Medium-Dependent Antibacterial Properties and Bacterial Filtration Ability of Reduced Graphene Oxide
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of rGO
2.2. Materials Characterization
2.3. E. coli Biofluorescence Tests
2.4. Investigation of Interactions of E. coli with rGO
3. Results
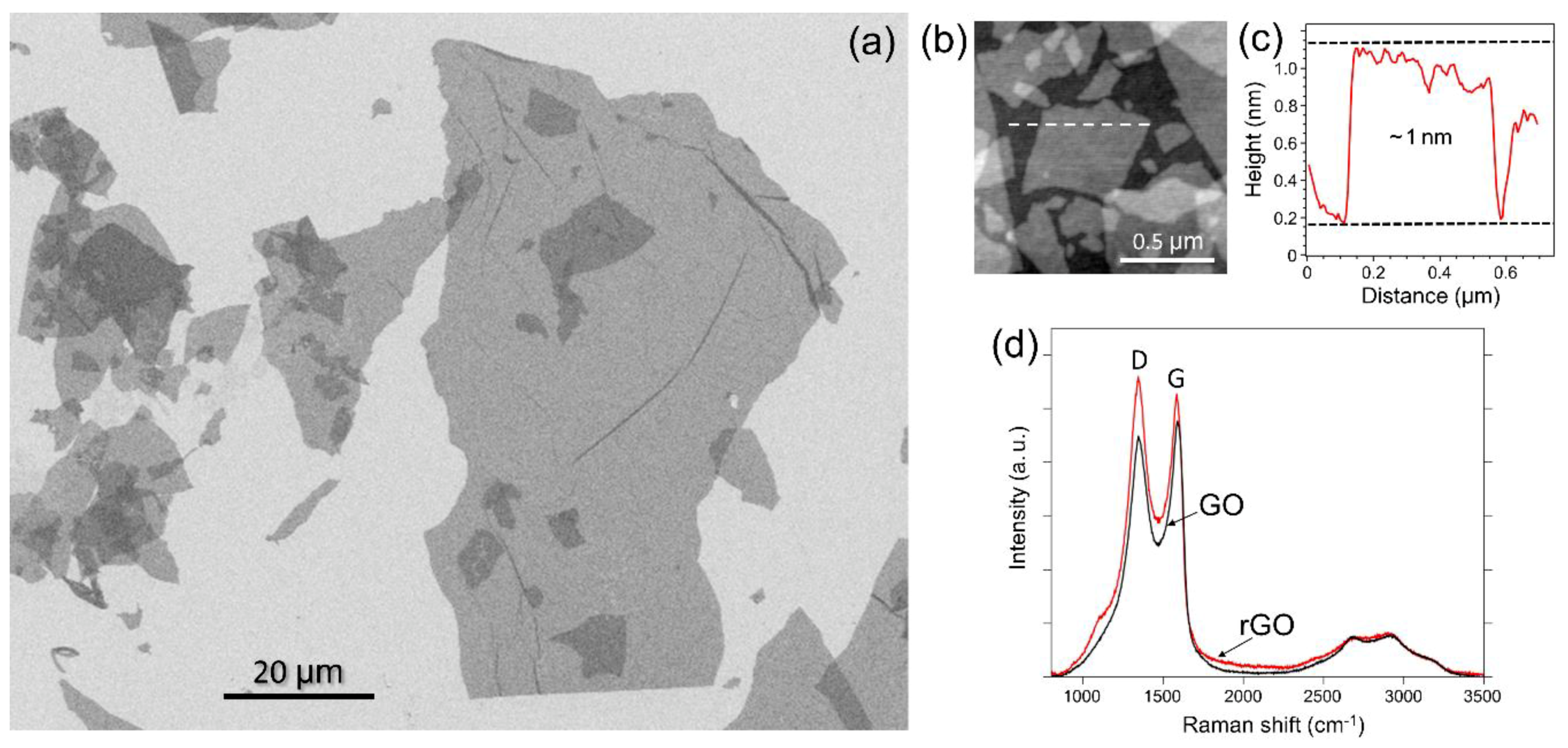
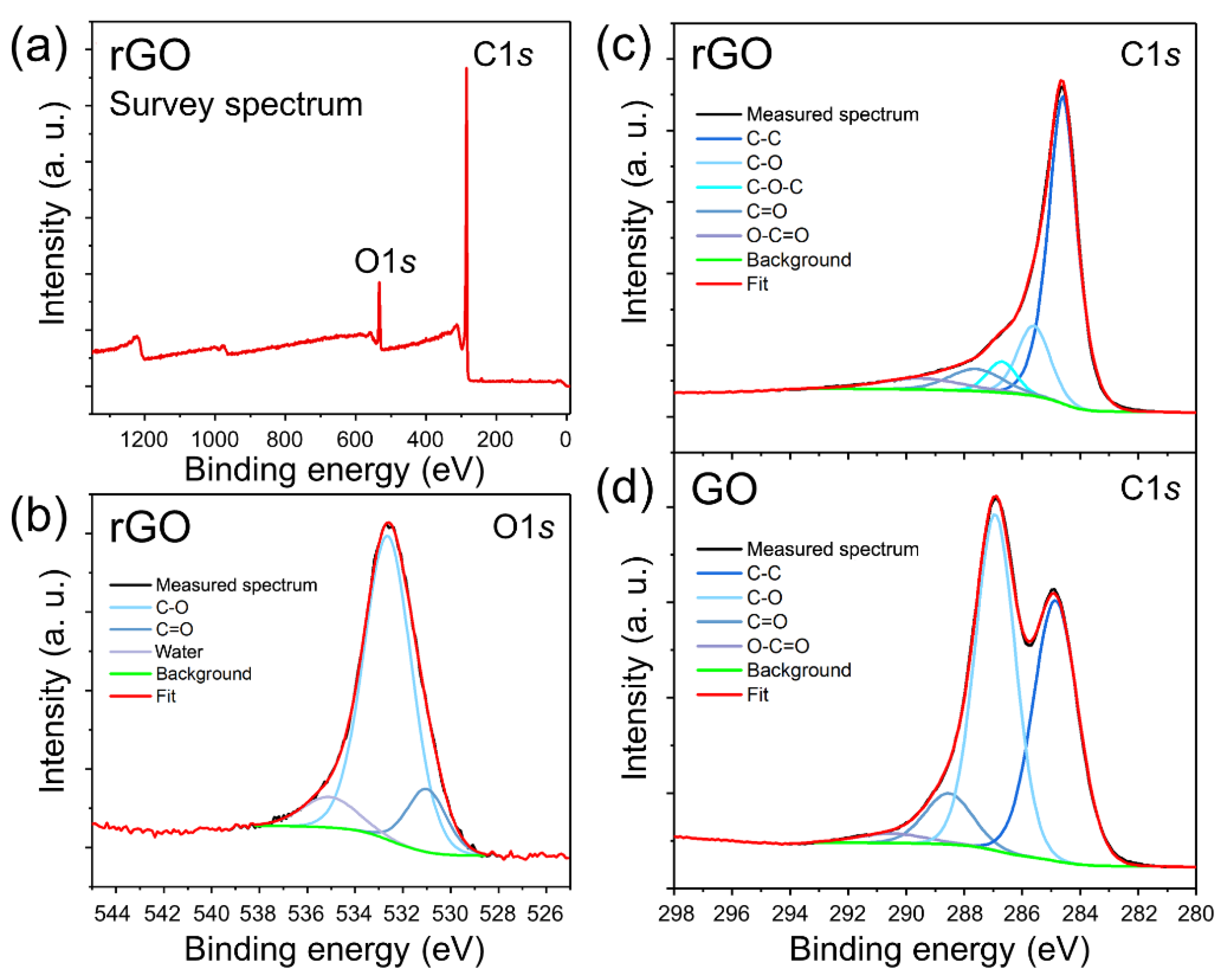
3.1. Characterization of rGO
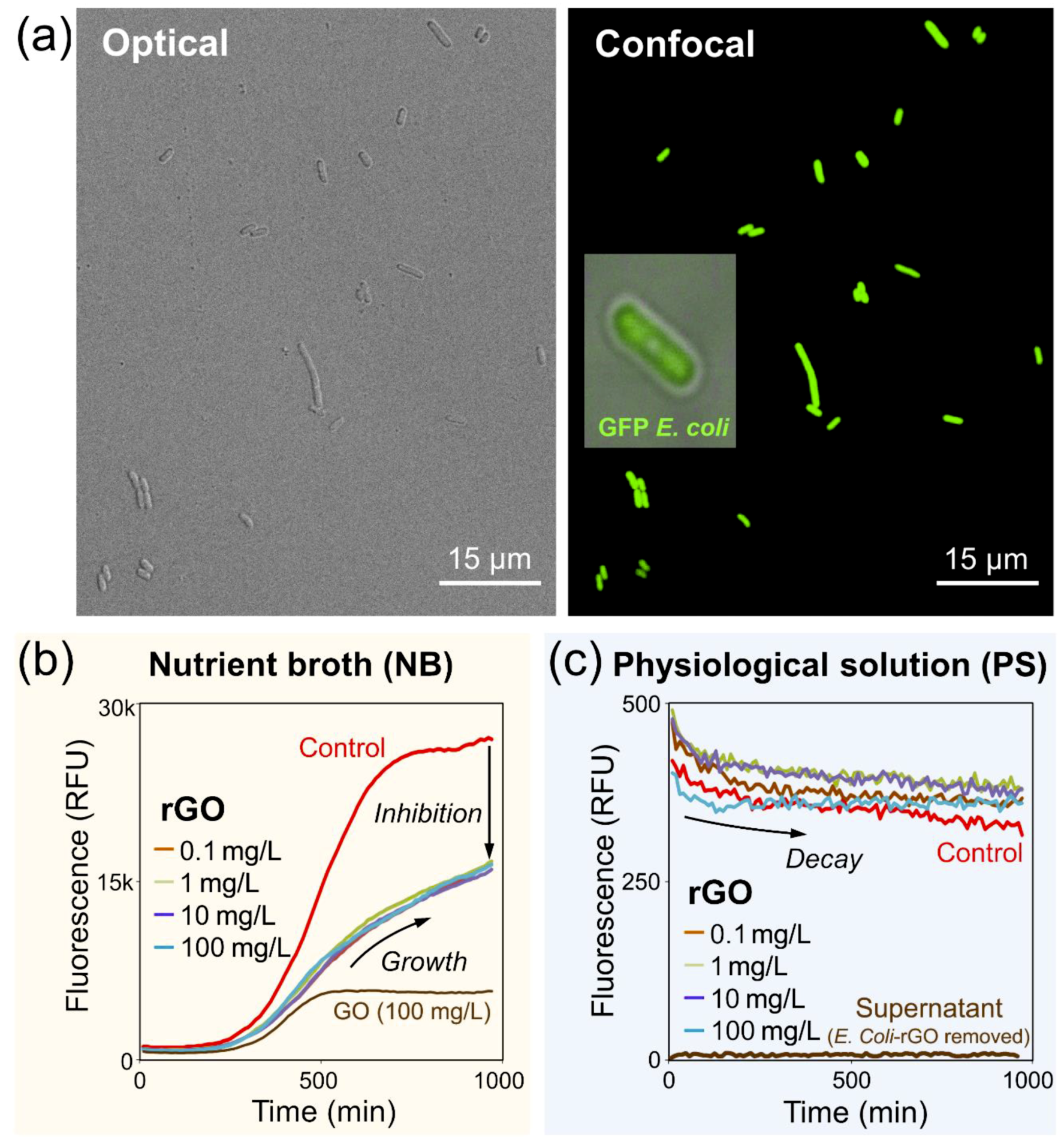
3.2. Bacterial Fluorescence Studies
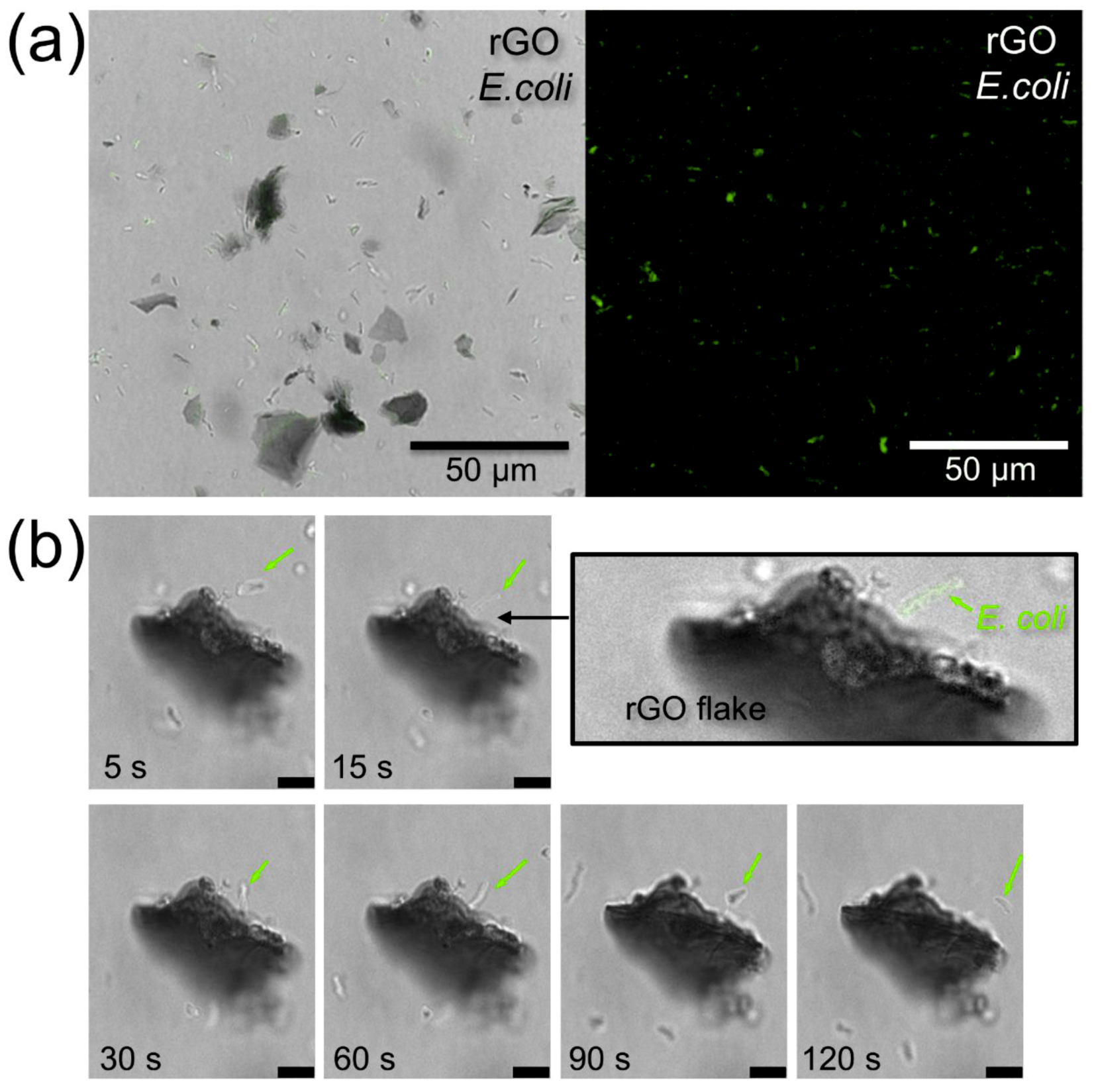
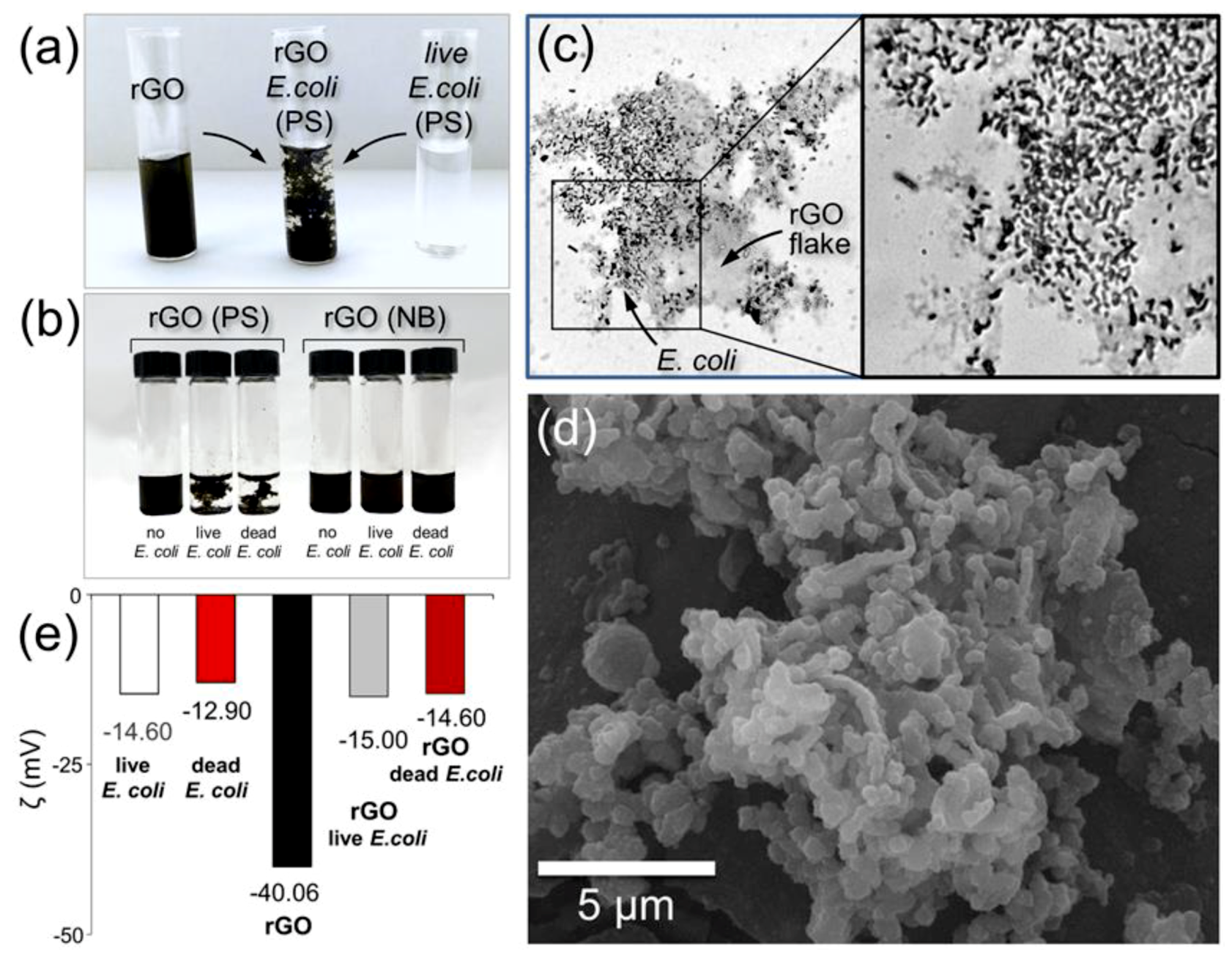
3.3. rGO-Bacteria Interactions
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Ray, S.C.; Saha, A.; Jana, N.R.; Sarkar, R. Fluorescent Carbon Nanoparticles: Synthesis, Characterization, and Bioimaging Application. J. Phys. Chem. C 2009, 113, 18546–18551. [Google Scholar] [CrossRef]
- Chandra, S.; Das, P.; Bag, S.; Laha, D.; Pramanik, P. Synthesis, functionalization and bioimaging applications of highly fluorescent carbon nanoparticles. Nanoscale 2011, 3, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhang, L.; Liu, M.; Zhang, Z. Biomedical Applications of Graphene. Theranostics 2012, 2, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Seabra, A.B.; Paula, A.J.; de Lima, R.; Alves, O.L.; Durán, N. Nanotoxicity of Graphene and Graphene Oxide. Chem. Res. Toxicol. 2014, 27, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A. Graphene: Safe or Toxic? The Two Faces of the Medal. Angew. Chem. Int. Ed. 2013, 52, 4986–4997. [Google Scholar] [CrossRef]
