Optimization of the Split-Spinach Aptamer for Monitoring Nanoparticle Assembly Involving Multiple Contiguous RNAs
Abstract
1. Introduction
2. Materials and Methods
2.1. Design and Synthesis of Split-Spinach Aptamer and Fluorophore
2.2. Monitoring Nanoring Assembly
2.3. Co-Transcriptional Assembly
3. Results & Discussion
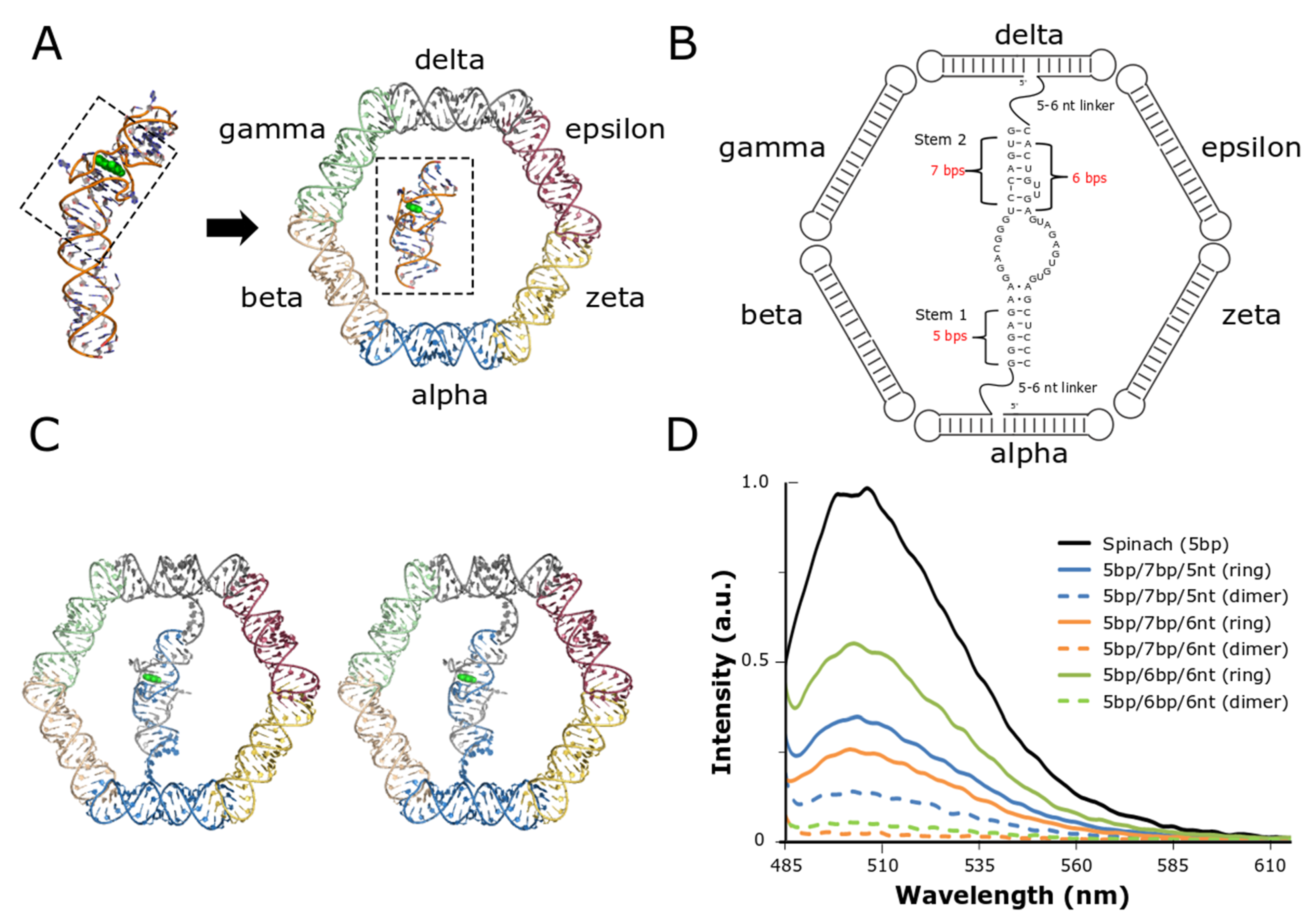
3.1. Initial Design of the Split-Spinach/Nanoring System
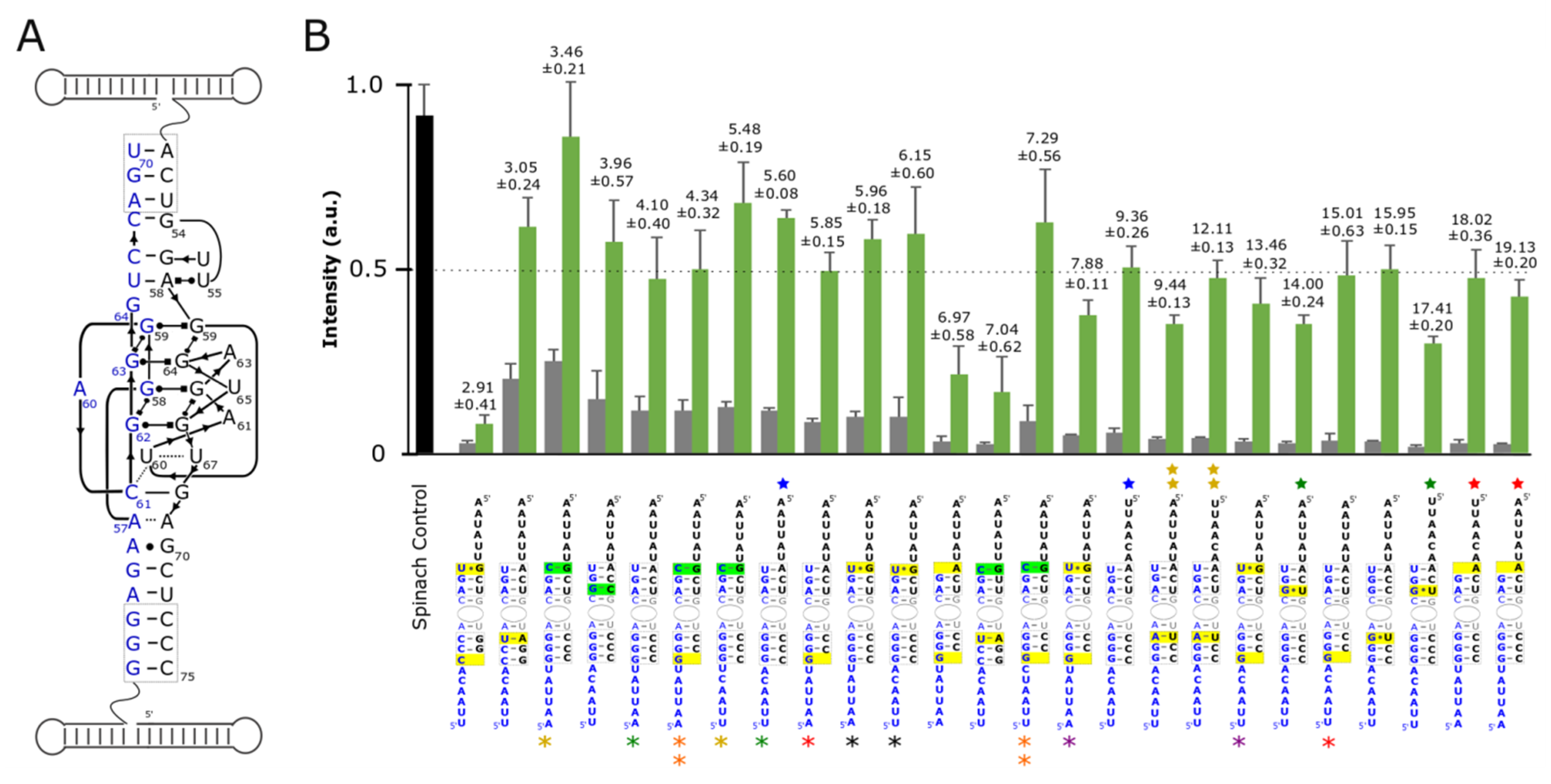
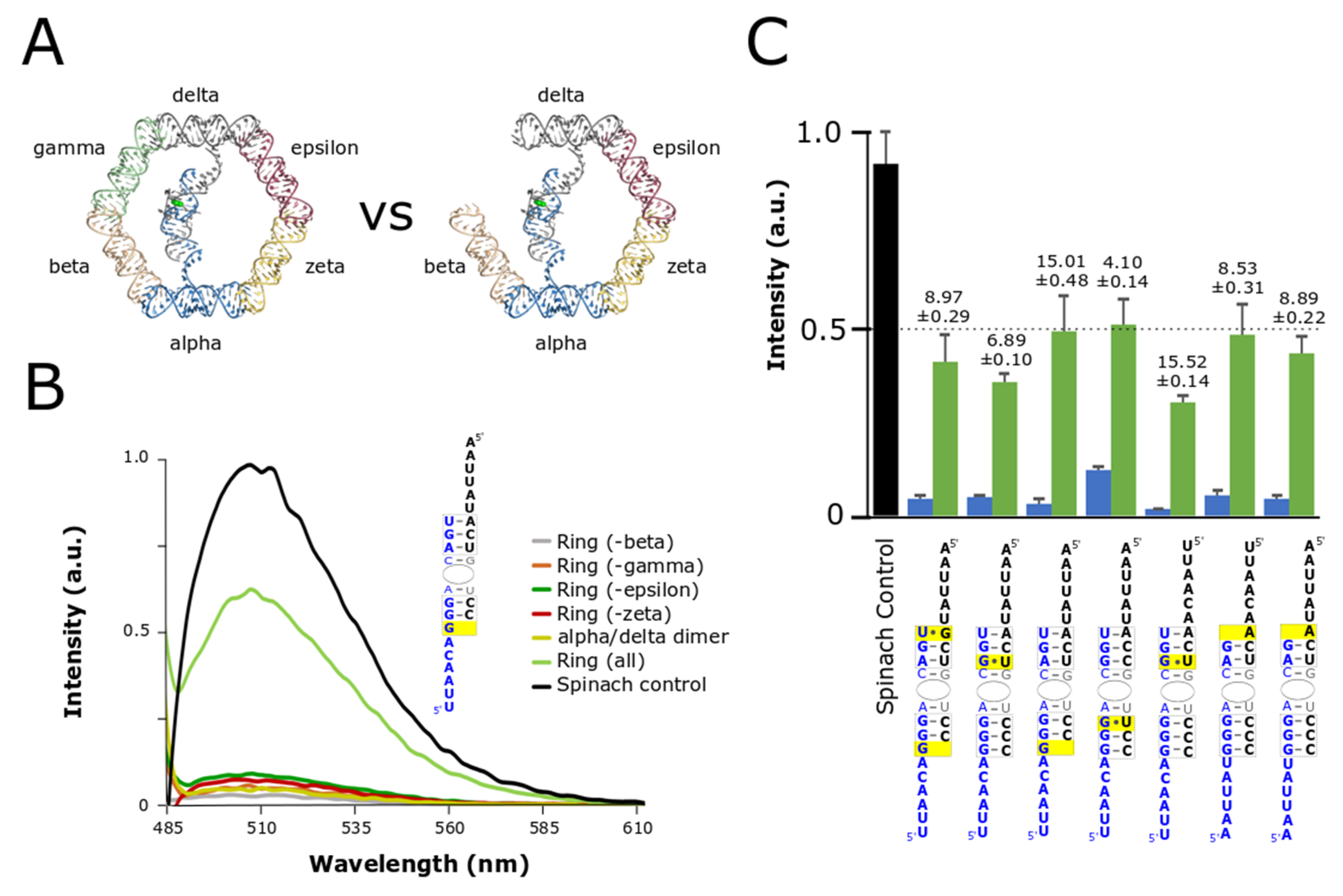
3.2. Optimization and Assessment of Split-Spinach Variants
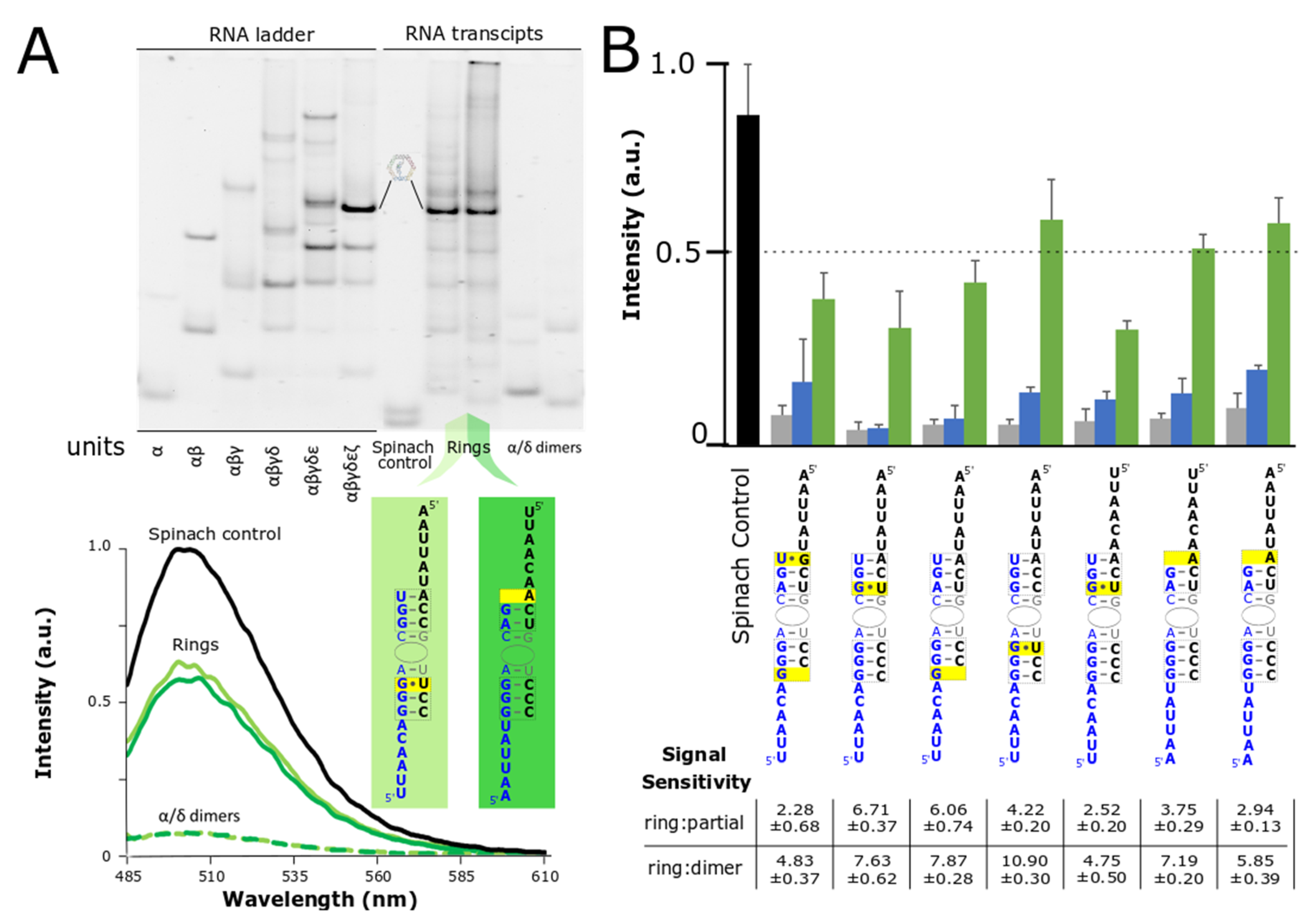
3.3. Co-Transcriptional Assembly
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jaeger, L.; Westhof, E.; Leontis, N.B. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001, 29, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Geary, C.; Chworos, A.; Verzemnieks, E.; Voss, N.R.; Jaeger, L. Composing RNA nanostructures from a syntax of RNA structural modules. Nano Lett. 2017, 17, 7095–7101. [Google Scholar] [CrossRef] [PubMed]
- Chworos, A.; Severcan, I.; Koyfman, A.Y.; Weinkam, P.; Oroudjev, E.; Hansma, H.G.; Jaeger, L. Building programmable jigsaw puzzles with RNA. Science 2004, 306, 2068–2072. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.; Moll, W.D.; Deng, Z.; Mao, C.; Guo, P. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett. 2004, 4, 1717–1723. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.W.; Jaeger, L. RNA self-assembly and RNA nanotechnology. Acc. Chem. Res. 2014, 47, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Viard, M.; Koyfman, A.Y.; Martins, A.N.; Kasprzak, W.K.; Panigaj, M.; Desai, R.; Santhanam, A.; Grabow, W.W.; Jaeger, L.; et al. Multifunctional RNA nanoparticles. Nano Lett. 2014, 16, 1097–1110. