Research Progress of M13 Bacteriophage-Based Biosensors
Abstract
1. Introduction
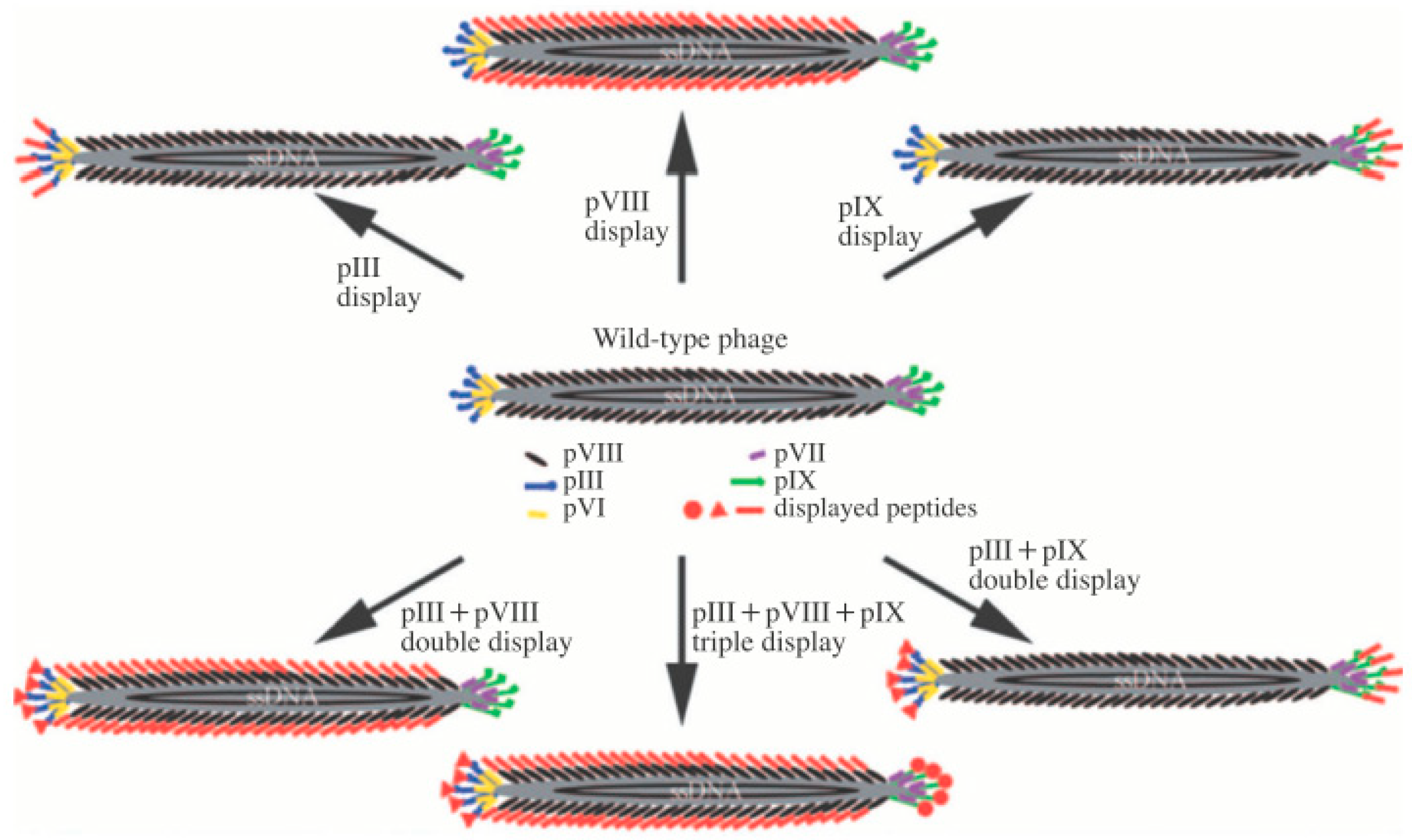
1.1. M13 Bacteriophage
1.2. M13 Bacteriophage-Based Biosensor
2. M13 Bacteriophage-Based Protein and Microorganism Sensing
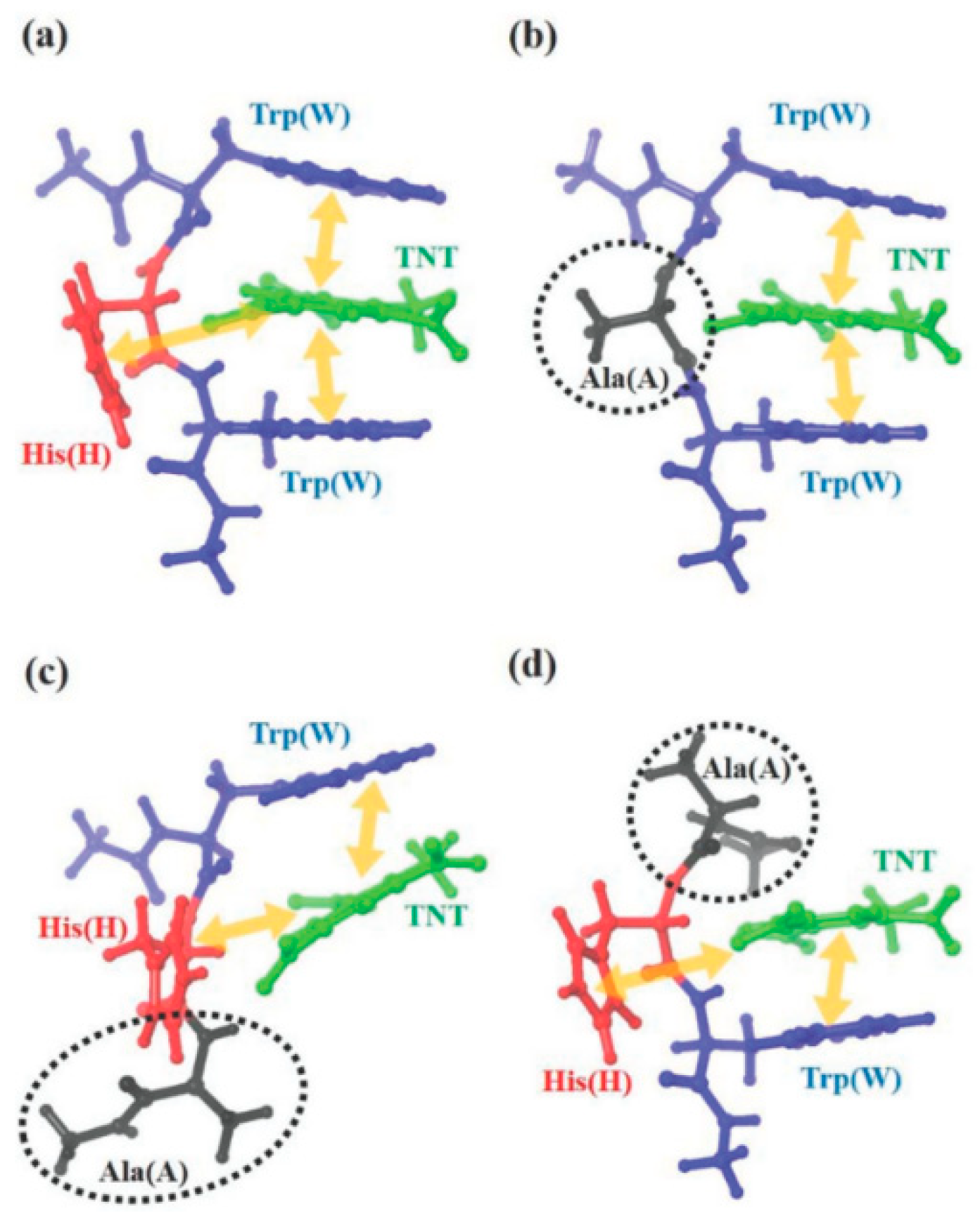
3. M13 Bacteriophage-Based Chemical Sensing
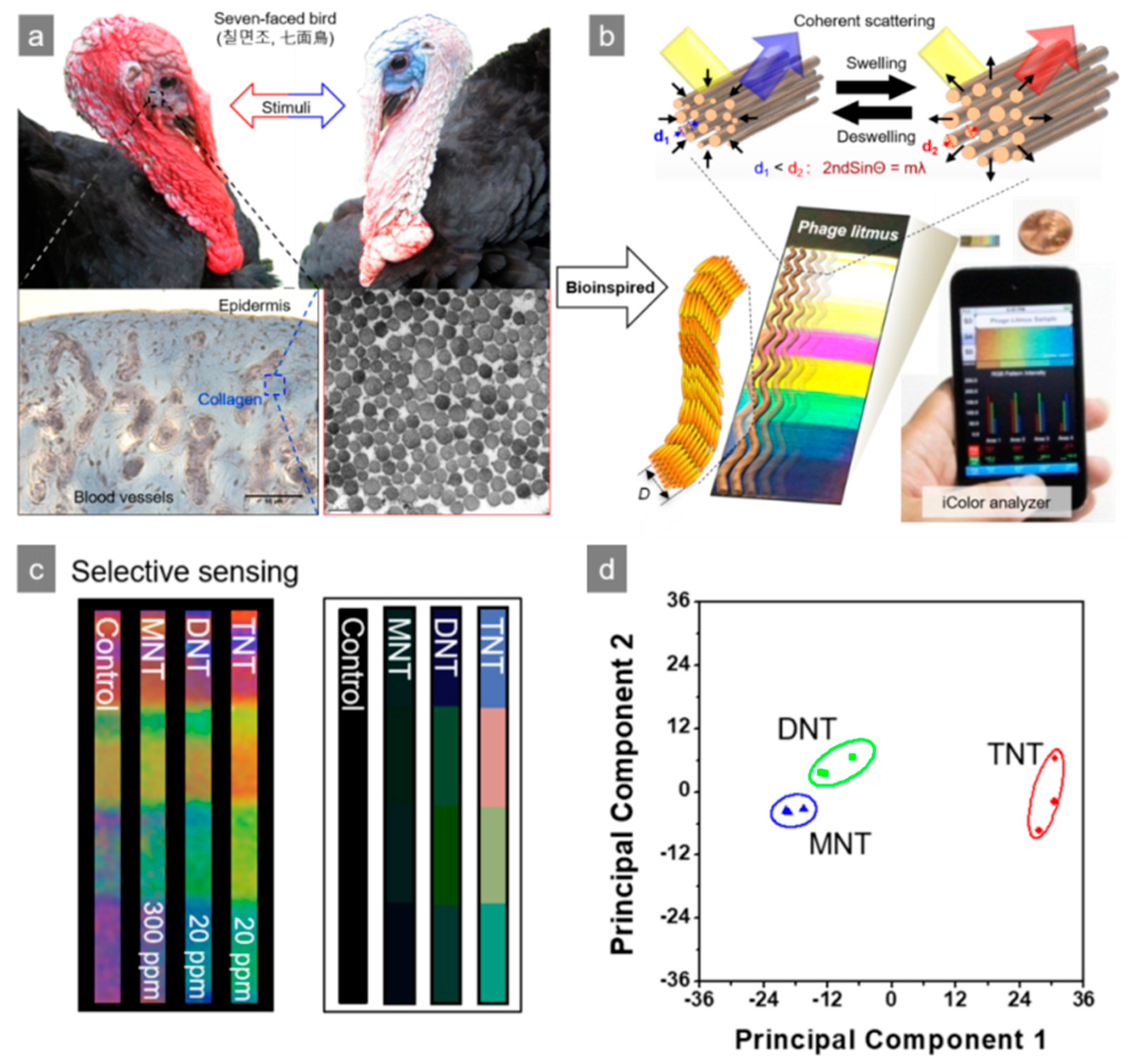
4. M13 Bacteriophage-Based Color Sensor
5. Expansion of Analytic Method from Direct Sensing Using M13 Bacteriophage
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Andersen, P.D.; Jørgensen, B.H.; Lading, L.; Rasmussen, B. Sensor foresight—Technology and market. Technovation 2004, 24, 311–320. [Google Scholar] [CrossRef]
- Hollingum, J. Foresight launches three sensor programmes. Sens. Rev. 1999, 19, 116–120. [Google Scholar] [CrossRef]
- Sensors: The next wave of infotech innovation. Ten-Year Forecast Institute for the future. 1997, 115–122.
- Oh, J.-W.; Chung, W.-J.; Heo, K.; Jin, H.-E.; Lee, B.Y.; Wang, E.; Zueger, C.; Wong, W.; Meyer, J.; Kim, C.; et al. Biomimetic virus-based colorimetric sensors. Nat. Commun. 2014, 5, 3043. [Google Scholar] [CrossRef]
- Martin-Herranz, A.; Ahmad, A.; Evans, H.M.; Ewert, K.; Schulze, U.; Safinya, C.R. Surface functionalized cationic lipid-DNA complexes for gene delivery: PEGylated lamellar complexes exhibit distinct DNA-DNA Interaction Regimes. Biophys. J. 2004, 86, 1160–1168. [Google Scholar] [CrossRef]
- Moon, J.S.; Kim, W.G.; Kim, C.; Park, G.-T.; Heo, J.; Yoo, S.Y.; Oh, J.-W. M13 bacteriophage-based self-assembly Structures and their functional capabili-ties. Mini Rev. Org. Chem. 2015, 12, 271–281. [Google Scholar] [CrossRef]
- Chung, W.J.; Lee, D.Y.; Yoo, S.Y. Chemical modulation of M13 bacteriophage and its functional opportunities for nanomedicine. Int. J. Nanomed. 2014, 9, 5825–5836. [Google Scholar]
- Lee, Y.J.; Yi, H.; Kim, W.-J.; Kang, K.; Yun, D.S.; Strano, M.S.; Ceder, G.; Belcher, A.M. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 2009, 324, 1051–1055. [Google Scholar]
- Lee, Y.; Kim, J.; Yun, D.S.; Nam, Y.S.; Yang, S.-H.; Belcher, A.M. Virus-templated Au and Au-Pt core-shell nanowires and their electrocatalytic activities for fuel cell applications. Energy Sci. 2012, 5, 8328–8334. [Google Scholar] [CrossRef]
- Mao, C.; Liu, A.; Cao, B. Virus-based chemical and biological sensing. Angew 2009, 48, 6790–6810. [Google Scholar] [CrossRef]
- Sundar, V.C.; Yablon, A.D.; Grazul, J.L.; Ilan, M.; Aizenberg, J. Fibre-optical features of a glass sponge. Nature 2003, 424, 899–900. [Google Scholar] [CrossRef]