- Tang, J.; Chen, Q.; Xu, L.; Zhang, S.; Feng, L.; Cheng, L.; Xu, H.; Liu, Z.; Peng, R. Graphene Oxide—Silver Nanocomposite as a Highly Effective Antibacterial Agent with Species-Specific Mechanisms. ACS Appl. Mater. Interfaces 2013, 5, 3867–3874. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E. Toxicity of Graphene and Graphene Oxide Nanowalls Against Bacteria. ACS Nano 2010, 4, 5731–5736. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Lin, M.; Mustapha, A. Toxicity of graphene oxide on intestinal bacteria and Caco-2 cells. J. Food Prot. 2015, 78, 996–1002. [Google Scholar] [CrossRef]
- Pham, V.T.H.; Truong, V.K.; Quinn, M.D.J.; Notley, S.M.; Guo, Y.; Baulin, V.A.; Al Kobaisi, M.; Crawford, R.J.; Ivanova, E.P. Graphene Induces Formation of Pores That Kill Spherical and Rod-Shaped Bacteria. ACS Nano 2015, 9, 8458–8467. [Google Scholar] [CrossRef]
- Nanda, S.S.; An, S.S.; Yi, D.K. Oxidative stress and antibacterial properties of a graphene oxide-cystamine nanohybrid. Int. J. Nanomed. 2015, 10, 549–556. [Google Scholar]
- Ruiz, O.N.; Fernando, K.A.S.; Wang, B.; Brown, N.A.; Luo, P.G.; McNamara, N.D.; Vangsness, M.; Sun, Y.-P.; Bunker, C.E. Graphene Oxide: A Nonspecific Enhancer of Cellular Growth. ACS Nano 2011, 5, 8100–8107. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, G.; Zhang, G.; Xu, X.; Yang, F. Using graphene oxide to enhance the activity of anammox bacteria for nitrogen removal. Bioresour. Technol. 2013, 131, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Yang, X.; Tan, X.; Xu, L.; Liu, Z.; Xiao, J.; Peng, R. Functionalized graphene oxide in microbial engineering: An effective stimulator for bacterial growth. Carbon 2016, 103, 172–180. [Google Scholar] [CrossRef]
- Barbolina, I.; Woods, C.R.; Lozano, N.; Kostarelos, K.; Novoselov, K.S.; Roberts, I.S. Purity of graphene oxide determines its antibacterial activity. 2D Mater. 2016, 3, 025025. [Google Scholar] [CrossRef]
