Abstract
Chemotaxonomic profiling based on secondary metabolites offers a reliable approach for identifying and authenticating medicinal plants, addressing limitations associated with traditional morphological and genetic methods. Recent advances in microfluidics and nanoengineered technologies—including lab-on-a-chip systems as well as nano-enabled optical and electrochemical sensors—enable the rapid, accurate, and portable detection of key metabolites, such as alkaloids, flavonoids, terpenoids, and phenolics. Integrating artificial intelligence and machine learning techniques further enhances the analytical capabilities of these technologies, enabling automated, precise plant identification in field-based applications. Therefore, this review aims to highlight the potential applications of micro- and nanoengineered devices in herbal medicine markets, medicinal plant authentication, and biodiversity conservation. We discuss strategies to address current challenges, such as biocompatibility and material toxicity, technical limitations in device miniaturization, and regulatory and standardization requirements. Furthermore, we outline future trends and innovations necessary to fully realize the transformative potential of these technologies in real-world chemotaxonomic applications.
1. Introduction
Chemotaxonomy—the classification of plants based on their chemical constituents—is an essential tool for accurately identifying medicinal plants [1]. This approach addresses key limitations associated with traditional morphological and genetic methods [2,3]. Traditional morphological classification is often challenged by phenotypic plasticity and environmental viability, leading to potential misidentification. Although genetic methods offer high precision, they are often expensive, time-consuming, and not well-suited for rapid, on-site analysis [4]. Consequently, research has increasingly focused on secondary metabolites—such as alkaloids, flavonoids, terpenoids, and phenolics—which serve as highly reliable chemical markers owing to their unique, species-specific, and relatively stable profiles [5,6]. These metabolites play essential roles in plant defense mechanisms and exhibit significant therapeutic properties, further highlighting their importance in identifying and authenticating medicinal plants [7,8].
Despite their analytical strengths, conventional methods for metabolite profiling—including high-performance liquid chromatography (HPLC), liquid chromatography–mass spectrometry (LC-MS), and nuclear magnetic resonance (NMR)—are limited by their lack of portability, speed, and user-friendliness in field-based contexts [9,10]. These methods typically require complex laboratory setups, expensive instrumentation, and extensive sample preparation, making them impractical for real-time, on-site applications, especially in resource-limited settings. Consequently, demand continues to rise for faster, more accurate, and portable analytical techniques capable of delivering rapid and reliable chemotaxonomic profiling directly in the field, where immediate results are critical for effective decision making in herbal medicine, biodiversity conservation, and detection of adulterated herbal products [11,12].
Recent advancements in microfluidics, lab-on-a-chip systems, and nanoengineered sensors have provided promising avenues for addressing these traditional limitations [13,14,15]. These miniaturized devices offer rapid, highly sensitive, and selective detection of secondary metabolites at reduced costs while requiring significantly reduced amounts of samples and reagents. Furthermore, integrating optical and electrochemical detection methods—enhanced by nanomaterials, such as gold nanoparticles, nanowires, and nanostructured surfaces—has significantly enhanced analytical sensitivity, facilitating trace-level detection of metabolites in complex plant extracts [16,17]. When coupled with artificial intelligence (AI) techniques—including machine learning and data fusion algorithms that integrate metabolomic, genomic, and environmental datasets—these devices enable sophisticated real-time analysis, automated classification, and enhanced accuracy in plant identification [18,19]. Despite their potential, the widespread adoption of these innovative technologies faces several challenges [20,21]. Key challenges—including concerns over the biocompatibility and potential toxicity of nanomaterials, technical limitations in maintaining high analytical throughput and sensitivity during miniaturization, as well as the absence of comprehensive regulatory guidelines and international standards—must be systematically addressed [22,23]. Overcoming these obstacles requires ongoing interdisciplinary collaboration among researchers, technologists, regulatory bodies, and industry stakeholders to ensure these innovative technologies achieve widespread adoption and effectiveness in practical applications.
This review aims to provide a comprehensive analysis of recent advances in micro- and nanoengineered devices for the chemotaxonomic profiling of medicinal plants. This review is distinct from earlier works by its focus on the integration of AI-based data analysis with micro- and nanoengineered devices, enhancing data processing capabilities for real-time, field-based plant identification. The review is organized into three main sections: (1) device types, (2) detection methods, and (3) applications. The device types section discusses the various nano-enabled devices used for plant metabolite analysis, such as microfluidic platforms, lab-on-a-chip systems, and nano-engineered sensors. The detection methods section focuses on the integration of optical and electrochemical sensors, enhanced by nanomaterials, to improve sensitivity and specificity in detecting secondary metabolites. The application of these technologies for field-based applications in herbal medicine, plant authentication, and biodiversity conservation is also emphasized, setting this review apart from those focused mainly on laboratory applications. Finally, this review addresses challenges related to portability, sensitivity, and cost-effectiveness and discusses strategies to overcome these barriers.
2. Target Metabolites and Analytical Needs
Analyzing plant metabolites is essential for classifying medicinal plants through chemotaxonomy. This approach helps identify and distinguish plant species based on their chemical composition [24]. Medicinal plants are rich in diverse secondary metabolites, each contributing to their biological functions and therapeutic potential. Compounds such as alkaloids, flavonoids, terpenoids, and phenolics are common markers used to classify plants into specific families or genera [25]. The chemical composition of these secondary metabolites is important not only for identifying the plant species but also for understanding their medicinal potential, as many of these compounds exhibit well-documented pharmacological activities [26]. As the need for fast and accurate plant identification grows, efficient profiling of metabolites has become increasingly important in areas such as pharmacognosy, biodiversity research, and herbal medicine development [26]. Traditional plant classification methods, which rely on morphological traits or genetic markers, have inherent limitations, especially when plants exhibit similar physical characteristics or require rapid identification [27]. Therefore, profiling these key metabolites using advanced analytical techniques has become an integral part of modern chemotaxonomy.
2.1. Overview of Major Phytochemicals in Medicinal Plants
Medicinal plants contain various secondary metabolites that serve as key indicators of their pharmacological potential and their relevance in chemotaxonomy. Among the most significant groups are alkaloids, flavonoids, terpenoids, and phenolics [28,29]. Alkaloids are nitrogen-containing compounds with strong pharmacological effects. For example, morphine from the opium poppy (Papaver somniferum) is used for pain relief, and quinine from Cinchona bark is effective against malaria [30,31,32,33]. These compounds frequently form the foundation of traditional medicines and modern pharmaceuticals, highlighting their importance in chemotaxonomic classification of plants. Flavonoids are common in many medicinal plants and are well-known for their antioxidant, anti-inflammatory, and anticancer properties. Flavonoids typically consist of two aromatic rings connected by a three-carbon chain, and they contribute to the coloration, flavor, and therapeutic properties of numerous plants [34,35]. Terpenoids represent the largest and most diverse group of plant metabolites. These compounds include essential oils, cannabinoids, and steroids, all of which exhibit therapeutic properties, such as antimicrobial, anti-inflammatory, and anticancer effects [36,37]. Furthermore, phenolic compounds, including tannins and lignans, are primarily responsible for the antioxidant and anti-inflammatory properties of many plants. They also play a key role in the defense mechanisms of the plant against environmental stressors [38,39].
The chemical structures of these metabolites are highly diverse, with each structure closely associated with its biological activity. Alkaloids contain nitrogen atoms within a heterocyclic ring and are often known for their potent effects on the human nervous system (Figure 1). Flavonoids have a basic 15-carbon skeleton and are further classified into various subgroups, such as flavones, flavonols, and isoflavones, based on their structural variations [40] (Figure 2). Their role in protecting plants from oxidative damage and their potential health benefits in humans have been extensively studied. Terpenoids are made up of isoprene units and show a wide range of structural complexity. They include simple monoterpenes like limonene and more complex compounds such as cannabinoids, which demonstrate various pharmacological effects [41,42] (Figure 3). Phenolic compounds consist of a hydroxyl group attached to an aromatic ring and include important subgroups like flavonoids, tannins, and lignans. These compounds contribute to several biological activities, including antimicrobial and anticancer effects [43,44] (Figure 4). Understanding the structural diversity of these compounds is essential for their application in chemotaxonomy, as they offer distinct chemical markers for differentiating plant species. Figure 5 illustrates a visual representation of the major groups of phytochemicals found in medicinal plants, showing types and examples from commonly used medicinal plants.
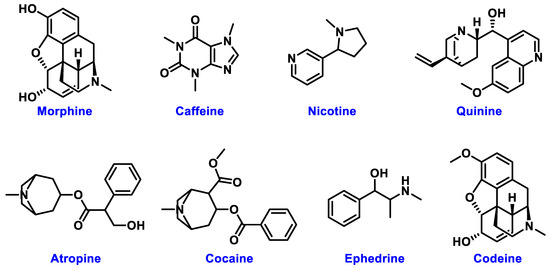
Figure 1.