- Espinosa, H.D.; Luster, A.L.; Latourte, F.J.; Loh, O.Y.; Gregoire, D.; Zavattieri, P.D. Tablet-level origin of toughening in abalone shells and translation to synthetic composite materials. Nat. Commun. 2011, 2, 173. [Google Scholar] [CrossRef] [PubMed]
- Aizenberg, J.; Weaver, J.C.; Thanawala, M.S.; Sundar, V.C.; Morse, D.E.; Fratzl, P. Skeleton of euplectella sp.: Structural hierarchy from the nanoscale to the macroscale. Science 2005, 309, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.-J.; Oh, J.-W.; Kwak, K.; Lee, B.Y.; Meyer, J.; Wang, E.; Hexemer, A.; Lee, S.-W. Biomimetic self-templating supramolecular structure. Naure 2011, 478, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Dogic, Z.; Fraden, S. Ordered phases of filamentous viruses. Curr. Opin. Colloid Interface Sci. 2006, 11, 47–55. [Google Scholar] [CrossRef]
- Yang, S.H.; Chung, W.J.; Mcfarland, S.; Lee, S.W. Assembly of Bacteriophage into Functional Materials. Chem. Rec. 2013, 13, 43–59. [Google Scholar] [CrossRef]
- Gimenez, S.; Lana-Villarreal, T.; Gómez, R.; Agouram, S.; Muñoz-Sanjosé, V.; Mora-Seró, I. Determination of limiting factors of photovoltaic efficiency in quantum dot sensitized solar cells: Correlation between cell performance and structural properties. J. Appl. Phys. 2010, 108, 064310. [Google Scholar] [CrossRef]
- Karpan, V.M.; Khomyakov, P.A.; Starikov, A.A.; Giovannetti, G.; Zwierzycki, M.; Talanana, M.; Brocks, G.; van den Brink, J.; Kelly, P.J. Theoretical prediction of perfect spin filtering at interfaces between close-packed surfaces of Ni or Co and graphite or graphene. Phys. Rev. B 2008, 78, 195419. [Google Scholar] [CrossRef]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Cao, B.; Mao, C. Phage Nanobiotechnology; Petrenko, V., Smith, G.P., Eds.; RSC publishing: Cambridge, UK, 2011. [Google Scholar]
- Brissette, R.; Goldstein, N.I. Methods in Molecular Biology; Fisher, P., Ed.; Humana: Totowa, NJ, USA, 2007. [Google Scholar]
- Flynn, C.E.; Lee, S.W.; Peelle, B.R.; Belcher, A.M. Viruses as vehicles for growth, organization and assembly of materials. Acta Mater. 2003, 51, 5867–5880. [Google Scholar] [CrossRef]
- Cui, Y.; Kim, S.N.; Jones, S.E.; Wissler, L.L.; Naik, R.R.; McAlpine, M.C. Chemical functionalization of graphene enabled by phage displayed peptides. Nano Lett. 2010, 10, 4559–4565. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.N.; Kuang, Z.; Slocik, J.M.; Jones, S.E.; Cui, Y.; Farmer, B.L.; McAlpine, M.C.; Naik, R.R. Preferential binding of peptides to graphene edges and planes. J. Am. Chem. Soc. 2011, 133, 14480–14483. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Solis, D.J.; Reiss, B.D.; Kottmann, S.T.; Sweeney, R.Y.; Hayhurst, A.; Georgiou, G.; Iverson, B.; Belcher, A.M. Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science 2004, 303, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Mio, C.; Flynn, C.E.; Hayhurst, A.; Sweeney, R.; Qi, J.; Georgiou, G.; Iverson, B.; Belcher, A.M. Viral assembly of oriented quantum dot nanowires. Proc. Natl. Acad. Sci. USA 2003, 100, 6946–6951. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chiang, C.-Y.; Lee, S.K.; Gao, Y.; Hu. E., L.; De Yoreo, J.; Belcher, A.M. Programmable assembly of nanoarchitectures using genetically engineered viruses. Nano Lett. 2005, 5, 1429–1434. [Google Scholar] [CrossRef] [PubMed]
- Deo, S.; Godwin, H.A. A selective, ratiometric fluorescent sensor for Pb2+. J. Am. Chem. Soc. 2000, 122, 174–175. [Google Scholar] [CrossRef]
- Chen, L.; Wu, Y.; Lin, Y.; Wang, Q. Virus-templated FRET platform for the rational design of ratiometric fluorescent nanosensors. Chem. Commun. 2015, 51, 10190–10193. [Google Scholar] [CrossRef]
- Yoo, S.Y.; Oh, J.-W.; Lee, S.W. Phage-chips for novel optically readable tissue engineering assays. Langmuir 2012, 28, 2166–2172. [Google Scholar] [CrossRef][Green Version]
- Lee, J.H.; Xu., P.F.; Domaille, D.W.; Choi, C.; Jin, S.; Cha, J.N. M13 Bacteriophage as materials for amplified surface enhanced Raman scattering protein sensing. Adv. Funct. Mater. 2014, 24, 2079–2084. [Google Scholar] [CrossRef]
- Kawakami, K.; Nishihara, Y.; Hirano, K. Effect of hydrophilic polymers on physical stability of liposome dispersions. J. Phys. Chem. B 2001, 105, 2374–2385. [Google Scholar] [CrossRef]
- Ruysschaert, T.; Germain, M.; Gomes, J.F.; Fournier, D.; Sukhorukov, G.B.; Meier, W.; Winterhalter, M. Liposome-based nanocapsules. IEEE Trans. Nanobiosci. 2004, 3, 49–55. [Google Scholar] [CrossRef]
- Ngweniform, P.; Abbineni, G.; Cao, B.; Mao, C. Self-assembly of drug-loaded liposomes on genetically engineered target-recognizing M13 phage: A novel nanocarrier for targeted drug delivery. Small 2009, 5, 1963–1969. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, J.W.; Raorane, D.; Huh, J.H.; Majumdar, A.; Lee, S.-W. Evolutionary screening of biomimetic coatings for selective detection of explosives. Langmuir 2008, 24, 4938–4943. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Won, N.; Ahn, B.; Kwang, J.; Heo, K.; Oh, J.-W.; Sun, Y.; Cho, S.G.; Lee, S.-W.; Kim., S. Quantum dot-engineered M13 virus layer-by-layer composite films for highly selective and sensitive turn-on TNT sensors. Chem. Commun. 2013, 49, 6045–6047. [Google Scholar] [CrossRef]