- Yan, L.; Zhao, F.; Li, S.; Hu, Z.; Zhao, Y. Low-toxic and safe nanomaterials by surface-chemical design, carbon nanotubes, fullerenes, metallofullerenes, and graphenes. Nanoscale 2011, 3, 362–382. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Wan, J.; Zhang, S.; Zhang, Y.; Lee, S.-T.; Liu, Z. In Vivo Pharmacokinetics, Long-Term Biodistribution, and Toxicology of PEGylated Graphene in Mice. ACS Nano 2011, 5, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Yang, S.-T.; Liu, J.-H.; Dong, E.; Wang, Y.; Cao, A.; Liu, Y.; Wang, H. In vitro toxicity evaluation of graphene oxide on A549 cells. Toxicol. Lett. 2011, 200, 201–210. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E.; Akhavan, A. Size-dependent genotoxicity of graphene nanoplatelets in human stem cells. Biomaterials 2012, 33, 8017–8025. [Google Scholar] [CrossRef]
- Kanchanapally, R.; Viraka Nellore, B.P.; Sinha, S.S.; Pedraza, F.; Jones, S.J.; Pramanik, A.; Chavva, S.R.; Tchounwou, C.; Shi, Y.; Vangara, A.; et al. Antimicrobial Peptide-Conjugated Graphene Oxide Membrane for Efficient Removal and Effective Killing of Multiple Drug Resistant Bacteria. RSC Adv. 2015, 5, 18881–18887. [Google Scholar] [CrossRef]
- Bao, Q.; Zhang, D.; Qi, P. Synthesis and characterization of silver nanoparticle and graphene oxide nanosheet composites as a bactericidal agent for water disinfection. J. Colloid Interface Sci. 2011, 360, 463–470. [Google Scholar] [CrossRef]