Structural Representation of Common Plant Alkaloids.
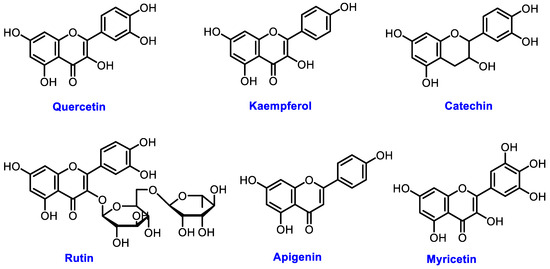
Figure 2.
Structural representation of common plant flavonoids.
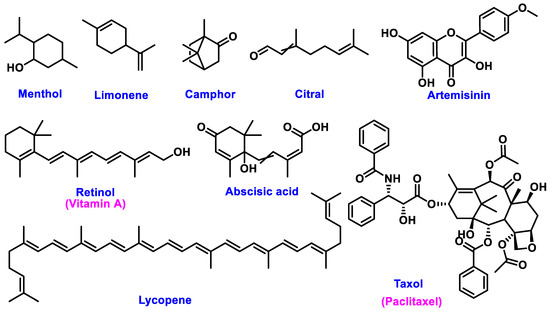
Figure 3.
Structural Representation of Common Plant Terpenoids.
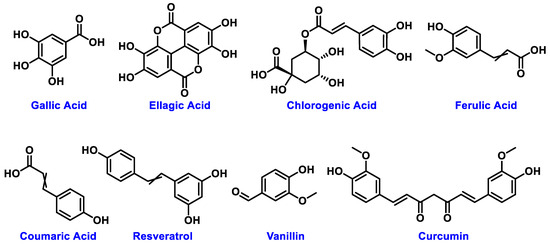
Figure 4.
Structural representation of common plant phenolics.
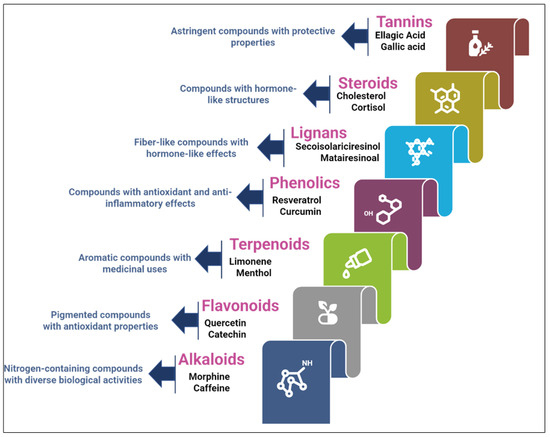
Figure 5.
Representation of major phytochemical groups in medicinal plants.
2.2. Challenges in Traditional Analytical Techniques
Despite the significance of secondary metabolites in plant classification and identification, traditional analytical methods, such as HPLC, LC-MS, and NMR, exhibit significant limitations, particularly in field-based or high-throughput applications [45,46,47]. HPLC remains a standard method for separating and identifying plant metabolites [48]. However, it is resource-intensive and requires advanced instruments, specialized reagents, and a stable power supply. These requirements pose challenges in remote areas where medicinal plants are often harvested [48,49]. LC-MS offers higher sensitivity and can analyze complex mixtures, but it also demands costly equipment and trained personnel, making it unsuitable for quick, on-site identification [49]. NMR, although highly effective for structural elucidation of metabolites, requires large sample quantities and expensive instrumentation. These limitations reduce its utility in field-based applications or real-time, high-throughput analyses [45,50].
Although traditional methods are highly effective in laboratory settings, they are not easily adapted for portable or on-site use. This creates significant challenges for applications in settings like herbal medicine markets or biodiversity conservation. As a result, there is an urgent need for compact and portable devices capable of performing rapid and accurate chemotaxonomic profiling without reliance on complex laboratory infrastructure. Emerging technologies such as lab-on-a-chip systems, microfluidics platforms, and portable spectroscopic tools offer promising solutions to address this challenge [51,52]. These miniaturized systems facilitate on-site analysis with precision comparable to those of traditional methods while offering advantages such as portability, low power consumption, and cost-effectiveness. By enabling chemical-profile-based identification, they can ensure accurate plant species identification even in remote locations [53]. Integrating AI and machine learning algorithms further enhances these systems by enabling the rapid and accurate processing of large, complex datasets. This advancement supports the development of real-time, high-throughput applications [54,55]. Consequently, developing portable and efficient analytical devices is essential to addressing the limitations of traditional techniques, enabling chemotaxonomic profiling to be conducted rapidly, efficiently, and accurately in field settings.
3. Microfluidics and Lab-on-a-Chip Platforms
Microfluidics and lab-on-a-chip platforms have transformed plant metabolite analysis by integrating sample preparation, separation, and detection into a single device. These miniaturized systems manipulate small fluid volumes through microchannels, offering significant advantages in speed, portability, and cost compared to those of traditional methods [13,52]. By consolidating multiple analytical steps on a single chip, microfluidics enables high-throughput, rapid, and on-site profiling of medicinal plants based on their chemical composition—which is crucial for chemotaxonomy [56]. The portability of these devices also makes them ideal for real-time, field-based plant identification and classification in remote areas.
3.1. Introduction to Microfluidic Systems
Microfluidic systems are designed to manipulate fluids at the microliter or nanoliter scale using microchannels that precisely control liquid movement. These systems rely on principles such as capillary action and pressure-driven flow to manage fluid dynamics within small-scale channels, often smaller than the width of a human hair [57,58]. A key advantage of microfluidics is its ability to integrate multiple laboratory functions—mixing, reaction, separation, and detection—into a single compact device. This integration enables high-throughput analysis with minimal sample and reagent consumption, making it highly efficient for plant metabolite analysis. The development of microfluidic systems has significantly affected fields such as clinical diagnostics, environmental monitoring, and chemotaxonomy, where rapid, cost-effective, and portable analytical techniques are essential.
Miniaturization in microfluidics not only reduces analysis costs but also enables real-time monitoring and rapid processing of large numbers of plant samples. This is particularly valuable in chemotaxonomy, as it enables the rapid screening of multiple plant species based on their secondary metabolite profiles [14,59]. However, metabolite levels in plants can vary significantly throughout their life cycle due to growth stages, environmental conditions, and stress responses. This variability presents a challenge when using lab-on-a-chip systems, which are primarily designed for liquid samples. To overcome this, sample preparation methods such as liquid extraction, pre-concentration, and effective extraction techniques are necessary to ensure metabolites are present at appropriate concentrations for analysis. Furthermore, due to the structural similarities of metabolites such as alkaloids, flavonoids, and terpenoids, a single detection method may not be sufficient to discriminate and quantify them. To address this, multi-detection systems that combine fluorescence, electrochemical, and SERS techniques within a single platform are proposed. These combined methods can enhance sensitivity and specificity, improving the ability to distinguish between similar compounds. The integration of artificial intelligence (AI) for data fusion across these different detection techniques also improves the accuracy and efficiency of the plant metabolite profiling. These portable systems are especially well-suited for field applications, where traditional methods, such as HPLC or LC-MS, are impractical owing to their size, cost, and complexity (Figure 6).
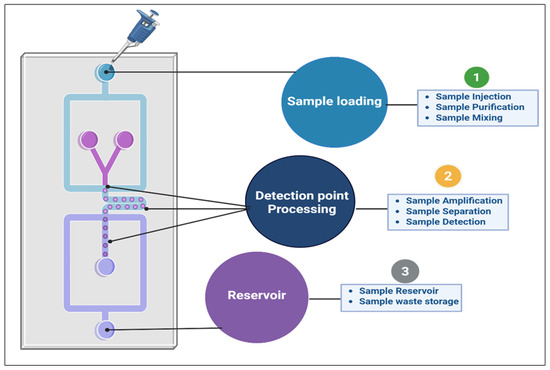
Figure 6.
Schematic diagram of a lab-on-a-chip device for phytochemical profiling.
3.2. Fabrication Techniques for Lab-on-a-Chip Devices
Lab-on-a-chip devices are commonly fabricated using materials such as polydimethylsiloxane (PDMS), which is preferred for its ease of use, optical transparency, and biocompatibility. PDMS-based devices are often produced using soft lithography, a rapid prototyping technique for microfluidic devices [60]. This process involves creating a mold using a photomask and then pouring PDMS over it to form microchannels, which can be used for separating and analyzing plant metabolites. While soft lithography is cost-effective and versatile, it has limitations, such as the potential absorption of certain small molecules and challenges in scaling up production [61,62]. An emerging fabrication method is three-dimensional printing, which enables the creation of custom-designed microfluidic devices by layering materials to build up the device structure of the device. This technique offers greater flexibility in design but may suffer from lower resolution than that of the traditional methods. Both fabrication techniques offer distinct advantages and limitations, depending on the specific requirements of chemotaxonomic profiling applications [63].