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Tschammer, N.; Mohammed, S.; Guo, P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum. Gene Ther. 2005, 16, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Haque, F.; Pi, F.; Zhao, Z.; Gu, S.; Hu, H.; Yu, H.; Guo, P. RNA versatility, flexibility, and thermostability for practice in RNA nanotechnology and biomedical applications. Wiley Interdiscip. Rev. RNA 2018, 9, e1452. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, J.; Furuta, H.; Ikawa, Y. RNA tectonics (tectoRNA) for RNA nanostructure design and its application in synthetic biology. Wiley Interdiscip. Rev. RNA 2013, 4, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.; Jaeger, L. RNA modularity for synthetic biology. F1000Prime Rep. 2013, 5, 46. Available online: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3816761&tool=pmcentrez&rendertype=abstract (accessed on 3 March 2019). [CrossRef] [PubMed]
- Binzel, D.W.; Shu, Y.; Li, H.; Sun, M.; Zhang, Q.; Shu, D.; Guo, B.; Guo, P. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther. 2016, 24, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Guo, P. RNA nanotechnology: Engineering, assembly and applications in detection, gene delivery and therapy. J. Nanosci. Nanotechnol. 2005, 5, 1964–1982. [Google Scholar] [CrossRef] [PubMed]
- Bouhedda, F.; Autour, A.; Ryckelynck, M. Light-up RNA aptamers and their cognate fluorogens: From their development to their applications. Int. J. Mol. Sci. 2018, 19, 44. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Veetil, A.T.; Jaffrey, S.R.; Krishnan, Y. Nucleic Acid–Based Nanodevices in Biological Imaging. Annu. Rev. Biochem. 2016, 85, 349–373. [Google Scholar] [CrossRef] [PubMed]