- Deng, C.-H.; Gong, J.-L.; Zeng, G.-M.; Niu, C.-G.; Niu, Q.-Y.; Zhang, W.; Liu, H.-Y. Inactivation performance and mechanism of Escherichia coli in aqueous system exposed to iron oxide loaded graphene nanocomposites. J. Hazard Mater. 2014, 276, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, O.; Ghaderi, E.; Esfandiar, A. Wrapping Bacteria by Graphene Nanosheets for Isolation from Environment, Reactivation by Sonication, and Inactivation by Near-Infrared Irradiation. J. Phys. Chem. B 2011, 115, 6279–6288. [Google Scholar] [CrossRef] [PubMed]
- Lipatov, A.; Varezhnikov, A.; Wilson, P.; Sysoev, V.; Kolmakov, A.; Sinitskii, A. Highly selective gas sensor arrays based on thermally reduced graphene oxide. Nanoscale 2013, 5, 5426–5434. [Google Scholar] [CrossRef] [PubMed]
- Marcano, D.C.; Kosynkin, D.V.; Berlin, J.M.; Sinitskii, A.; Sun, Z.Z.; Slesarev, A.; Alemany, L.B.; Lu, W.; Tour, J.M. Improved Synthesis of Graphene Oxide. ACS Nano 2010, 4, 4806–4814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, H.; Shen, G.; Cheng, P.; Zhang, J.; Guo, S. Reduction of graphene oxide via L-ascorbic acid. Chem. Commun. 2010, 46, 1112–1114. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Merino, M.J.; Guardia, L.; Paredes, J.I.; Villar-Rodil, S.; Solís-Fernández, P.; Martínez-Alonso, A.; Tascón, J.M.D. Vitamin C Is an Ideal Substitute for Hydrazine in the Reduction of Graphene Oxide Suspensions. J. Phys. Chem. C 2010, 114, 6426–6432. [Google Scholar] [CrossRef]
- Garcia, A.; Recillas, S.; Sanchez, A.; Font, X. The luminescent bacteria test to determine the acute toxicity of nanoparticle suspensions. Methods Mol. Biol. 2012, 926, 255–259. [Google Scholar]
- Mogilnaya, O.A.; Puzyr’, A.P.; Bondar’, V.S. Growth and bioluminescence of luminous bacteria under the action of aflatoxin B1 before and after its treatment with nanodiamonds. Appl. Biochem. Microbiol. 2010, 46, 33–37. [Google Scholar] [CrossRef]