3.3. Integrating Detection Methods in Lab-on-a-Chip Systems
Lab-on-a-chip devices combine multiple detection methods for analyzing plant metabolites, making them versatile tools in chemotaxonomy (Table 1). Optical detection methods, such as fluorescence and absorbance, are widely used owing to their sensitivity and seamless integration with microfluidic systems. Fluorescence detection, for instance, is highly effective for identifying low-abundance metabolites by utilizing fluorescent tags or the natural fluorescence of the compounds [64]. Absorbance detection, typically used for compounds that absorb ultraviolet or visible light, provides reliable results for many plant metabolites, including phenolics and flavonoids. Electrochemical detection methods, in contrast, measure the changes in electrical properties caused by the oxidation or reduction of metabolites, offering high selectivity and sensitivity. These electrochemical sensors are particularly effective for detecting specific plant metabolites, such as alkaloids and terpenoids [65,66].
Real-world case studies reveal the successful integration of optical and electrochemical detection in lab-on-a-chip devices for metabolite analysis. For example, fluorescence-based lab-on-a-chip devices have been used by researchers to identify and quantify flavonoids in various plant species, while electrochemical sensors are employed to detect alkaloids in Cinchona bark [67]. These technologies offer rapid, low-cost, and portable solutions for field-based plant identification, which are crucial for herbal medicine markets, conservation efforts, and biodiversity monitoring.

Table 1.
Comparison of lab-on-a-chip platforms for phytochemical analysis.
Table 1.
Comparison of lab-on-a-chip platforms for phytochemical analysis.
| Detection Technique | Advantages | Limitations | Real-World Case Study | Examples of Plant Metabolites Detected |
|---|---|---|---|---|
| Fluorescence detection | High sensitivity for low-abundance metabolites. Non-invasive, rapid, and real-time detection. Easily integrated with microfluidic systems. | Requires fluorescent tagging or natural fluorescence. May not be applicable to all plant metabolites. | Fluorescence-based lab-on-a-chip devices are employed to detect and quantify flavonoids in plant species, such as Citrus and Ginkgo biloba, aiding in the identifying secondary metabolites in medicinal plants [68]. | Flavonoids, phenolic acids, and anthocyanins |
| Absorbance detection | Simple, cost-effective, and widely used. Applicable to UV or visible light-absorbing compounds. | Lower sensitivity than that of fluorescence. May require extensive sample preparation for complex mixtures. | Commonly used to analyze phenolics and flavonoids, especially in agricultural and food safety applications. For example, used for polyphenol analysis in tea and grape samples [69]. | Phenolic compounds, flavonoids, and tannins |
| Electrochemical detection | High selectivity and sensitivity for low concentrations. Suitable for real-time monitoring. Highly specific for certain compounds (such as alkaloids and terpenoids). | Requires specialized electrodes and systems. Limited to metabolites that can undergo redox reactions. Potential interference from other electroactive substances. | Electrochemical sensors integrated into lab-on-a-chip devices have been employed to detect alkaloids in Cinchona bark (for quinine) and terpenoids in aromatic plants such as lavender and peppermint. These sensors are particularly useful in herbal medicine research and conservation [70,71] | Alkaloids, terpenoids, cinchonine, and quinine |
| SPR | Provides real-time detection without labeling. Sensitive to changes in refractive index near the sensor surface. Non-destructive to samples. | Sensitive to surface conditions and requires highly specialized equipment. | Used in lab-on-a-chip devices to detect polyphenols and flavonoids by measuring refractive index changes at the sensor surface, often used for profiling complex plant mixtures [72]. | Polyphenols, flavonoids, and antioxidants |
| CE | High resolution, fast, and effective for separating various metabolites. Can be combined with detection methods (UV, fluorescence, and electrochemical). | Requires more complex sample preparation and sophisticated equipment. May not be suitable for large-scale screening. | Used for separating and quantifying carotenoids and fatty acids in various plant extracts, especially in food quality control and metabolite profiling [73,74]. | Carotenoids, fatty acids, and lipids |
SPR, surface plasmon resonance; CE, capillary electrophoresis; UV, ultraviolet.
4. Nano-Enabled Optical and Electrochemical Sensors
Nano-enabled optical and electrochemical sensors significantly enhance plant metabolite analysis by utilizing the distinct properties of nanomaterials, such as nanoparticles, nanowires, and nanostructured surfaces [75,76]. These materials enhance sensitivity and selectivity, which are crucial for detecting low concentrations of metabolites, such as alkaloids, flavonoids, terpenoids, and phenolics. In optical methods such as surface-enhanced Raman spectroscopy (SERS), nanomaterials enhance performance by increasing surface area and enabling more efficient molecular interactions [77,78]. These nano-enabled sensors are typically portable, cost-effective, and well-suited for on-site applications. This makes them valuable tools in chemotaxonomy and medicinal plant identification, especially in field-based or high-throughput applications.
4.1. Introducing Nano-Enabled Sensors
Nano-enabled sensors function based on optical and electrochemical detection principles, where nanomaterials enhance the sensitivity and specificity of traditional methods (Table 2). In optical sensors, such as those based on fluorescence or absorbance, materials like gold nanoparticles, silver nanoparticles, and quantum dots amplify optical signals. This amplification makes it possible to detect plant metabolites at extremely low concentrations. For instance, gold nanoparticles are widely used in SERS. They significantly amplify Raman signals, enabling highly sensitive detection of trace compounds [79,80]. Electrochemical sensors, in contrast, measure current or voltage changes caused by redox reactions between metabolites and the sensor interface. Nanomaterials such as carbon nanotubes and gold nanowires are often employed to improve conductivity and signal transduction. These enhancements allow electrochemical sensors to detect specific plant metabolites with high precision. The progress in nanotechnology enables rapid, on-site analysis of plant metabolites, especially benefiting chemotaxonomy and identifying medicinal plants.

Table 2.
Types of nano-enabled sensors for medicinal plant profiling.
4.2. Surface-Enhanced Raman Spectroscopy for Plant Metabolites
SERS utilizes nanomaterials, typically metallic nanoparticles such as gold or silver, to enhance the Raman signal of plant metabolites. This enhancement arises from localized surface plasmon resonance, which occurs when metabolites interact with nanostructures, such as gold nanoparticle surfaces. The resulting signal amplification enables highly sensitive detection of low-abundance compounds within complex plant extracts [92]. SERS is particularly effective for identifying secondary metabolites, such as terpenoids, alkaloids, and phenolics, which are vital in plant chemotaxonomy. For example, SERS can be used to detect terpenoids in essential oils or alkaloids in plant extracts, providing valuable chemical markers for plant identification. Additionally, SERS requires minimal sample preparation, making it an efficient and non-destructive technique for real-time plant metabolite profiling [93,94]. Figure 7 illustrates the operating principle of a SERS-based nanosensor for plant metabolite detection, showing how the enhanced Raman signal enables compound-specific identification.
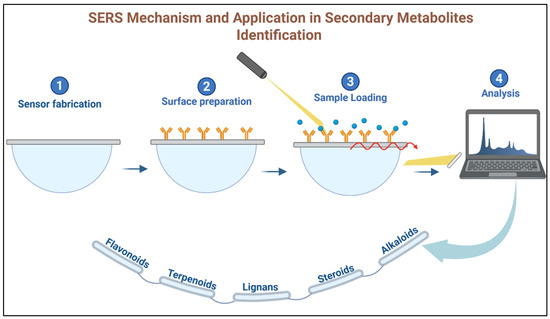
Figure 7.
SERS mechanism and application in secondary metabolites identification.
4.3. Field-Effect Transistor FET-Based Sensors for Phytochemical Detection
FET-based sensors are highly effective in detecting plant metabolites because they measure electrical changes that occur when metabolites interact with the sensor surface [95]. These sensors detect variations in current or voltage when specific molecules bind to the active surface of the sensor. Integrating nanomaterials, such as carbon nanotubes and gold nanowires, significantly enhances the sensitivity and selectivity of FETs for plant metabolites. Carbon nanotubes, for instance, offer excellent conductivity and a large surface area for metabolite adsorption, making FET-based sensors particularly efficient for detecting trace metabolites in plant extracts. FET-based sensors offer several advantages, including high sensitivity, miniaturization, and real-time operation, which are essential for on-site applications. They are particularly useful for detecting key metabolites, such as alkaloids and terpenoids, which are crucial for chemotaxonomy. Furthermore, their integration with microfluidic devices enables high-throughput analysis, making them valuable tools for large-scale plant metabolite profiling in the field [95].