- Lee, B.Y.; Zhang, J.; Zueger, C.; Chung, W.-J.; Yoo, S.Y.; Wang, E.; Meyer, J.; Ramesh, R.; Lee, S.-W. Virus-based piezoelectric energy generation. Nat. Nanotechnol. 2012, 7, 351–356. [Google Scholar] [CrossRef]
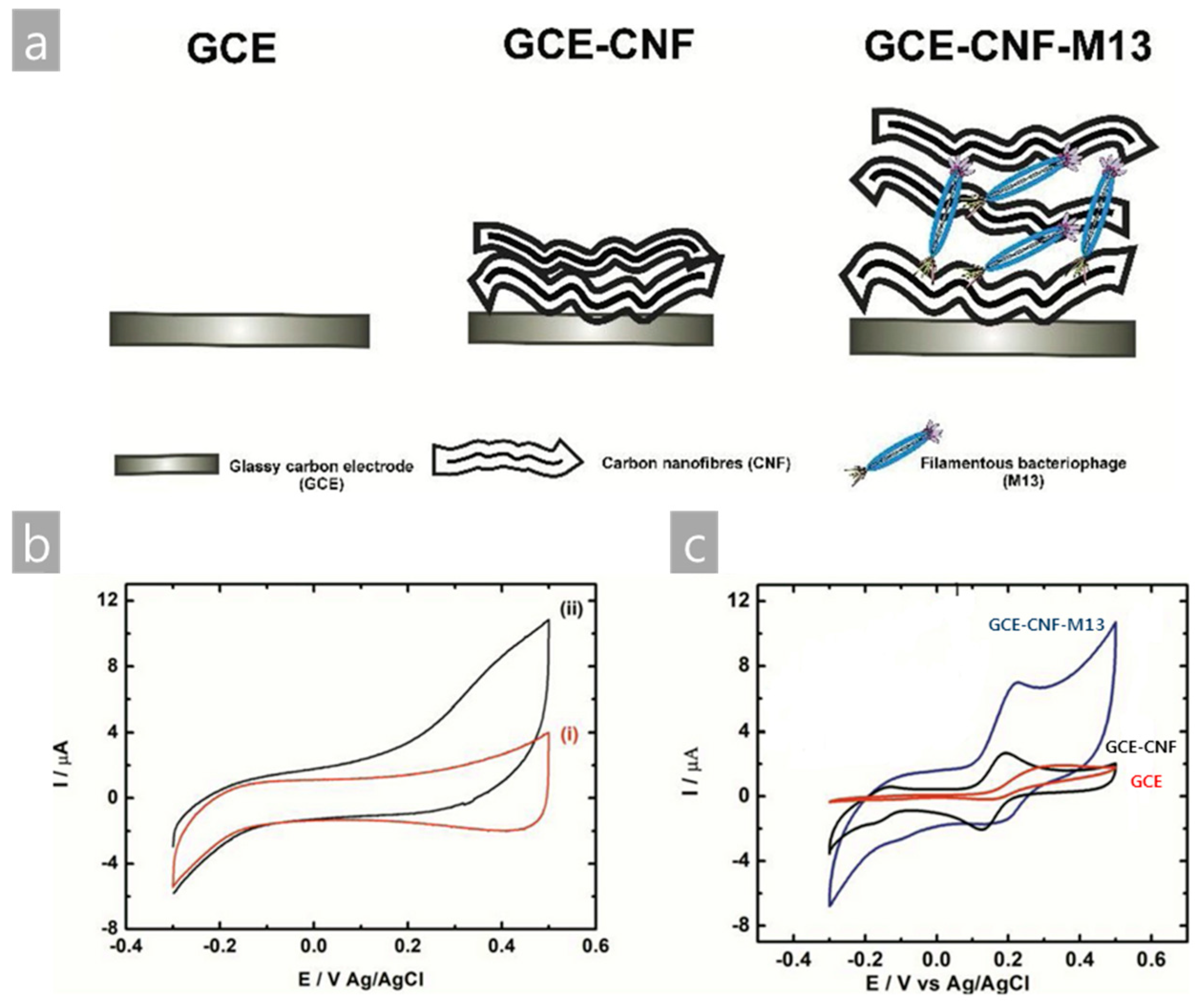
- Szot-Karpińska, K.; Leśniewski, A.; Jönsson-Niedziółka, M.; Marken, F.; Niedziółka-Jönsson, J. Electrodes modified with bacteriophages and carbon nanofibres for cysteine detection. Sens. Actuat B Chem. 2019, 287, 78–85. [Google Scholar] [CrossRef]
- Szot, K.; Lesniewski, A.; Niedziolka, J.; Jönsson, M.; Rizzi, C.; Gaillon, L.; Marken, F.; Rogalski, J.; Opallo, M. Sol–gel processed ionic liquid—Hydrophilic carbon nanoparticles multilayer film electrode prepared by layer-by-layer method. J. Electroanal. Chem. 2008, 623, 170–176. [Google Scholar] [CrossRef]
- Macdonald, S.; Szot, K.; Niedziolka, J.; Marken, F.; Opallo, M. Introducing hydrophilic carbon nanoparticles into hydrophilic sol-gel film electrodes. J. Solid State Electr. 2008, 12, 287–293. [Google Scholar] [CrossRef]
- Bajenova, O.; Chaika, N.; Tolkunova, E.; Davydov-Sinitsyn, A.; Gapon, S.; Thomas, P.; O’Brien, S. Carcinoembryonic antigen promotes colorectal cancer progression by targeting adherens junction complexes. Exp. Cell Res. 2014, 324, 115–123. [Google Scholar] [CrossRef]
- Bajenova, O.; Gorbunova, A.; Evsyukov, I.; Rayko, M.; Gapon, S.; Bozhokina, E.; Shishkin, A.; O’Brien, S.J. The genome-wide analysis of carcinoembryonic antigen signaling by colorectal cancer cells using RNA sequencing. PLoS ONE 2016, 11, e0161256. [Google Scholar] [CrossRef]
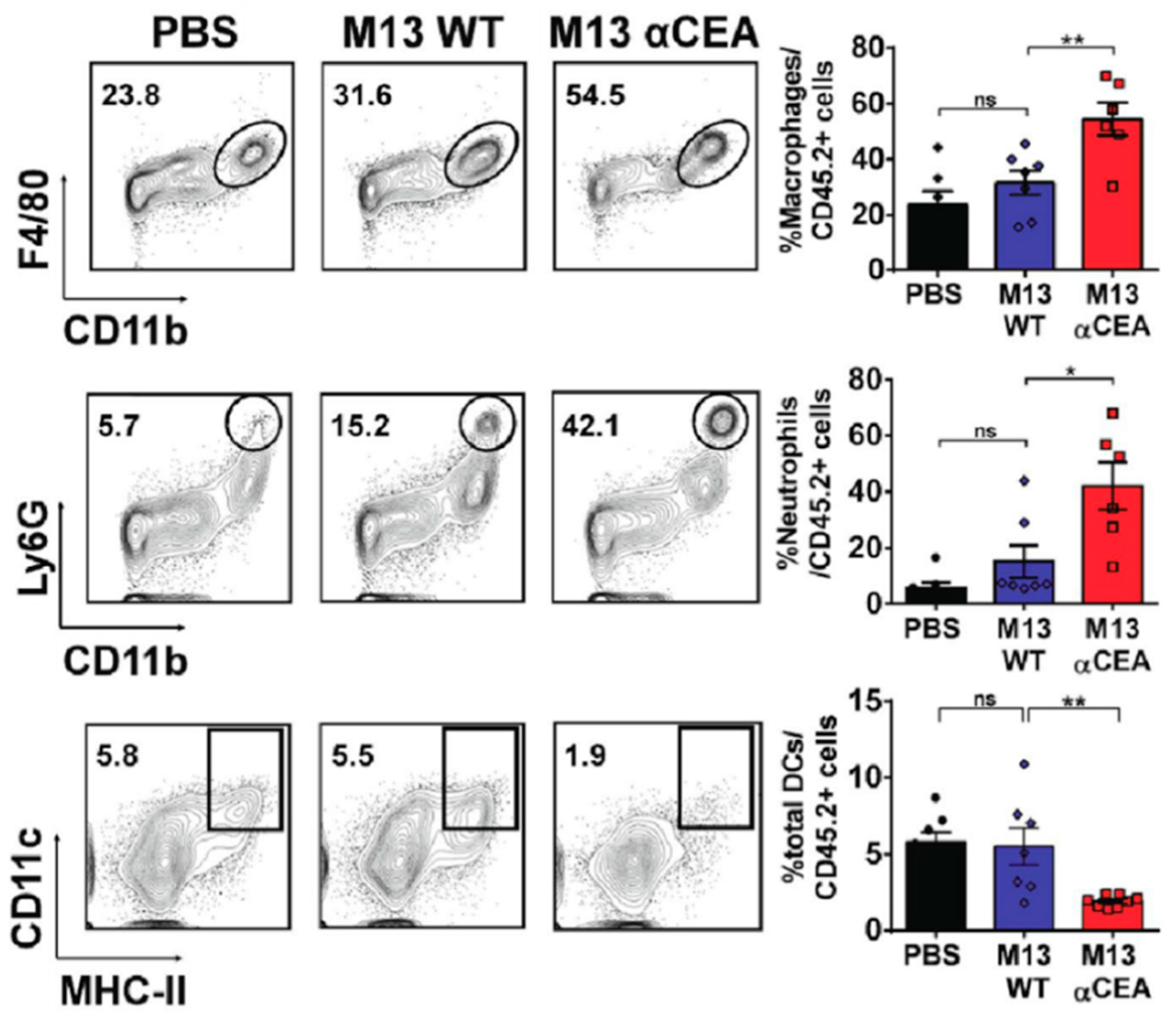
- Murgas, P.; Bustamante, N.; Araya, N.; Cruz-Gómez, S.; Durán, E.; Gaete, D.; Oyarce, C.; López, E.; Herrada, A.A.; Ferreira, N.; et al. A filamentous bacteriophage targeted to carcinoembryonic antigen induces tumor regression in mouse models of colorectal cancer. Cancer Immunol. Immun. 2018, 67, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Ordoñez, C.; Screaton, R.A.; Ilantzis, C.; Stanners, C.P. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Can. Res. 2000, 60, 3419–3424. [Google Scholar]
- Taheri, M.; Saragovi, H.U.; Stanners, C.P. The adhesion and differentiation-inhibitory activities of the immunoglobulin super-family member, carcinoembryonic antigen, can be independently blocked. J. Biol. Chem. 2003, 278, 14632–14639. [Google Scholar] [CrossRef] [PubMed]