- Zarubina, A.P.; Lukashev, E.P.; Deev, L.I.; Parkhomenko, I.M.; Rubin, A.B. Biotesting the biological effects of single-wall carbon nanotubes using bioluminescent bacteria test-system. Nanotechnol. Russ. 2009, 4, 871–875. [Google Scholar] [CrossRef]
- Sinitskii, A.; Tour, J.M. Chemical Approaches to Produce Graphene Oxide and Related Materials; Springer: New York, NY, USA, 2012; pp. 205–234. [Google Scholar]
- Romanchuk, A.Y.; Slesarev, A.S.; Kalmykov, S.N.; Kosynkin, D.V.; Tour, J.M. Graphene oxide for effective radionuclide removal. Phys. Chem. Chem. Phys. 2013, 15, 2321–2327. [Google Scholar] [CrossRef]
- Li, D.; Müller, M.B.; Gilje, S.; Kaner, R.B.; Wallace, G.G. Processable aqueous dispersions of graphene nanosheets. Nat. Nanotechnol. 2008, 3, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Sinitskii, A.; Kosynkin, D.V.; Dimiev, A.; Tour, J.M. Corrugation of Chemically Converted Graphene Monolayers on SiO2. ACS Nano 2010, 4, 3095–3102. [Google Scholar] [CrossRef] [PubMed]
- Stankovich, S.; Dikin, D.A.; Piner, R.D.; Kohlhaas, K.A.; Kleinhammes, A.; Jia, Y.; Wu, Y.; Nguyen, S.T.; Ruoff, R.S. Synthesis of graphene-based nanosheets via chemical reduction of exfoliated graphite oxide. Carbon 2007, 45, 1558–1565. [Google Scholar] [CrossRef]
- Yang, D.; Velamakanni, A.; Bozoklu, G.; Park, S.; Stoller, M.; Piner, R.D.; Stankovich, S.; Jung, I.; Field, D.A.; Ventrice, C.A.; et al. Chemical analysis of graphene oxide films after heat and chemical treatments by X-ray photoelectron and Micro-Raman spectroscopy. Carbon 2009, 47, 145–152. [Google Scholar] [CrossRef]
- Sinitskii, A.; Dimiev, A.; Kosynkin, D.V.; Tour, J.M. Graphene Nanoribbon Devices Produced by Oxidative Unzipping of Carbon Nanotubes. ACS Nano 2010, 4, 5405–5413. [Google Scholar] [CrossRef] [PubMed]