5. Integration with Artificial Intelligence and Data Processing
Integrating AI and data processing with chemotaxonomy offers a transformative approach to plant identification [96]. Machine learning algorithms and data fusion techniques enable AI to analyze complex plant metabolic profiles alongside genomic and environmental data [97,98]. This integration enables rapid, accurate, and automated plant identification, enhancing the functionality of micro- and nanoengineered devices. AI-driven platforms can process vast amounts of data efficiently, offering high-throughput analysis vital for applications such as biodiversity conservation, herbal medicine, and counterfeit detection [98]. With ongoing advancements, AI systems are becoming increasingly capable of real-time, field-based plant identification, making the process more efficient and accessible than ever before.
5.1. Role of Machine Learning in Chemotaxonomy
Machine learning algorithms have become essential tools in chemotaxonomy, providing powerful methods to analyze and classify plants based on their metabolic profiles (Table 3). Algorithms such as neural networks, support vector machines (SVM), and decision trees are increasingly used to interpret the complex chemical data generated by advanced analytical techniques like HPLC, LC-MS, and SERS. These algorithms can be trained to recognize patterns in the chemical profiles of plant metabolites, allowing for accurate identification and classification of plant species [99]. Machine learning models are particularly effective at identifying non-linear relationships between metabolites, making them ideal for complex datasets with overlapping or intricate chemical signatures. Neural networks, in particular, are well-suited for handling large datasets and identifying non-linear relationships between metabolites, making them ideal for chemotaxonomy applications. Support vector machines, on the other hand, excel in classification tasks, especially when dealing with high-dimensional data such as plant metabolite profiles. By integrating these machine learning techniques, AI can significantly enhance the speed and accuracy of plant species identification, even with complex or overlapping chemical signatures [100,101].

Table 3.
AI algorithms used for chemotaxonomic profiling.
5.2. Data Fusion Techniques: Integrating Metabolomics and Genomics
Data fusion techniques integrate metabolomics with genomic and environmental data to enhance plant species identification (Figure 8). This integration enables more comprehensive insights into plant characteristics by leveraging their chemical and genetic profiles. For instance, combining metabolomic data with genomic markers enhances plant identification reliability by offering phenotypic and genotypic data points. This approach is particularly valuable when traditional morphological traits are insufficient to distinguish closely related species [114,115]. Incorporating environmental variables, such as soil conditions or climatic factors, allows AI systems to account for ecological variations that might influence metabolite profiles. These data fusion techniques enable the development of automated plant identification systems capable of processing and analyzing large datasets, thereby improving the efficiency and accessibility of plant classification.
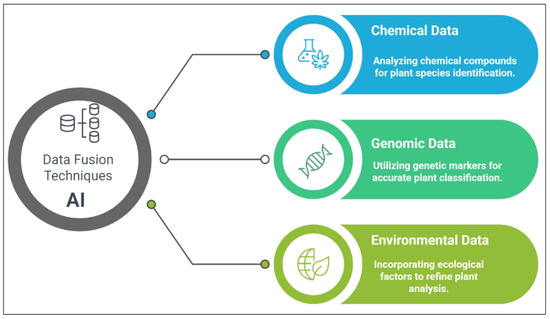
Figure 8.
Data fusion techniques for plant identification.
5.3. Real-Time Data Processing for Field-Based Applications
Integrating AI with micro- and nanoengineered devices enables real-time, on-site data processing for field-based plant identification. These devices, often coupled with portable sensors and lab-on-a-chip platforms, can rapidly analyze plant samples based on their chemical profiles and deliver immediate identification results. Real-time data processing is particularly beneficial in remote areas with limited laboratory access, facilitating rapid, accurate in situ classification [116]. Additionally, developing portable, AI-driven plant identification platforms holds significant potential for enhancing the ability to monitor and conserve plant biodiversity, detect counterfeit medicinal products, and promote the sustainable application of plant resources. The potential for AI to process large datasets in real time and deliver immediate feedback is transforming the application of chemotaxonomy in research and field settings [117,118].
6. Applications and Case Studies
Nanoengineered devices have transformed plant metabolite profiling, with significant applications in herbal medicine, medicinal plant authentication, and biodiversity conservation. These technologies enable rapid, sensitive, and portable real-time plant identification, supporting the authenticity of herbal products and protecting endangered species [119]. Nano-enabled sensors, such as those based on SERS and FET technologies, have demonstrated high accuracy in plant species identification and adulteration detection, enhancing quality control and enabling efficient biodiversity monitoring [120].
6.1. Applications in Herbal Medicine
Nanoengineered devices are transforming herbal medicine by enabling precise detection of counterfeit herbal products and preventing species adulteration (Table 4). Their ability to detect subtle molecular differences is crucial for verifying the authenticity of herbal medicines [121,122]. By leveraging nanoscale technologies, these devices can distinguish between genuine herbal products from counterfeit or adulterated alternatives, ensuring the safety and efficacy of medicinal plant-based treatments. Given that counterfeit herbal products and species adulteration pose significant risks to public health, accurate detection methods are crucial. Nanoengineered devices enable molecular-level analysis of herbal products by detecting unique chemical signatures specific to each plant species. These devices, such as nanofluidic biosensors and molecular imaging tools, can detect trace levels of adulterants or fraudulent substitutions [123,124]. This rapid, accurate detection preserves the integrity of herbal medicine markets and safeguards consumer health [125]. For instance, Geetha et al. [126] demonstrate the application of nanoengineered devices to detect adulteration in ginseng products. Their study applied nanoparticle-based sensors to identify adulterants by comparing the chemical markers of authentic ginseng with those of counterfeit substitutes, providing a non-invasive, efficient method for quality control in the herbal industry.

Table 4.
Case studies on nanoengineered devices for herbal medicine applications.
6.2. Medicinal Plant Authentication
Authenticating medicinal plants is crucial for consumer safety and species conservation (Figure 9). Nano-enabled devices offer efficient, cost-effective authentication methods. Benedetti et al. [134] demonstrated that SERS-based sensors could differentiate between Ginseng species, successfully distinguishing Panax ginseng from other species, such as Panax quinquefolius, based on their unique chemical profiles [135]. Similarly, Tavakoli et al. [136] employed FET-based sensors to authenticate Withania somnifera (ashwagandha), proving that nanosensors could accurately identify the plant, even within complex mixtures [137]. These studies highlight the crucial role of nano-enabled sensors in medicinal plant authentication and improving regulatory standards in the herbal medicine industry.
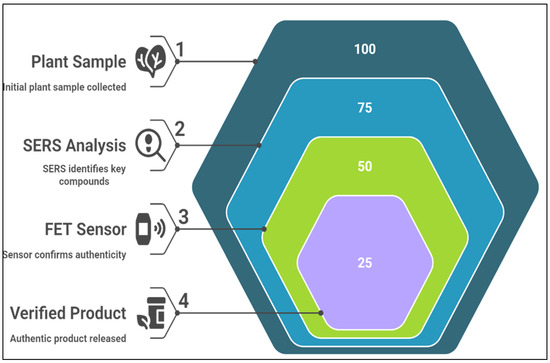
Figure 9.
Process of medicinal plant authentication using nano-enabled devices.
6.3. Biodiversity and Conservation Studies
Nanoengineered devices play a crucial role in biodiversity conservation by monitoring and protecting endangered plant species. Efficient, real-time plant species identification is crucial for assessing population health and directing conservation efforts on the right plants. Zhou et al. employed SERS-based sensors to monitor wild Ginseng populations, enabling non-invasive tracking of this highly valuable medicinal plant and aiding in its conservation [138]. This example highlights the crucial role of nano-enabled devices in real-time biodiversity monitoring and conservation of medicinal plants.
7. Challenges and Future Outlook
Despite significant advancements in micro- and nanoengineered devices for chemotaxonomic profiling, several challenges must be addressed before these technologies can be widely adopted in herbal medicine, plant authentication, and biodiversity conservation [122,139]. Current limitations include biocompatibility, material toxicity, technical reliability, and portability. Future research and development should focus on overcoming these barriers through innovative nanoengineering approaches, device miniaturization, and the integration of multi-sensor systems with mobile platforms. Additionally, addressing regulatory and standardization issues will ensure the broad acceptance and adoption of these technologies in chemotaxonomy and related fields [140].
7.1. Challenges in Current Micro- and Nanoengineered Devices
A key challenge in micro- and nanoengineered devices is ensuring biocompatibility and safety. Numerous nanomaterials, such as metallic nanoparticles or carbon-based nanostructures, may pose toxicity risks or interact adversely with biological systems, affecting human health and environmental safety. Concerns include nanoparticle accumulation, environmental contamination, and potential cytotoxicity, particularly in devices designed for repeated or direct plant contact, necessitating extensive safety testing and careful material selection [141,142].