- Rondot, S.; Koch, J.; Breitling, F.; Dubel, S. A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol. 2001, 19, 75–78. [Google Scholar] [CrossRef]
- Niyomdecha, S.; Limbut, W.; Numnuam, A.; Kanatharana, P.; Charlermroj, R.; Karoonuthaisiri, N.; Thavarungkul, P. Phage-based capacitive biosensor for Salmonella detection. Talanta 2018, 188, 658–664. [Google Scholar] [CrossRef]
- Wolffs, P.F.G.; Glencross, K.; Thibaudeau, R.; Griffths, M.W. Direct quantitation and detection of salmonellae in biological samples without enrichment, using two-step filtration and real-time PCR. Appl. Environ. Microbiol. 2006, 72, 3896–3900. [Google Scholar] [CrossRef]
- Bennett, A.R.; Greenwood, D.; Tennant, C.; Banks, J.G.; Betts, R.P. Rapid and definitive detection of Salmonella in foods by PCR. Lett. Appl. Microbiol. 1998, 26, 437–441. [Google Scholar] [CrossRef]
- Herikstad, H.; Motarjemi, Y.; Tauxe, R.V. Salmonella surveillance: A global survey of public health serotyping. Epidemiol. Infect. 2002, 129, 1–8. [Google Scholar] [CrossRef]
- Guliy, O.; Zaitsev, B.D.; Borodina, I.A.; Shikhabudinov, A.M.; Teplykh, A.A.; Staroverov, S.A.; Fomin, A.S. The biological acoustic sensor to record the interactions of the microbial cells with the phage antibodies in conducting suspensions. Talanta 2018, 178, 569–576. [Google Scholar] [CrossRef]
- Kanoatov, M.; Krylov, S.N. Analysis of DNA in phosphate buffered saline using kinetic capillary electrophoresis. Anal. Chem. 2016, 88, 7421–7428. [Google Scholar] [CrossRef]
- Smith, G.P.; Scott, J.K. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993, 217, 228–257. [Google Scholar] [PubMed]
- Yan, Y.; Zhang, M.; Moon, C.H.; Su, H.C.; Myung, N.V.; Haberer, E.D. Viral-templated gold/polypyrrole nanopeapods for an ammonia gas sensor. Nanotechnology 2016, 27, 325502. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Dang, X.; Yi, H.; Allen, M.A.; Xu, K.; Lee, Y.J.; Belcher, A.M. Graphene sheets stabilized on genetically engineered M13 viral templates as conducting frameworks for hybrid energy-storage materials. Small 2012, 8, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, G.; Lundström, I. The effect of ammonia on the physical properties of polypyrrole. Synth. Met. 1987, 21, 203–208. [Google Scholar] [CrossRef]
- Gustafsson, G.; Lundström, B.; Liedberg, I.; Wu, C.R.; Inganäs, O.; Wennerström, O. The interaction between ammonia and poly(pyrrole). Synth. Met. 1989, 31, 163–179. [Google Scholar]
- Choi, J.; Hormes, J.; Kahol, P.K. Study of ammonia-gas-induced irreversibility in polypyrrole films. Appl. Phys. Lett. 2003, 83, 2288–2290. [Google Scholar] [CrossRef]