- Lipatov, A.; Guinel, M.J.-F.; Muratov, D.S.; Vanyushin, V.O.; Wilson, P.M.; Kolmakov, A.; Sinitskii, A. Low-temperature thermal reduction of graphene oxide: In situ correlative structural, thermal desorption, and electrical transport measurements. Appl. Phys. Lett. 2018, 112, 053103. [Google Scholar] [CrossRef]
- Ferrari, A.C.; Robertson, J. Interpretation of Raman spectra of disordered and amorphous carbon. Phys. Rev. B 2000, 61, 14095–14107. [Google Scholar] [CrossRef]
- Ganguly, A.; Sharma, S.; Papakonstantinou, P.; Hamilton, J. Probing the Thermal Deoxygenation of Graphene Oxide Using High-Resolution in Situ X-ray-Based Spectroscopies. J. Phys. Chem. C 2011, 115, 17009–17019. [Google Scholar] [CrossRef]
- Stankovich, S.; Piner, R.D.; Chen, X.; Wu, N.; Nguyen, S.T.; Ruoff, R.S. Stable aqueous dispersions of graphitic nanoplatelets via the reduction of exfoliated graphite oxide in the presence of poly (sodium 4-styrenesulfonate). J. Mater. Chem. 2006, 16, 155–158. [Google Scholar] [CrossRef]
- Perreault, F.; de Faria, A.F.; Nejati, S.; Elimelech, M. Antimicrobial Properties of Graphene Oxide Nanosheets: Why Size Matters. ACS Nano 2015, 9, 7226–7236. [Google Scholar] [CrossRef]
- Chen, J.; Peng, H.; Wang, X.; Shao, F.; Yuan, Z.; Han, H. Graphene oxide exhibits broad-spectrum antimicrobial activity against bacterial phytopathogens and fungal conidia by intertwining and membrane perturbation. Nanoscale 2014, 6, 1879–1889. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Piao, J.-G.; Auletta, J.; Hu, K.; Zhu, Y.; Meyer, T.; Liu, H.; Yang, L. Availability of the basal planes of graphene oxide determines whether it is antibacterial. ACS Appl. Mater. Interfaces 2014, 6, 13183–13190. [Google Scholar] [CrossRef] [PubMed]