Another major challenge is achieving consistently high throughput, sensitivity, and portability in nano-enabled analytical tools. Miniaturization often compromises sensitivity, limiting the performance of portable devices compared to those of laboratory-based instruments. Additionally, developing devices that can simultaneously analyze multiple metabolites within complex biological samples remains technically challenging [143]. To fully realize the benefits of nanoengineered chemotaxonomic profiling, future technologies must balance portability, sensitivity, robustness, and cost-effectiveness for effective field deployment.
7.2. Innovations in Nanoengineering for Field-Based Chemotaxonomy
Overcoming current limitations will require continued advancements in nanoengineering, particularly in device miniaturization, enhanced sensitivity, and multi-sensor integration. Emerging innovations include multifunctional “lab-on-a-chip” platforms capable of simultaneously detecting multiple metabolites or genetic markers, enhancing the accuracy and speed of plant identification [144]. Additionally, integrating these sensors with mobile devices such as smartphones or tablets further enables the development of portable, user-friendly, and cost-effective tools for field-based chemotaxonomic analysis [145,146].
Incorporating AI into nano-enabled systems also presents a significant opportunity for innovation. AI-based algorithms can rapidly process complex chemical data, enabling real-time, automated plant species identification even under challenging field conditions [76,147]. These innovations promise to deliver intelligent, miniaturized devices that can transform on-site plant authentication and biodiversity monitoring with broad applications in herbal medicine, agriculture, and conservation efforts.
7.3. Regulatory and Standardization Issues
With the growing application of micro- and nanoengineered devices in chemotaxonomy, addressing regulatory and standardization issues becomes crucial. Currently, regulatory frameworks for nano-based analytical devices are fragmented and often unclear, creating uncertainty for developers, manufacturers, and end-users. Regulatory bodies must establish well-defined guidelines for the safety, performance, and quality assurance of these devices to support commercialization and broad adoption [148,149]. In addition to regulatory issues, scalability remains a significant challenge. Transitioning from laboratory-based systems to large-scale, field-deployed devices presents obstacles related to cost, performance, and robustness. These devices must be miniaturized without compromising on sensitivity, which is essential for real-time, on-site plant identification in diverse environments. Furthermore, field validation is essential to ensure that nano-enabled devices perform reliably under variable environmental conditions such as temperature, humidity, and soil composition. Standardized testing protocols must be established to validate device performance across various environments and ensure consistency. Additionally, establishing globally recognized standards and validation protocols for plant identification using chemotaxonomic profiling is essential to ensure consistency, accuracy, and result comparability across devices and laboratories [150]. Standardized protocols will enhance the reliability of nanoengineered devices and support their acceptance by regulatory bodies, herbal medicine industries, and conservation organizations. This standardization is crucial for safeguarding consumers, advancing biodiversity conservation, and enabling the global adoption of effective nano-enabled plant authentication devices [151].
8. Conclusions
Micro- and nanoengineered devices are transforming chemotaxonomic profiling in medicinal plants by providing rapid and highly sensitive means of plant identification and authentication. Their potential to analyze complex plant metabolite profiles using innovative approaches—such as microfluidics, lab-on-a-chip systems, nano-enabled optical and electrochemical sensors, as well as integration with artificial intelligence—is advancing the development of more accurate, accessible, and efficient chemotaxonomic methodologies. These technologies facilitate real-time, portable, and high-throughput analyses, strengthening the reliability of medicinal plant identification in herbal medicine markets. They detect counterfeit and adulterated products and support biodiversity conservation by accurately identifying and monitoring endangered plant species. However, to fully harness the capabilities of these advanced technologies, several challenges must be systematically addressed. These include biocompatibility concerns, nanomaterial toxicity, and technical limitations in achieving optimal sensitivity, throughput, and balance between miniaturization and performance. Additionally, there is a pressing need for further integration of multi-sensor systems, AI-enabled mobile platforms, and data fusion techniques to improve real-time plant identification, particularly in field conditions. Clinical validation of these devices is also critical to ensure their accuracy and reliability under real-world, diverse environmental conditions. Innovations in nanoengineering, including multi-sensor system integration, enhanced miniaturization, and AI-enabled mobile platforms, present viable solutions. Continued innovations in these areas, along with standardized protocols for data interpretation, will be crucial for the effective deployment of these technologies in real-world applications. Equally crucial is the establishment of clear regulatory guidelines and globally standardized protocols for device validation and operation. Overcoming these barriers through coordinated research, industry collaboration, and regulatory oversight will facilitate the safe and effective adoption of these novel technologies across diverse real-world scenarios. Addressing the technical gaps in scalability, portability, and device integration will be essential to achieving widespread adoption. Continued advancements and interdisciplinary collaboration among scientists, engineers, regulatory agencies, and the herbal medicine industry will be crucial to advancing micro- and nanoengineered devices for accurate medicinal plant identification, public health protection, biodiversity conservation, and ecological sustainability. As these technologies mature and their adoption expands, they are poised to become indispensable tools, ushering in a new era of precision and sustainability in chemotaxonomic research and practice.
Author Contributions
Conceptualization, writing—original draft preparation, resources, software, validation, visualization, A.A., S.A. and W.Z.; writing—review and editing, M.S.A. and W.Z.; supervision, W.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Acknowledgments
During the preparation of this manuscript, the authors used ChatGPT (GPT-4o and GPT-4.5) (developed by OpenAI) to assist in drafting, organizing content, and refining sections of the text to improve clarity and readability. Furthermore, the figures included in this review were created using the following tools: (1) Napkin.ai and Biorender for diagram preparation. (2) Chemdraw.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Marne, P.A.; Pawar, A.T.; Tagalpallewar, A.A.; Baheti, A.M. Comparative Phytochemistry and Chemotaxonomy. In Pharmacognosy and Phytochemistry: Principles, Techniques, and Clinical Applications; Wiley: Hoboken, NJ, USA, 2025; pp. 333–346. [Google Scholar]