- Koh, E.H.; Mun, C.; Kim, C.; Park, S.G.; Choi, E.J.; Kim, S.H.; Dang, J.; Choo, J.; Oh, J.-W.; Kim, D.H.; et al. M13 bacteriophage/silver nanowire surface-enhanced Raman scattering sensor for sensitive and selective pesticide detection. ACS Appl. Mater. Interfaces 2018, 10, 10388–10397. [Google Scholar] [CrossRef]
- Park, S.-G.; Mun, C.W.; Lee, M.K.; Jeon, T.Y.; Shim, H.-S.; Lee, Y.-J.; Kwon, J.-D.; Kim, C.S.; Kim, D.-H. 3D Hybrid Plasmonic Nanomaterials for Highly Efficient Optical Absorbers and Sensors. Adv. Mater. 2015, 27, 4290–4295. [Google Scholar] [CrossRef]
- Nagy-Simon, T.; Tatar, A.-S.; Craciun, A.-M.; Vulpoi, A.; Jurj, M.-A.; Florea, A.; Tomuleasa, C.; Berindan-Neagoe, I.; Astilean, S.; Boca, S. Antibody Conjugated, Raman Tagged Hollow Gold−Silver Nanospheres for Specific Targeting and Multimodal Dark-Field/SERS/Two Photon-FLIM Imaging of CD19(+) B Lymphoblasts. ACS Appl. Mater. Interfaces 2017, 9, 21155–21168. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, K.; Li, Y.; Ji, J.; Liu, B. High-Resolution and universal visualization of latent fingerprints based on aptamer-functionalized core−shell nanoparticles with embedded SERS reporters. ACS Appl. Mater. Interfaces 2016, 8, 14389–14395. [Google Scholar] [CrossRef]
- Fang, H.; Zhang, X.; Zhang, S.J.; Liu, L.; Zhao, Y.M.; Xu, H.J. Ultrasensitive and quantitative detection of paraquat on fruits skins via surface-enhanced Raman spectroscopy. Sens. Actuators B 2015, 213, 452–456. [Google Scholar] [CrossRef]
- Moon, J.-S.; Park, M.; Kim, W.-G.; Kim, C.; Hwang, J.; Seol, D.; Kim, C.-S.; Sohn, J.-R.; Chung, H.; Oh, J.-W. M-13 bacteriophage based structural color sensor for detecting antibiotics. Sens. Actuators B 2017, 240, 757–762. [Google Scholar] [CrossRef]
- Slavik, R.; Homola, J. Ultrahigh resolution long range surface plasmon-based sensor. Sens. Actuators B 2007, 123, 10–12. [Google Scholar] [CrossRef]
- Ferber, D. WHO advises kicking the livestock antibiotic habit. Science 2003, 301, 1027. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.T.; Kim, D.-W.; Yoo, P.J.; Chiang, C.-Y.; Meethong, N.; Hammond, P.T.; Chiang, Y.-M.; Belcher, A.M. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science 2006, 312, 885–888. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.S.; Lee, Y.; Shin, D.M.; Kim, C.; Kim, W.G.; Park, M.; Han, J.; Song, H.; Kim, K.; Oh, J.-W. Identification of endocrine disrupting chemicals using a virus-based colorimetric sensor. Chem. Asian J. 2016, 11, 3097–3101. [Google Scholar] [CrossRef]