- Mangadlao, J.D.; Santos, C.M.; Felipe, M.J.L.; de Leon, A.C.C.; Rodrigues, D.F.; Advincula, R.C. On the antibacterial mechanism of graphene oxide (GO) Langmuir-Blodgett films. Chem. Commun. 2015, 51, 2886–2889. [Google Scholar] [CrossRef] [PubMed]
- Medhekar, N.V.; Ramasubramaniam, A.; Ruoff, R.S.; Shenoy, V.B. Hydrogen Bond Networks in Graphene Oxide Composite Paper: Structure and Mechanical Properties. ACS Nano 2010, 4, 2300–2306. [Google Scholar] [CrossRef] [PubMed]
- Perkins, H.R. Chemical structure and biosynthesis of bacterial cell walls. Bacteriol. Rev. 1963, 27, 18–55. [Google Scholar]
- Braun, V. Covalent lipoprotein from the outer membrane of escherichia coli. Biochim. Biophys. Acta BBA Rev. Biomembr. 1975, 415, 335–377. [Google Scholar] [CrossRef]
- Boggs, J.M. Lipid intermolecular hydrogen bonding: Influence on structural organization and membrane function. Biochim. Biophys. Acta BBA Rev. Biomembr. 1987, 906, 353–404. [Google Scholar] [CrossRef]
- Chouhan, R.S.; Pandey, A.; Qureshi, A.; Ozguz, V.; Niazi, J.H. Nanomaterial resistant microorganism mediated reduction of graphene oxide. Colloids Surf. B Biointerfaces 2016, 146, 39–46. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Kim, J.-H. Microbial reduction of graphene oxide by Escherichia coli: A green chemistry approach. Colloids Surf. B Biointerfaces 2013, 102, 772–777. [Google Scholar] [CrossRef]
- Zou, X.; Zhang, L.; Wang, Z.; Luo, Y. Mechanisms of the Antimicrobial Activities of Graphene Materials. J. Am. Chem. Soc. 2016, 138, 2064–2077. [Google Scholar] [CrossRef]