- Arceusz, A.; Radecka, I.; Wesołowski, M. Identification of diversity in elements content in medicinal plants belonging to different plant families. Food Chem. 2010, 120, 52–58. [Google Scholar] [CrossRef]
- Singh, R. Chemotaxonomy of medicinal plants: Possibilities and limitations. In Natural Products and Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2018; pp. 119–136. [Google Scholar]
- Wang, M.; Lin, H.; Lin, H.; Du, P.; Zhang, S. From species to varieties: How modern sequencing technologies are shaping Medicinal Plant Identification. Genes 2024, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Qiao, X.; Chen, K.; Wang, Y.; Ji, S.; Feng, J.; Li, K.; Lin, Y.; Ye, M. Biosynthesis-Based Quantitative Analysis of 151 Secondary Metabolites of Licorice To Differentiate Medicinal Glycyrrhiza Species and Their Hybrids. Anal. Chem. 2017, 89, 3146–3153. [Google Scholar] [CrossRef] [PubMed]
- Roaa, M.H. A review article: The importance of the major groups of plants secondary metabolism phenols, alkaloids, and terpenes. Int. J. Res. Appl. Sci. Biotechnol. (IJRASB) 2020, 7, 354–358. [Google Scholar]
- Hao, D.-C.; Gu, X.-J.; Xiao, P. Chemotaxonomy: A Phylogeny-Based Approach; Woodhead Publishing: Cambridge, UK, 2015; pp. 1–48. [Google Scholar]
- Salam, U.; Ullah, S.; Tang, Z.-H.; Elateeq, A.A.; Khan, Y.; Khan, J.; Khan, A.; Ali, S. Plant metabolomics: An overview of the role of primary and secondary metabolites against different environmental stress factors. Life 2023, 13, 706. [Google Scholar] [CrossRef]
- Farag, M.A.; Porzel, A.; Wessjohann, L.A. Comparative metabolite profiling and fingerprinting of medicinal licorice roots using a multiplex approach of GC–MS, LC–MS and 1D NMR techniques. Phytochemistry 2012, 76, 60–72. [Google Scholar] [CrossRef]
- Gashaw, A.D.; Desta, M.A.; Yaya, E.E. A comprehensive review-current development in spectroscopic and chromatographic techniques for natural product analysis. Results Chem. 2025, 16, 102341. [Google Scholar] [CrossRef]
- Porzel, A.; Farag, M.A.; Mülbradt, J.; Wessjohann, L.A. Metabolite profiling and fingerprinting of Hypericum species: A comparison of MS and NMR metabolomics. Metabolomics 2014, 10, 574–588. [Google Scholar] [CrossRef]
- Farag, M.A.; Porzel, A.; Schmidt, J.; Wessjohann, L.A. Metabolite profiling and fingerprinting of commercial cultivars of Humulus lupulus L. (hop): A comparison of MS and NMR methods in metabolomics. Metabolomics 2012, 8, 492–507. [Google Scholar] [CrossRef]
- Crevillen, A.; Pumera, M.; González, M.C.; Escarpa, A. Towards lab-on-a-chip approaches in real analytical domains based on microfluidic chips/electrochemical multi-walled carbon nanotube platforms. Lab A Chip 2009, 9, 346–353. [Google Scholar] [CrossRef]
- Maisch, J.; Kreppenhofer, K.; Büchler, S.; Merle, C.; Sobich, S.; Görling, B.; Luy, B.; Ahrens, R.; Guber, A.E.; Nick, P. Time-resolved NMR metabolomics of plant cells based on a microfluidic chip. J. Plant Physiol. 2016, 200, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Yuan, Y.; Han, Q.; Wang, Y.; Liang, Q. Lab-on-a-chip: An advanced technology for the modernization of traditional Chinese medicine. Chin. Med. 2024, 19, 80. [Google Scholar] [CrossRef] [PubMed]
- Aboulthana, W.M.K.; Refaat, E.; Khaled, S.E.; Ibrahim, N.E.-S.; Youssef, A.M. Metabolite profiling and biological activity assessment of Casuarina equisetifolia bark after incorporating gold nanoparticles. Asian Pac. J. Cancer Prev. APJCP 2022, 23, 3457. [Google Scholar] [CrossRef] [PubMed]
- Kruszka, D.; Selvakesavan, R.K.; Kachlicki, P.; Franklin, G. Untargeted metabolomics analysis reveals the elicitation of important secondary metabolites upon treatment with various metal and metal oxide nanoparticles in Hypericum perforatum L. cell suspension cultures. Ind. Crops Prod. 2022, 178, 114561. [Google Scholar] [CrossRef]
- Cembrowska-Lech, D.; Krzemińska, A.; Miller, T.; Nowakowska, A.; Adamski, C.; Radaczyńska, M.; Mikiciuk, G.; Mikiciuk, M. An integrated multi-omics and artificial intelligence framework for advance plant phenotyping in horticulture. Biology 2023, 12, 1298. [Google Scholar] [CrossRef]
- Varghese, R.; Shringi, H.; Efferth, T.; Ramamoorthy, S. Artificial intelligence driven approaches in phytochemical research: Trends and prospects. Phytochem. Rev. 2025. [Google Scholar] [CrossRef]
- Labroo, P.; Cui, Y. Graphene nano-ink biosensor arrays on a microfluidic paper for multiplexed detection of metabolites. Anal. Chim. Acta 2014, 813, 90–96. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, Y.; Jiang, S.; Sun, X.; Wang, X.; Wang, Z.; Wu, Y.; Wu, J.; Li, Y. A breakthrough in phytochemical profiling: Ultra-sensitive surface-enhanced Raman spectroscopy platform for detecting bioactive components in medicinal and edible plants. Mikrochim. Acta 2024, 191, 286. [Google Scholar] [CrossRef]
- Babamiri, B.; Sadri, R.; Farrokhnia, M.; Hassani, M.; Kaur, M.; Roberts, E.; Ashani, M.M.; Nezhad, A.S. Molecularly Imprinted Polymer Biosensor Based on Nitrogen-Doped Electrochemically Exfoliated Graphene/Ti3 CNTX MXene Nanocomposite for Metabolites Detection. ACS Appl. Mater. Interfaces 2024, 16, 27714–27727. [Google Scholar] [CrossRef]
- Kumar, A.; Jain, D.; Bahuguna, J.; Bhaiyya, M.; Dubey, S.; Javed, A.; Goel, S. Machine learning assisted and smartphone integrated homogeneous electrochemiluminescence biosensor platform for sample to answer detection of various human metabolites. Biosens. Bioelectron. 2023, 238, 115582. [Google Scholar] [CrossRef]
- Palacio, E.-D.; Iaz-Navarro, C.; Algieri, F.; Iguez-Cabezas, E.R.; Genilloud, O.; Vicente, F. Metabolomic analysis of Lavandula dentata L. and Lavandula stoechas L. extracts by LC-QTOF/MS experiments and multivariate analysis techniques as a chemotaxonomical tool. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2020, 154, 231–240. [Google Scholar]
- Zhao, S.-Y.; Liu, Z.-L.; Shu, Y.; Wang, M.; He, D.; Song, Z.-Q.; Zeng, H.; Ning, Z.; Lu, C.; Lu, A.; et al. Chemotaxonomic Classification Applied to the Identification of Two Closely-Related Citrus TCMs Using UPLC-Q-TOF-MS-Based Metabolomics. Mol. A J. Synth. Chem. Nat. Prod. Chem. 2017, 22, 1721. [Google Scholar] [CrossRef] [PubMed]
- Mannochio-Russo, H.; De Almeida, R.; Nunes, W.; Bueno, P.; Caraballo-Rodríguez, A.; Bauermeister, A.; Dorrestein, P.; Bolzani, V. Untargeted Metabolomics Sheds Light on the Diversity of Major Classes of Secondary Metabolites in the Malpighiaceae Botanical Family. Front. Plant Sci. 2022, 13, 854842. [Google Scholar] [CrossRef]
- Qiu, F.; Fine, D.; Wherritt, D.; Lei, Z.; Sumner, L. PlantMAT: A Metabolomics Tool for Predicting the Specialized Metabolic Potential of a System and for Large-Scale Metabolite Identifications. Anal. Chem. 2016, 88, 11373–11383. [Google Scholar] [CrossRef]
- Gupta, N. Therapeutic Efficacy of the Plant Bioactive Phytochemicals with Special Reference to Alkaloids, Terpenoids, Phenolics and Cardiac Glycosides. Int. J. Plant Environ. 2024, 10, 22–30. [Google Scholar] [CrossRef]
- Fatima, M.; Dar, M.; Dhanavade, M.; Abbas, S.Z.; Bukhari, M.N.; Arsalan, A.; Liao, Y.; Wan, J.; Bukhari, J.S.S.; Zhen, O. Biosynthesis and Pharmacological Activities of the Bioactive Compounds of White Mulberry (Morus alba): Current Paradigms and Future Challenges. Biology 2024, 13, 506. [Google Scholar] [CrossRef]
- Kumaravel, S.; Muthukumaran, P.; Thomas, N. Phytochemical, GC-MS and FT-IR analysis of Papver somniferum L. J. Pharm. Biol. Sci. 2019, 7, 1–8. [Google Scholar]
- Gupcsó, K.; Kókai, Z.; Bálint, M.; Tavaszi-Sárosi, S.; Németh, É.Z. Studies on Sensory and Phytochemical Characteristics of Poppy (Papaver somniferum L.) Varieties for Their Oil Utilisation. Foods 2023, 12, 3165. [Google Scholar] [CrossRef]