- Sadik, O.A.; Witt, D.M. Peer reviewed: Monitoring endocrine-disrupting chemicals. Environ. Sci. Technol. 1999, 33, 368A–374A. [Google Scholar] [CrossRef]
- Yoon, Y.; Westerhoff, P.; Snyder, S.A.; Esparza, M. HPLC-fluorescence detection and adsorption of bisphenol A, 17beta-estradiol, and 17alpha-ethynyl estradiol on powdered activated carbon. Water Res. 2003, 37, 3530–3537. [Google Scholar] [CrossRef]
- Moon, J.-S.; Kim, W.-G.; Shin, D.-M.; Lee, S.Y.; Kim, C.; Lee, Y.; Han, J.; Kim, K.; Yoo, S.Y.; Oh, J.-W. Bioinspired M-13 bacteriophage-based photonic nose for differential cell recognition. Chem. Sci. 2017, 8, 921–927. [Google Scholar] [CrossRef]
- Peng, G.; Tisch, U.; Adams, O.; Hakim, M.; Shehada, N.; Broza, Y.Y.; Billan, S.; Bortnyak, R.R.; Kuten, A.; Haick, H. Diagnosing lung cancer in exhaled breath using gold nanoparticles. Nat. Nanotechnol. 2009, 4, 669–673. [Google Scholar] [CrossRef]
- Yun, J.M.; Kim, K.N.; Kim, J.Y.; Shin, D.O.; Lee, W.J.; Lee, S.H.; Lieberman, M.; Kim, S.O. DNA origami nanopatterning on chemically modified graphene. Angew 2011, 51, 912–915. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.M.; Ganesan, R.; Choi, J.-H.; Kim, J.-B. Local pH-Responsive Diazoketo-Functionalized Photoresist for Multicomponent Protein Patterning. ACS Appl. Mater. Interfaces 2013, 5, 10253–10259. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.G.; Zueger, C.; Kim, C.; Wong, W.; Devaraj, V.; Yoo, H.W.; Hwang, S.; Oh, J.-W.; Lee, S.W. Experimental and numerical evaluation of a genetically engineered M13 bacteriophage with high sensitivity and selectivity for 2,4,6-trinitrotoluene. Org. Biomol. Chem. 2019, 17, 5666–5670. [Google Scholar] [CrossRef] [PubMed]
- Blaik, R.A.; Lan, E.; Huang, Y.; Dunn, B. gold-coated m13 bacteriophage as a template for glucose oxidase biofuel cells with direct electron transfer. ACS Nano 2015, 10, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.D.; Forzani, E.S.; Leright, M.; Tsow, F.; Cagan, A.; Iglesias, R.A.; Nagahara, L.A.; Amlani, I. A hybrid nanosensor for tnt vapor detection. Nano Lett. 2010, 10, 380–384. [Google Scholar] [CrossRef]
- Rose, A.; Zhu, Z.; Madigan, C.F.; Swager, T.M.; Bulović, V. Sensitivity gains in chemosensing by lasing action in organic polymers. Nature 2005, 434, 876–879. [Google Scholar] [CrossRef]
- Riskin, M.; Tel-Vered, R.; Lioubashevski, O.; Willner, I. Ultrasensitive surface plasmon resonance detection of trinitrotoluene by a bis-aniline-cross-linked au nanoparticles composite. J. Am. Chem. Soc. 2009, 131, 7368–7378. [Google Scholar] [CrossRef]