- Dolgosheina, E.V.; Unrau, P.J. Fluorophore-binding RNA aptamers and their applications. Wiley Interdiscip. Rev. RNA 2016, 7, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, J. RNA Fluorescence with Light-Up Aptamers. Front. Chem. 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R. RNA-Based Fluorescent Biosensors for Detecting Metabolites in vitro and in Living Cells. Adv. Pharmacol. 2018, 82, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Alam, K.K.; Tawiah, K.D.; Lichte, M.F.; Porciani, D.; Burke, D.H. A Fluorescent Split Aptamer for Visualizing RNA-RNA Assembly in Vivo. ACS Synth. Biol. 2017, 6, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Baugh, C.; Grate, D.; Wilson, C. 2.8 Å crystal structure of the malachite green aptamer. J. Mol. Biol. 2000, 301, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Dolgosheina, E.V.; Jeng, S.C.; Panchapakesan, S.S.; Cojocaru, R.; Chen, P.S.; Wilson, P.D.; Hawkins, N.; Wiggins, P.A.; Unrau, P.J. RNA Mango Aptamer-Fluorophore: A Bright, High-Affinity Complex for RNA Labeling and Tracking. ACS Chem. Biol. 2014, 9, 2412–2420. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Pothoulakis, G.; Ceroni, F.; Reeve, B.; Ellis, T. The spinach RNA aptamer as a characterization tool for synthetic biology. ACS Synth. Biol. 2014, 3, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Benenson, Y. RNA-based computation in live cells. Curr. Opin. Biotechnol. 2009, 20, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, C.A.; Wilson, S.C.; Sales-Lee, J.; Hammond, M.C. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messengers Cyclic di-GMP and Cyclic AMP-GMP. J. Am. Chem. Soc. 2013, 135, 35. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Hickey, S.F.; Keyser, S.G.L.; Hammond, M.C. In Vitro and in Vivo Enzyme Activity Screening via RNA-Based Fluorescent Biosensors for S-Adenosyl-l-homocysteine (SAH). J. Am. Chem. Soc. 2016, 138, 7040–7047. [Google Scholar] [CrossRef] [PubMed]
- Sunbul, M.; Arora, A.; Jäschke, A. Visualizing RNA in Live Bacterial Cells Using Fluorophore- and Quencher-Binding Aptamers. In RNA Detection; Humana Press: New York, NY, USA, 2018; pp. 289–304. [Google Scholar] [CrossRef]
- Yerramilli, V.S.; Kim, K.H. Labeling RNAs in Live Cells Using Malachite Green Aptamer Scaffolds as Fluorescent Probes. ACS Synth. Biol. 2018, 7, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Nakata, E.; Morii, T. Facile conversion of RNA aptamers to modular fluorescent sensors with tunable detection wavelengths. Bioorg. Med. Chem. Lett. 2011, 21, 4503–4506. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, D.L.; Khisamutdinov, E.F.; Lyubchenko, Y.L.; Guo, P. Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano 2014, 8, 7620–7629. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; Lyalina, T.; Halman, J.; Rackley, L.; Lee, L.; Dang, D.; Ke, W.; Sajja, S.; Woods, S.; Acharya, S. Broccoli Fluorets: Split Aptamers as a User-Friendly Fluorescent Toolkit for Dynamic RNA Nanotechnology. Molecules 2018, 23, 3178. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, N.; Kolpashchikov, D.M. A universal split spinach aptamer (USSA) for nucleic acid analysis and DNA computation. Chem. Commun. 2017, 53, 4977–4980. [Google Scholar] [CrossRef] [PubMed]