- Sundowo, A.; Artanti, N.; Hanafi, M.; Minarti, M.; Primahana, G. Phytochemical screening, total phenolic, total flavonoids contents and antioxidant activity of Cinchona ledgeriana leaves ethanol extract. In Proceedings of the 3rd International Symposium on Applied Chemistry, Jakarta, Indonesia, 23–24 October 2017. [Google Scholar]
- Yadav, R.; Sahu, M.; Yadav, P.K.; Thakur, S.S.; Rathi, J. Phytochemical Estimation and Antioxidant Potential of Cinchona officinalis L. Stem Bark Extracts. Int. J. Med. Sci. Pharma Res. 2023, 9, 32–35. [Google Scholar] [CrossRef]
- Almatroodi, S.; Alsahli, M.; Almatroudi, A.; Verma, A.; Aloliqi, A.; Allemailem, K.; Khan, A.; Rahmani, A. Potential Therapeutic Targets of Quercetin, a Plant Flavonol, and Its Role in the Therapy of Various Types of Cancer through the Modulation of Various Cell Signaling Pathways. Molecules 2021, 26, 1315. [Google Scholar] [CrossRef]
- Ullah, A.; Munir, S.; Badshah, S.; Khan, N.; Ghani, L.; Poulson, B.; Emwas, A.; Jaremko, M. Important Flavonoids and Their Role as a Therapeutic Agent. Molecules 2020, 25, 5243. [Google Scholar] [CrossRef] [PubMed]
- Cox-Georgian, D.; Ramadoss, N.; Dona, C.; Basu, C. Therapeutic and Medicinal Uses of Terpenes. In Medicinal Plants; Springer: Berlin/Heidelberg, Germany, 2019; pp. 333–359. [Google Scholar]
- Borges, A.; Mandim, F.; Heleno, S.; Calhelha, R. Application and Medicinal of Terpenoids. Austin J. Anal. Pharm. Chem. 2024, 11, 1167. [Google Scholar] [CrossRef]
- Shrivastava, A.K.; Keshari, M.; Neupane, M.; Chaudhary, S.; Dhakal, P.K.; Shrestha, L.; Palikhey, A.; Yadav, C.; Lamichhane, G.; Shekh, M.U.; et al. Evaluation of Antioxidant and Anti-Inflammatory Activities, and Metabolite Profiling of Selected Medicinal Plants of Nepal. J. Trop. Med. 2023, 2023, 6641018. [Google Scholar] [CrossRef]
- Osmakov, D.; Kalinovskii, A.; Belozerova, O.; Andreev, Y.; Kozlov, S. Lignans as Pharmacological Agents in Disorders Related to Oxidative Stress and Inflammation: Chemical Synthesis Approaches and Biological Activities. Int. J. Mol. Sci. 2022, 23, 6031. [Google Scholar] [CrossRef]
- Gadéa, A.; Khazem, M.; Gaslonde, T. Current knowledge on chemistry of Proteaceae family, and biological activities of their bis-5-alkylresorcinol derivatives. Phytochem. Rev. 2022, 21, 1969–2005. [Google Scholar] [CrossRef]
- Tanaka, N.; Kashiwada, Y. Characteristic metabolites of Hypericum plants: Their chemical structures and biological activities. J. Nat. Med. 2021, 75, 423–433. [Google Scholar] [CrossRef]
- Zager, J.; Lange, I.; Srividya, N.; Smith, A.; Lange, B. Gene Networks Underlying Cannabinoid and Terpenoid Accumulation in Cannabis1[OPEN]. Plant Physiol. 2019, 180, 1877–1897. [Google Scholar] [CrossRef]
- Ahmed, E.; Arshad, M.; Khan, M.Z.; Amjad, M.; Sadaf, H.; Riaz, I.; Sabir, S.; Ahmad, N. Secondary metabolites and their multidimensional prospective in plant life. J. Pharmacogn. Phytochem. 2017, 6, 205–214. [Google Scholar]
- Heblinski, M.; Santiago, M.; Fletcher, C.; Stuart, J.; Connor, M.; McGregor, I.; Arnold, J. Terpenoids Commonly Found in Cannabis sativa Do Not Modulate the Actions of Phytocannabinoids or Endocannabinoids on TRPA1 and TRPV1 Channels. Cannabis Cannabinoid Res. 2020, 5, 305–317. [Google Scholar] [CrossRef]
- Gathungu, R.M.; Kautz, R.; Kristal, B.S.; Bird, S.S.; Vouros, P. The integration of LC-MS and NMR for the analysis of low molecular weight trace analytes in complex matrices. Mass Spectrom. Rev. 2020, 39, 35–54. [Google Scholar] [CrossRef]
- Seger, C.; Sturm, S.; Stuppner, H. Mass spectrometry and NMR spectroscopy: Modern high-end detectors for high resolution separation techniques–state of the art in natural product HPLC-MS, HPLC-NMR, and CE-MS hyphenations. Nat. Prod. Rep. 2013, 30, 970–987. [Google Scholar] [CrossRef] [PubMed]
- Gika, H.G.; Wilson, I.D.; Theodoridis, G.A. LC–MS-based holistic metabolic profiling. Problems, limitations, advantages, and future perspectives. J. Chromatogr. B 2014, 966, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mlynek, F.; Himmelsbach, M.; Buchberger, W.; Klampfl, C. A new analytical workflow using HPLC with drift-tube ion-mobility quadrupole time-of-flight/mass spectrometry for the detection of drug-related metabolites in plants. Anal. Bioanal. Chem. 2020, 412, 1817–1824. [Google Scholar] [CrossRef]
- Wojtanowski, K.; Mroczek, T. Study of a complex secondary metabolites with potent anti-radical activity by two dimensional TLC/HPLC coupled to electrospray ionization time-of-flight mass spectrometry and bioautography. Anal. Chim. Acta 2018, 1029, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhao, X.; Bai, C.; Zhao, C.; Lu, G.; Xu, G. LC–MS-based metabonomics analysis. J. Chromatogr. B 2008, 866, 64–76. [Google Scholar] [CrossRef]
- Pires, N.M.M.; Dong, T.; Hanke, U.; Hoivik, N. Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors 2014, 14, 15458–15479. [Google Scholar] [CrossRef]
- Iyer, V.; Issadore, D.A.; Aflatouni, F. The next generation of hybrid microfluidic/integrated circuit chips: Recent and upcoming advances in high-speed, high-throughput, and multifunctional lab-on-IC systems. Lab A Chip 2023, 23, 2553–2576. [Google Scholar] [CrossRef]
- Sarwar, H.; Rahman, M. A Systematic Short Review of Machine Learning and Artificial Intelligence Integration in Current Project Management Techniques. In Proceedings of the 2024 IEEE 4th International Conference on Software Engineering and Artificial Intelligence (SEAI), Xiamen, China, 21–23 June 2024; pp. 262–270. [Google Scholar]
- Kumar, K.; Kumar, V. Integration of Artificial Intelligence and Machine Learning for Internet of Things. In Proceedings of the 2023 International Conference on Sustainable Communication Networks and Application (ICSCNA), Theni, India, 15–17 November 2023; pp. 491–497. [Google Scholar]
- Islam, S. Future Trends in SQL Databases and Big Data Analytics: Impact of Machine Learning and Artificial Intelligence. Int. J. Sci. Eng. 2024. [Google Scholar] [CrossRef]
- Beaton, A.D.; Cardwell, C.L.; Thomas, R.S.; Sieben, V.J.; Legiret, F.-E.; Waugh, E.M.; Statham, P.J.; Mowlem, M.C.; Morgan, H. Lab-on-chip measurement of nitrate and nitrite for in situ analysis of natural waters. Environ. Sci. Technol. 2012, 46, 9548–9556. [Google Scholar] [CrossRef]
- Roshan, U.; Dai, Y.; Yadav, A.S.; Hettiarachchi, S.; Mudugamuwa, A.; Zhang, J.; Nguyen, N.-T. Flexible droplet microfluidic devices for tuneable droplet generation. Sens. Actuators B Chem. 2025, 422, 136617. [Google Scholar] [CrossRef]
- Das, A.; Prajapati, P. Navigating pharmaceuticals: Microfluidic devices in analytical and formulation sciences. Discov. Chem. 2025, 2, 49. [Google Scholar] [CrossRef]
- Zhao, X.; Zhai, L.; Chen, J.; Zhou, Y.; Gao, J.; Xu, W.; Li, X.; Liu, K.; Zhong, T.; Xiao, Y. Recent advances in microfluidics for the early detection of plant diseases in vegetables, fruits, and grains caused by bacteria, fungi, and viruses. J. Agric. Food Chem. 2024, 72, 15401–15415. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, A.A.S.; Jothimuthu, P.; Papautsky, I. Photodefinable polydimethylsiloxane (PDMS) for rapid lab-on-a-chip prototyping. Lab A Chip 2007, 7, 1192–1197. [Google Scholar] [CrossRef] [PubMed]
- Dkhar, D.S.; Kumari, R.; Malode, S.J.; Shetti, N.P.; Chandra, P. Integrated lab-on-a-chip devices: Fabrication methodologies, transduction system for sensing purposes. J. Pharm. Biomed. Anal. 2023, 223, 115120. [Google Scholar] [CrossRef]
- Whitesides, G.M.; Ostuni, E.; Takayama, S.; Jiang, X.; Ingber, D.E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 2001, 3, 335–373. [Google Scholar] [CrossRef]
