grabow, W.W.; Kashlev, M.; Jaeger, L.; Shapiro, B.A. Co-transcriptional Assembly of Chemically Modified RNA Nanoparticles Functionalized with siRNAs. Nano Lett. 2012, 12, 5192–5195. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.; Khisamutdinov, E.F.; Zhang, L.; Guo, P. Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor. Nucleic Acids Res. 2014, 42, e10. [Google Scholar] [CrossRef]
- Shu, D.; Shu, Y.; Haque, F.; Abdelmawla, S.; Guo, P. Thermodynamically Stable RNA three-way junctions as platform for constructing multi-functional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011, 6, 658–667. [Google Scholar] [CrossRef]
- Jepsen, M.D.E.; Sparvath, S.M.; Nielsen, T.B.; Langvad, A.H.; Grossi, G.; Gothelf, K.V.; Andersen, E.S. Development of a genetically encodable FRET system using fluorescent RNA aptamers. Nat. Commun. 2018, 9, 18. [Google Scholar] [CrossRef]
- Li, M.; Zheng, M.; Wu, S.; Tian, C.; Liu, D.; Weizmann, Y.; Jiang, W.; Wang, G.; Mao, C. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs. Nat. Commun. 2018, 9, 2196. [Google Scholar] [CrossRef]
- Schwarz-Schilling, M.; Dupin, A.; Chizzolini, F.; Krishnan, S.; Mansy, S.S.; Simmel, F.C. Optimized Assembly of a Multifunctional RNA-Protein Nanostructure in a Cell-Free Gene Expression System. Nano Lett. 2018, 18, 2650–2657. [Google Scholar] [CrossRef]
- Lukavsky, P.J. Structure and function of HCV IRES domains. Virus Res. 2009, 139, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Baugh, C.; Grate, D.; Wilson, C. 2.8 A crystal structure of the malachite green aptamer. J. Mol. Biol. 2000, 301, 117–128. [Google Scholar] [CrossRef]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA Mimics of Green Fluorescent Protein. Science 2011, 333, 642. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Suslov, N.B.; Li, N.S.; Shelke, S.A.; Evans, M.E.; Koldobskaya, Y.; Rice, P.A.; Piccirilli, J.A. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.; Peabody, D.S. RNA recognition site of PP7 coat protein. Nucleic Acids Res. 2002, 30, 4138–4144. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.A.; Patskovsky, Y.; Almo, S.C.; Singer, R.H. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol. 2008, 15, 103–105. [Google Scholar] [CrossRef]
- Sparvath, S.L.; Geary, C.W.; Andersen, E.S. Computer-Aided Design of RNA Origami Structures. Methods Mol. Biol. 2017, 1500, 51–80. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Ennifar, E.; Walter, P.; Ehresmann, B.; Ehresmann, C.; Dumas, P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001, 8, 1064–1068. [Google Scholar] [CrossRef]
- Ennifar, E.; Nikulin, A.; Tishchenko, S.; Serganov, A.; Nevskaya, N.; Garber, M.; Ehresmann, B.; Ehresmann, C.; Nikonov, S.; Dumas, P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000, 304, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Crothers, D.M. The solution structure of an RNA loop-loop complex: The ColE1 inverted loop sequence. Structure 1998, 6, 993–1005. [Google Scholar] [CrossRef]
- Lukavsky, P.J.; Kim, I.; Otto, G.A.; Puglisi, J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003, 10, 1033–1038. [Google Scholar] [CrossRef]
- Zhao, Q.; Han, Q.; Kissinger, C.R.; Hermann, T.; Thompson, P.A. Structure of hepatitis C virus IRES subdomain IIa. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Andersen Lab for Biomolecular Design. Available online: http://bion.au.dk/ (accessed on 27 March 2019).
- Jossinet, F.; Ludwig, T.E.; Westhof, E. Assemble: An interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics 2010, 26, 2057–2059. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Tm Calculator v 1.10.2. Available online: http://tmcalculator.neb.com/ (accessed on 27 March 2019).
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chopra, A.; Sagredo, S.; Grossi, G.; Andersen, E.S.; Simmel, F.C. Out-of-Plane Aptamer Functionalization of RNA Three-Helix Tiles. Nanomaterials 2019, 9, 507. https://doi.org/10.3390/nano9040507
Chopra A, Sagredo S, Grossi G, Andersen ES, Simmel FC. Out-of-Plane Aptamer Functionalization of RNA Three-Helix Tiles. Nanomaterials. 2019; 9(4):507. https://doi.org/10.3390/nano9040507
Chicago/Turabian StyleChopra, Aradhana, Sandra Sagredo, Guido Grossi, Ebbe S. Andersen, and Friedrich C. Simmel. 2019. "Out-of-Plane Aptamer Functionalization of RNA Three-Helix Tiles" Nanomaterials 9, no. 4: 507. https://doi.org/10.3390/nano9040507
APA StyleChopra, A., Sagredo, S., Grossi, G., Andersen, E. S., & Simmel, F. C. (2019). Out-of-Plane Aptamer Functionalization of RNA Three-Helix Tiles. Nanomaterials, 9(4), 507. https://doi.org/10.3390/nano9040507