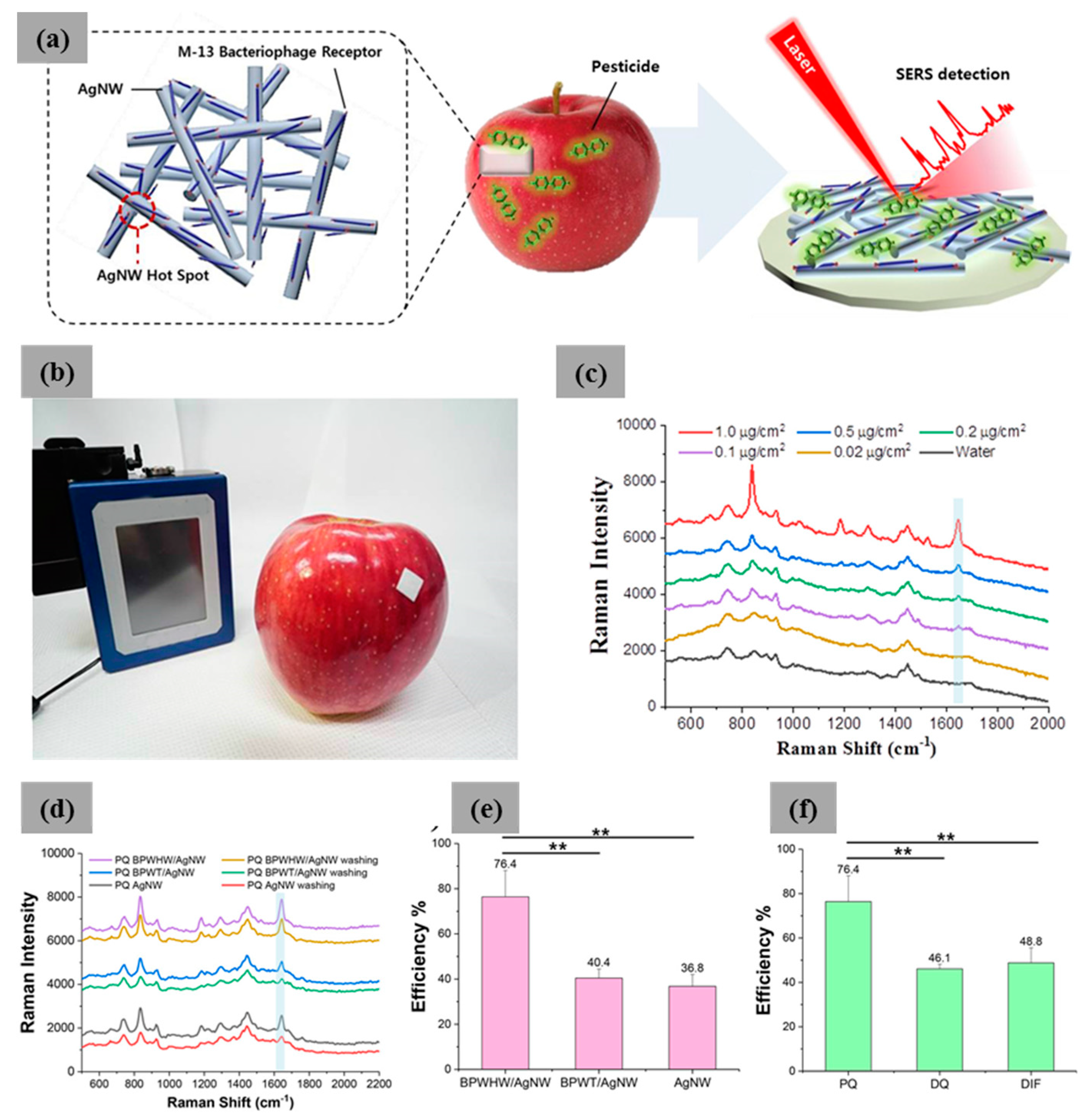
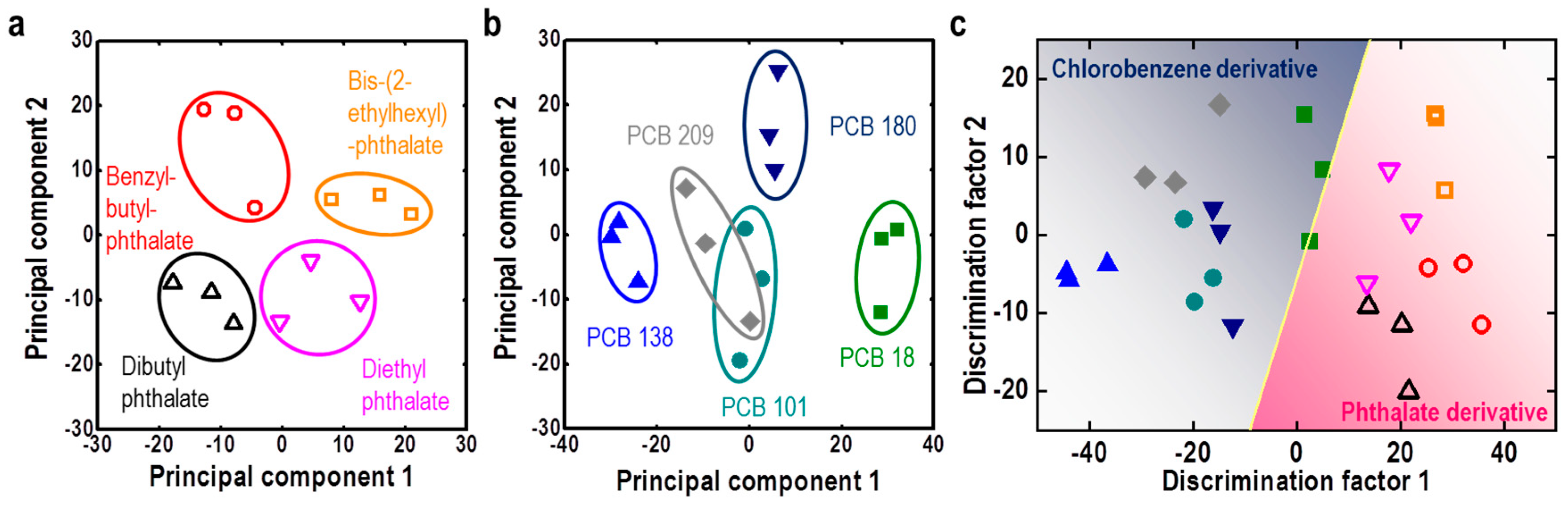
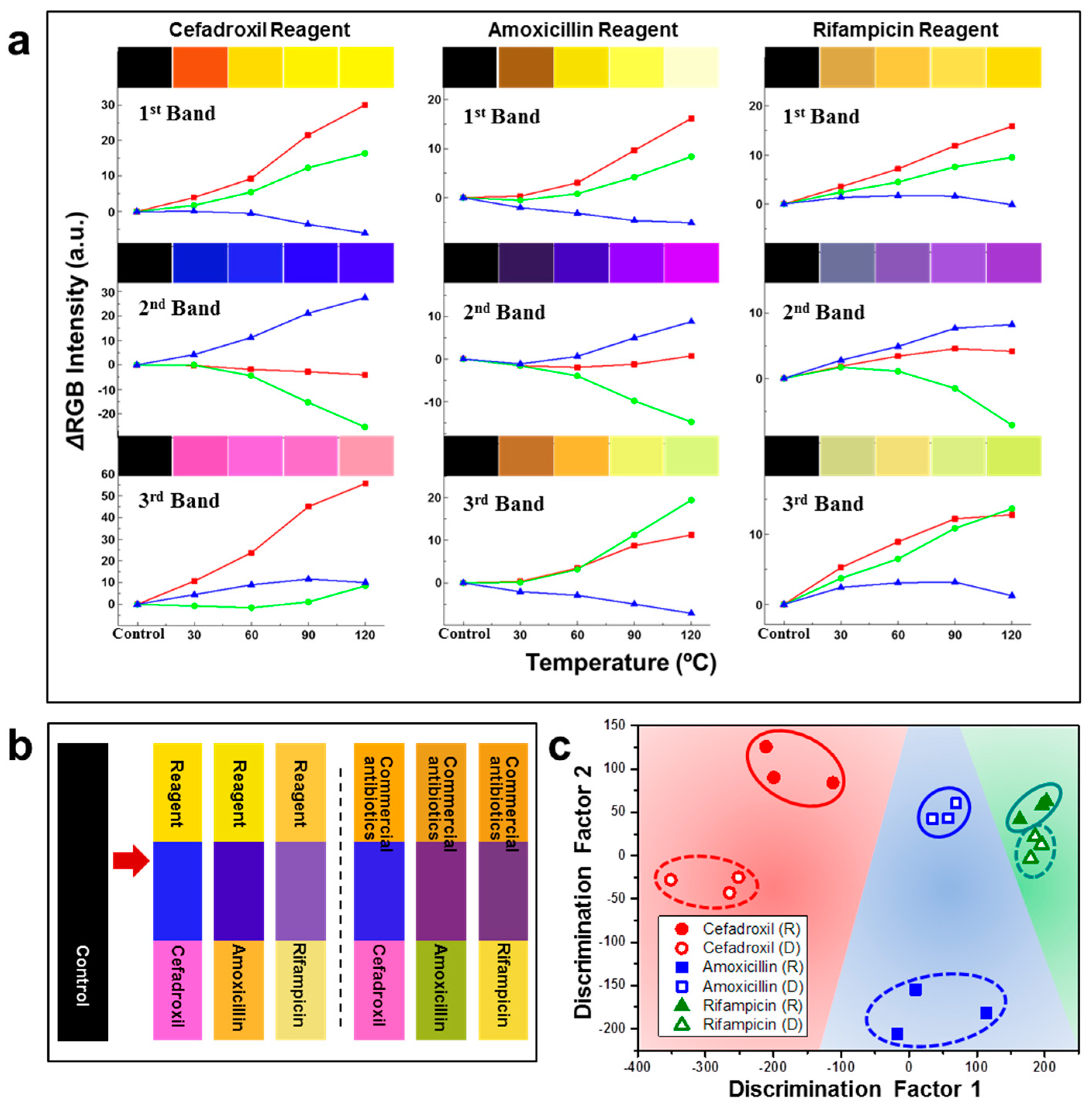
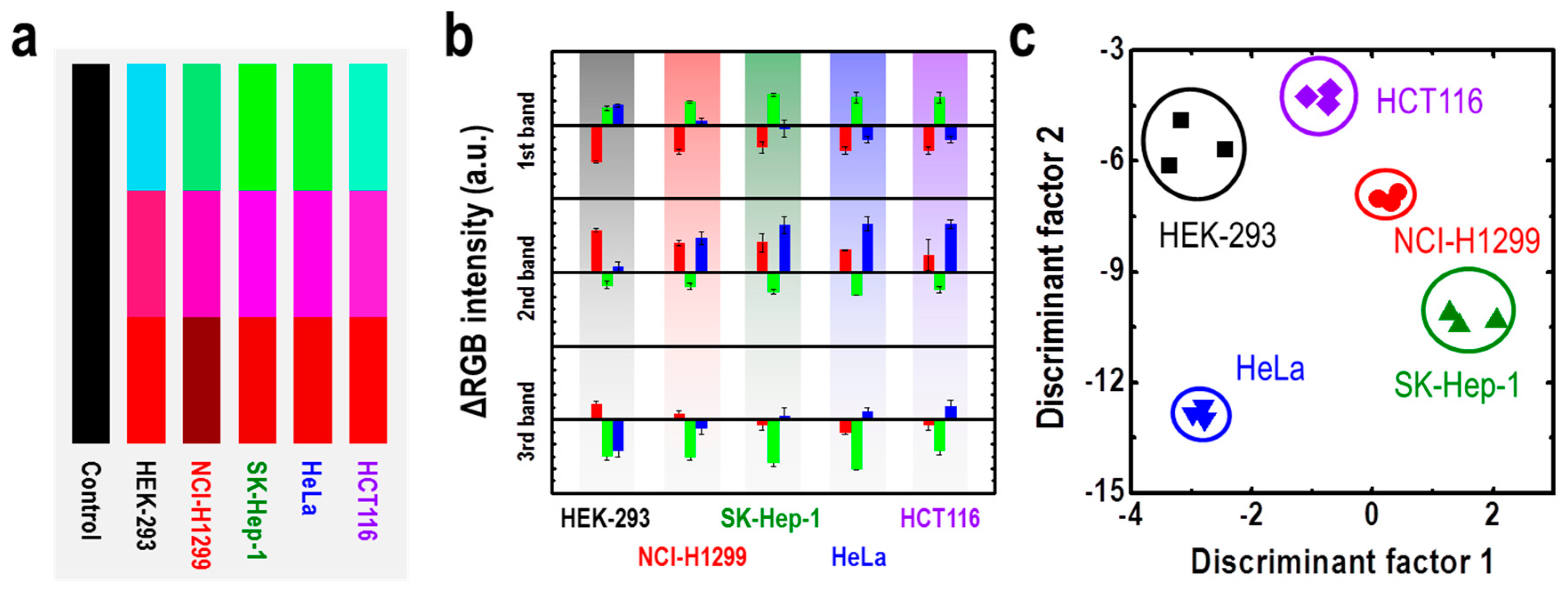
| M13 Bacteriophage Immobilizations | Detection Technique | Analytes | Real Sample | Ref. |
|---|---|---|---|---|
| M13/Au coated Si wafer | Color analysis | Nitrotoluene | TNT, DNT, MNT | [4] |
| M13/donor and acceptor dye | FRET | Intracellular pH | RAW264.7 macrophage | [30] |
| M13/Au coated glass | SPR | Cell proliferation signal | NIH3T3 mouse fibroblast | [31] |
| M13/Au@Ag NPs/Raman dye/DNA | SERS | Antibody concentration | Sterptavidin/anti-goat IgG | [32] |
| M13/zinc phthalocyanine/methyl viologen | Fluorescence image | Breast cancer cells | SKBR-3 cell line | [35] |
| M13/CNF/GCE | Electrochemical | Cysteine | L-Cysteine solution in PBS | [39] |
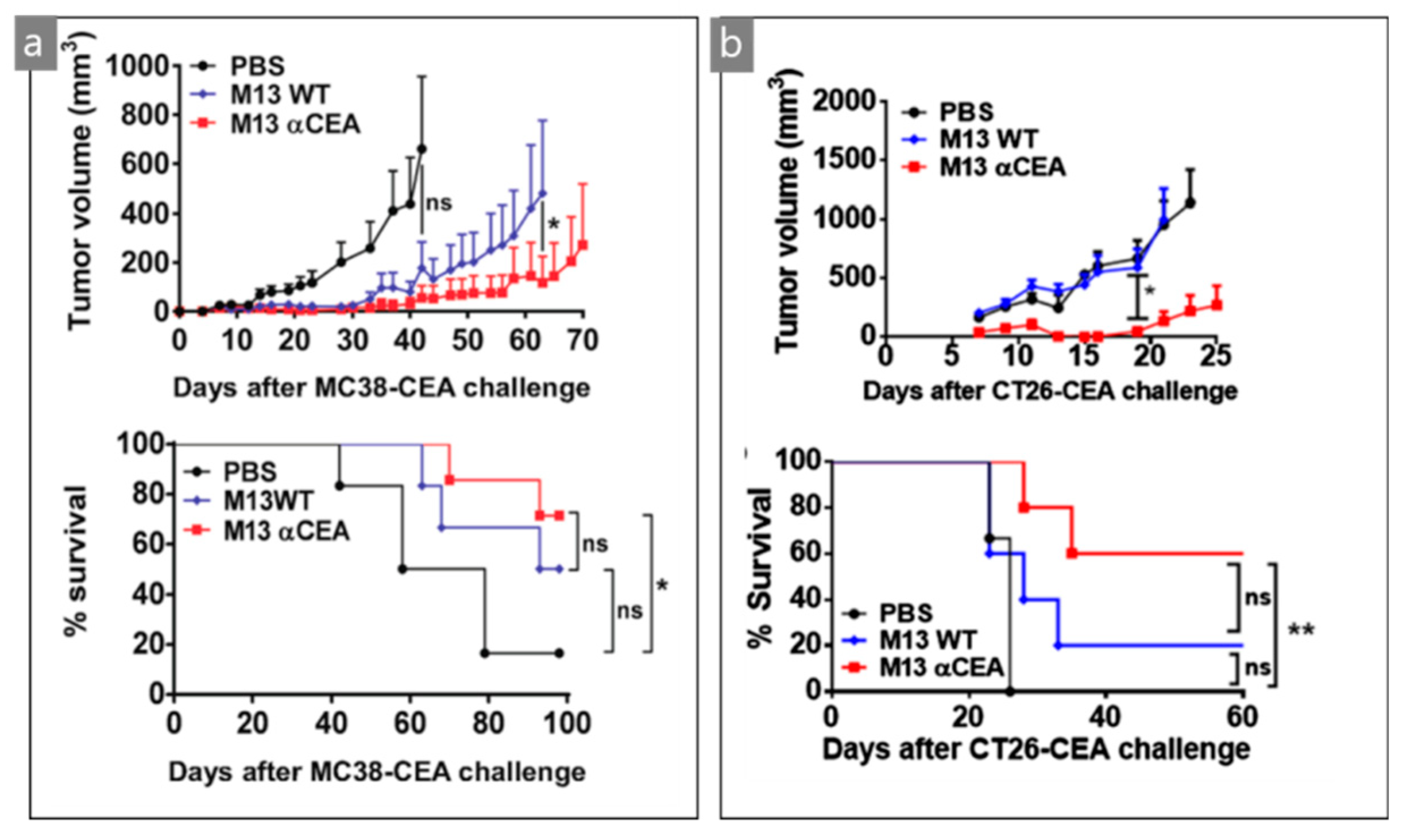
| M13/scFv | Optical/surgical | CEA tumor cells | MC38- and CT26-CEA | [44] |
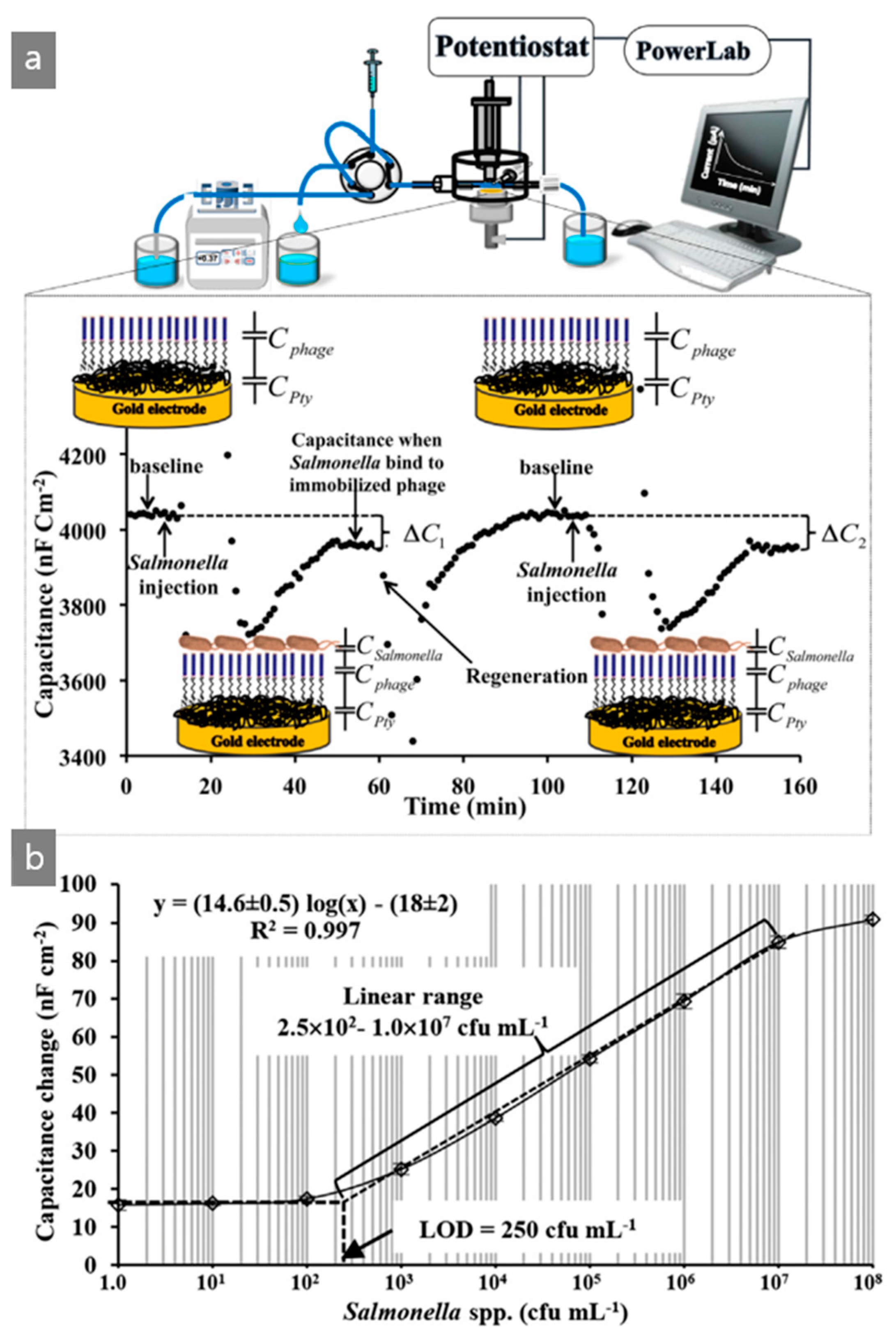
| M13/Pty/Au electrode | Capacitive | Salmonella | Chicken | [48] |
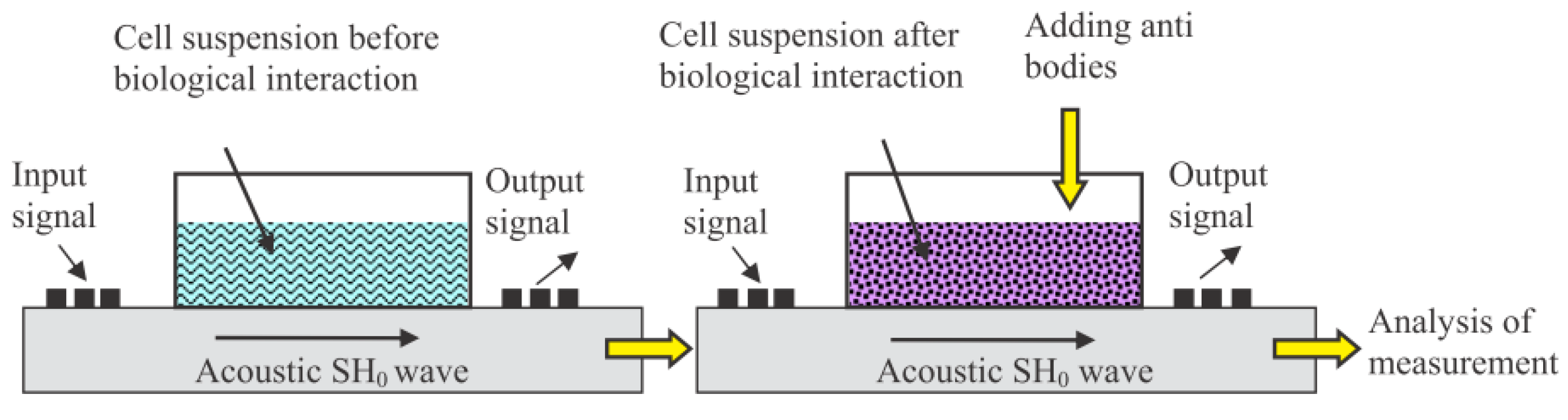
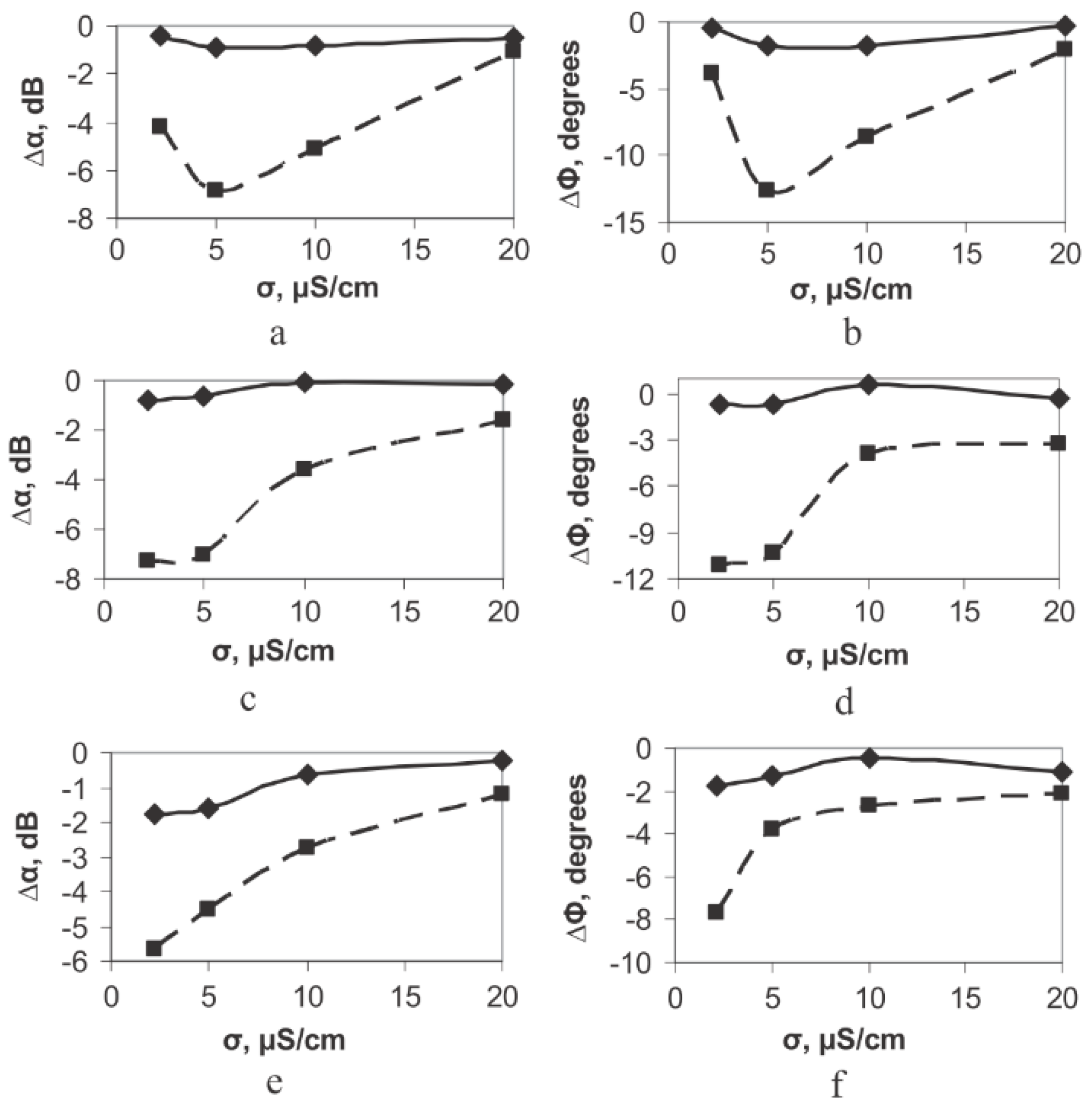
| M13/antibody | Electroacoustic | Cell number | Sp245 cell line | [52] |
| M13/Ag nanowire | SERS | Pesticides | PQ, DQ, and DIF | [60] |
| M13/Au coated Si wafer | Color analysis | Endocrine disrupting chemicals | Phthalate and PCB derivatives | [65] |
| M13/Au coated Si wafer | Color analysis | Antibiotics | Commercial and reagent antibiotics | [69] |
| M13/Au coated Si wafer | Color analysis | Cancer cells | HEK-293, NCI-H1299, SK-Hep-1, HeLa, HCT116 cancer cell lines | [72] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, J.-S.; Choi, E.J.; Jeong, N.-N.; Sohn, J.-R.; Han, D.-W.; Oh, J.-W. Research Progress of M13 Bacteriophage-Based Biosensors. Nanomaterials 2019, 9, 1448. https://doi.org/10.3390/nano9101448
Moon J-S, Choi EJ, Jeong N-N, Sohn J-R, Han D-W, Oh J-W. Research Progress of M13 Bacteriophage-Based Biosensors. Nanomaterials. 2019; 9(10):1448. https://doi.org/10.3390/nano9101448
Chicago/Turabian StyleMoon, Jong-Sik, Eun Jung Choi, Na-Na Jeong, Jong-Ryeul Sohn, Dong-Wook Han, and Jin-Woo Oh. 2019. "Research Progress of M13 Bacteriophage-Based Biosensors" Nanomaterials 9, no. 10: 1448. https://doi.org/10.3390/nano9101448
APA StyleMoon, J.-S., Choi, E. J., Jeong, N.-N., Sohn, J.-R., Han, D.-W., & Oh, J.-W. (2019). Research Progress of M13 Bacteriophage-Based Biosensors. Nanomaterials, 9(10), 1448. https://doi.org/10.3390/nano9101448