- Wang, J.; Wei, Y.; Shi, X.; Gao, H. Cellular entry of graphene nanosheets: The role of thickness, oxidation and surface adsorption. RSC Adv. 2013, 3, 15776–15782. [Google Scholar] [CrossRef]
- Liao, K.H.; Lin, Y.S.; Macosko, C.W.; Haynes, C.L. Cytotoxicity of graphene oxide and graphene in human erythrocytes and skin fibroblasts. ACS Appl. Mater. Interfaces 2011, 3, 2607–2615. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gusev, A.; Zakharova, O.; Muratov, D.S.; Vorobeva, N.S.; Sarker, M.; Rybkin, I.; Bratashov, D.; Kolesnikov, E.; Lapanje, A.; Kuznetsov, D.V.; et al. Medium-Dependent Antibacterial Properties and Bacterial Filtration Ability of Reduced Graphene Oxide. Nanomaterials 2019, 9, 1454. https://doi.org/10.3390/nano9101454
Gusev A, Zakharova O, Muratov DS, Vorobeva NS, Sarker M, Rybkin I, Bratashov D, Kolesnikov E, Lapanje A, Kuznetsov DV, et al. Medium-Dependent Antibacterial Properties and Bacterial Filtration Ability of Reduced Graphene Oxide. Nanomaterials. 2019; 9(10):1454. https://doi.org/10.3390/nano9101454
Chicago/Turabian StyleGusev, Alexander, Olga Zakharova, Dmitry S. Muratov, Nataliia S. Vorobeva, Mamun Sarker, Iaroslav Rybkin, Daniil Bratashov, Evgeny Kolesnikov, Aleš Lapanje, Denis V. Kuznetsov, and et al. 2019. "Medium-Dependent Antibacterial Properties and Bacterial Filtration Ability of Reduced Graphene Oxide" Nanomaterials 9, no. 10: 1454. https://doi.org/10.3390/nano9101454
APA StyleGusev, A., Zakharova, O., Muratov, D. S., Vorobeva, N. S., Sarker, M., Rybkin, I., Bratashov, D., Kolesnikov, E., Lapanje, A., Kuznetsov, D. V., & Sinitskii, A. (2019). Medium-Dependent Antibacterial Properties and Bacterial Filtration Ability of Reduced Graphene Oxide. Nanomaterials, 9(10), 1454. https://doi.org/10.3390/nano9101454