- Kolpashchikov, D.M. Binary Malachite Green Aptamer for Fluorescent Detection of Nucleic Acids. J. Am. Chem. Soc. 2005, 127, 12442–12443. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.A.; Andrews, G.E.; Jaeger, L.; Grabow, W.W. Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer. ACS Synth. Biol. 2015, 4, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Autour, A.; Westhof, E.; Ryckelynck, M. iSpinach: A fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 2016, 44, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Goldsworthy, V.; LaForce, G.; Abels, S.; Khisamutdinov, E. Fluorogenic RNA Aptamers: A Nano-platform for Fabrication of Simple and Combinatorial Logic Gates. Nanomaterials 2018, 8, 984. [Google Scholar] [CrossRef] [PubMed]
- Yingling, Y.G.; Shapiro, B.A. Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett. 2007, 7, 2328–2334. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.W.; Zakrevsky, P.; Afonin, K.A.; Chworos, A.; Shapiro, B.A.; Jaeger, L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011, 11, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Warner, K.D.; Chen, M.C.; Song, W.; Strack, R.L.; Thorn, A.; Jaffrey, S.R.; Ferré-D’Amaré, A.R. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014, 21, 658. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Diamend, A.; Peitsch, M.C.; Schwede, T. DeepView—Swiss-PdbViewer; Swiss Institute of Bioinformatics: Geneva, Switzerland, 2017. [Google Scholar]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 30, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Suslov, N.B.; Li, N.S.; Shelke, S.A.; Evans, M.E.; Koldobskaya, Y.; Rice, P.A.; Piccirilli, J.A. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Torelli, E.; Kozyra, J.W.; Gu, J.Y.; Stimming, U.; Piantanida, L.; Voïtchovsky, K.; Krasnogor, N. Isothermal folding of a light-up bio-orthogonal RNA origami nanoribbon. Nature 2018, 8, 6989. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Khisamutdinov, E.; Zhao, Z.; Pan, C.; Choi, J.W.; Leontis, N.B.; Guo, P. RNA nanotechnology for computer design and in vivo computation. Philos. Trans. A Math. Phys. Eng. Sci. 2013, 371, 20130310. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Grabow, W.W.; Walker, F.M.; Bindewald, E.; Dobrovolskaia, M.A.; Shapiro, B.A.; Jaeger, L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 2011, 6, 2022–2034. Available online: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3498981&tool=pmcentrez&rendertype=abstract (accessed on 3 March 2019). [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Hara, J.M.; Marashi, D.; Morton, S.; Jaeger, L.; Grabow, W.W. Optimization of the Split-Spinach Aptamer for Monitoring Nanoparticle Assembly Involving Multiple Contiguous RNAs. Nanomaterials 2019, 9, 378. https://doi.org/10.3390/nano9030378
O’Hara JM, Marashi D, Morton S, Jaeger L, Grabow WW. Optimization of the Split-Spinach Aptamer for Monitoring Nanoparticle Assembly Involving Multiple Contiguous RNAs. Nanomaterials. 2019; 9(3):378. https://doi.org/10.3390/nano9030378
Chicago/Turabian StyleO’Hara, Jack M., Dylan Marashi, Sean Morton, Luc Jaeger, and Wade W. Grabow. 2019. "Optimization of the Split-Spinach Aptamer for Monitoring Nanoparticle Assembly Involving Multiple Contiguous RNAs" Nanomaterials 9, no. 3: 378. https://doi.org/10.3390/nano9030378
APA StyleO’Hara, J. M., Marashi, D., Morton, S., Jaeger, L., & Grabow, W. W. (2019). Optimization of the Split-Spinach Aptamer for Monitoring Nanoparticle Assembly Involving Multiple Contiguous RNAs. Nanomaterials, 9(3), 378. https://doi.org/10.3390/nano9030378