- Huang, H.; Liu, H.; Ma, W.; Qin, L.; Chen, L.; Guo, H.; Xu, H.; Li, J.; Yang, C.; Hu, H.; et al. High-throughput MALDI-MSI metabolite analysis of plant tissue microarrays. Plant Biotechnol. J. 2023, 21, 2574–2584. [Google Scholar] [CrossRef]
- Schumacher, S.; Nestler, J.; Otto, T.; Wegener, M.; Ehrentreich-Förster, E.; Michel, D.; Wunderlich, K.; Palzer, S.; Sohn, K.; Weber, A. Highly-integrated lab-on-chip system for point-of-care multiparameter analysis. Lab A Chip 2012, 12, 464–473. [Google Scholar] [CrossRef]
- Dervisevic, E.; Tuck, K.; Voelcker, N.; Cadarso, V. Recent Progress in Lab-On-a-Chip Systems for the Monitoring of Metabolites for Mammalian and Microbial Cell Research. Sensors 2019, 19, 5027. [Google Scholar] [CrossRef]
- Alghannam, F.; Alayed, M.; Alfihed, S.; Sakr, M.A.; Almutairi, D.; Alshamrani, N.; Al Fayez, N. Recent Progress in PDMS-Based Microfluidics Toward Integrated Organ-on-a-Chip Biosensors and Personalized Medicine. Biosensors 2025, 15, 76. [Google Scholar] [CrossRef]
- Wang, X.; Zeng, S. Stereoselective metabolic and pharmacokinetic analysis of the chiral active components from herbal medicines. Curr. Pharm. Anal. 2010, 6, 39–52. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Wen, Y.; Serre, N.B.C.; Laursen, C.C.H.; Dietz, A.G.; Taylor, B.R.; Drobizhev, M.; Molina, R.S.; Aggarwal, A.; Rancic, V. A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life. PLoS Biol. 2022, 20, e3001772. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, M.T.; Sajid, Z.A.; Khilji, S.A. Graphene Oxide Nanoparticle-Assisted Promotion of Stevioside, Rebaudioside A, and Selected Biochemical Attributes in Stevia rebaudiana Bertoni. Scientifica 2024, 2024, 6693085. [Google Scholar] [CrossRef] [PubMed]
- Vaishampayan, V.; Dahake, R.; Athira, G.K.; Tyagi, M.; Kapoor, A.; Gumfekar, S. Current and emerging techniques for the detection of environmental contaminants. In Molecularly Imprinted Polymers for Environmental Monitoring: Fundamentals and Applications; IOP Publishing Ltd.: Bristol, UK, 2023; pp. 5-1–5-17. [Google Scholar]
- Kapoor, A.; Ramamoorthy, S.; Sundaramurthy, A.; Vaishampayan, V.; Sridhar, A.; Balasubramanian, S.; Ponnuchamy, M. Based lab-on-a-chip devices for detection of agri-food contamination. Trends Food Sci. Technol. 2024, 147, 104476. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Alshallash, K.S.; Eid, A.M.; Hassan, S.E.-D.; Salih, M.; Hamza, M.F.; Fouda, A. Exploring the Antimicrobial, Antioxidant, and Antiviral Potential of Eco-Friendly Synthesized Silver Nanoparticles Using Leaf Aqueous Extract of Portulaca oleracea L. Pharmaceuticals 2024, 17, 317. [Google Scholar] [CrossRef]
- Singh, G.; Kumar, P.; Mandal, P.; Aziz, D.; Rafiq, S.; Saini, P.; Gudi, S.; Saini, D.K. Plant metabolomic profiling to better understand biotic and abiotic stress resistance in cereals. In Omics and System Biology Approaches for Delivering Better Cereals; CRC Press: Boca Raton, FL, USA, 2024; pp. 268–292. [Google Scholar]
- Helmeczi, E.; Kroezen, Z.; Shanmuganathan, M.; Stanciu, A.R.; Martinez, V.; Kurysko, N.; Normando, P.; Castro, I.s.R.R.d.; Schincaglia, R.M.; Kac, G. A Software Tool for Rapid and Automated Preprocessing of Large-Scale Serum Metabolomic Data by Multisegment Injection-Capillary Electrophoresis-Mass Spectrometry. Anal. Chem. 2024, 97, 175–184. [Google Scholar] [CrossRef]
- Singh, A.P.; Palani, H.; Kumari, A.; Kushwaha, J.K.; Das, U. Nano (Bio) Sensor Technologies: Fostering the Renaissance of Horticulture. In Contemporary Suitability of Nanobionics in Agriculture: Nanotechnology in Horticultural Crops; Springer: Berlin/Heidelberg, Germany, 2025; pp. 255–273. [Google Scholar]
- Bharti, A.; Jain, U.; Chauhan, N. From lab to field: Nano-biosensors for real-time plant nutrient tracking. Plant Nano Biol. 2024, 9, 100079. [Google Scholar] [CrossRef]
- Goel, R.; Chakraborty, S.; Awasthi, V.; Bhardwaj, V.; Dubey, S.K. Exploring the various aspects of Surface enhanced Raman spectroscopy (SERS) with focus on the recent progress: SERS-active substrate, SERS-instrumentation, SERS-application. Sens. Actuators A Phys. 2024, 376, 115555. [Google Scholar] [CrossRef]
- Lee, H.K.; Lee, Y.H.; Koh, C.S.L.; Phan-Quang, G.C.; Han, X.; Lay, C.L.; Sim, H.Y.F.; Kao, Y.-C.; An, Q.; Ling, X.Y. Designing surface-enhanced Raman scattering (SERS) platforms beyond hotspot engineering: Emerging opportunities in analyte manipulations and hybrid materials. Chem. Soc. Rev. 2019, 48, 731–756. [Google Scholar] [CrossRef]
- Chugh, V.; Basu, A.; Kaushik, A.; Bhansali, S.; Basu, A.K. Employing nano-enabled artificial intelligence (AI)-based smart technologies for prediction, screening, and detection of cancer. Nanoscale 2024, 16, 5458–5486. [Google Scholar] [CrossRef]
- Khondakar, K.R.; Kaushik, A.K. Next-Generation Smart Biosensing: Nano-Platforms, Nano-Microfluidics Interfaces, and Emerging Applications of Quantum Sensing; Elsevier: Amsterdam, The Netherlands, 2024. [Google Scholar]
- Hatami, M.; Naghdi Badi, H.; Ghorbanpour, M. Nano-elicitation of secondary pharmaceutical metabolites in plant cells: A review. J. Med. Plants 2019, 18, 6–36. [Google Scholar] [CrossRef]
- Dzhagan, V.; Smirnov, O.; Kovalenko, M.; Mazur, N.; Hreshchuk, O.; Taran, N.; Plokhovska, S.; Pirko, Y.; Yemets, A.; Yukhymchuk, V. Spectroscopic study of phytosynthesized Ag nanoparticles and their activity as SERS substrate. Chemosensors 2022, 10, 129. [Google Scholar] [CrossRef]
- Zhao, L.; Hu, J.; Huang, Y.; Wang, H.; Adeleye, A.; Ortiz, C.; Keller, A.A. 1H NMR and GC–MS based metabolomics reveal nano-Cu altered cucumber (Cucumis sativus) fruit nutritional supply. Plant Physiol. Biochem. 2017, 110, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Lee, H.J. Carbon nanomaterials and metallic nanoparticles-incorporated electrochemical sensors for small metabolites: Detection methodologies and applications. Curr. Opin. Electrochem. 2020, 22, 234–243. [Google Scholar] [CrossRef]
- Carrara, S.; Bolomey, L.; Boero, C.; Cavallini, A.; Meurville, E.; De Micheli, G.; Rezzonico, T.; Proietti, M.; Grassi, F. Single-metabolite bio-nano-sensors and system for remote monitoring in animal models. In Proceedings of the SENSORS, 2011 IEEE, Limerick, Ireland, 28–31 October 2011; pp. 716–719. [Google Scholar]
- Javed, R.; Yucesan, B.; Zia, M.; Gurel, E. Elicitation of secondary metabolites in callus cultures of Stevia rebaudiana Bertoni grown under ZnO and CuO nanoparticles stress. Sugar Tech 2018, 20, 194–201. [Google Scholar] [CrossRef]
- Dong, B.-R.; Jiang, R.; Chen, J.-F.; Xiao, Y.; Lv, Z.-Y.; Chen, W.-S. Strategic nanoparticle-mediated plant disease resistance. Crit. Rev. Biotechnol. 2023, 43, 22–37. [Google Scholar] [CrossRef]
- Bag, B.G.; Garai, C.; Majumdar, R.; Laguerre, M. Natural triterpenoids as renewable nanos. Struct. Chem. 2012, 23, 393–398. [Google Scholar] [CrossRef]
- Wu, H.; Li, Z. Recent advances in nano-enabled agriculture for improving plant performance. Crop J. 2022, 10, 1–12. [Google Scholar] [CrossRef]
- Segneanu, A.-E.; Bradu, I.A.; Calinescu, M.S.; Vlase, G.; Vlase, T.; Herea, D.-D.; Buema, G.; Mihailescu, M.; Grozescu, I. Novel Nanocomposites and Biopolymer-Based Nanocomposites for Hexavalent Chromium Removal from Aqueous Media. Polymers 2024, 16, 3469. [Google Scholar] [CrossRef]
- Xu, Y.; Hassan, M.M.; Sharma, A.S.; Li, H.; Chen, Q. Recent advancement in nano-optical strategies for detection of pathogenic bacteria and their metabolites in food safety. Crit. Rev. Food Sci. Nutr. 2023, 63, 486–504. [Google Scholar] [CrossRef]
- Son, W.K.; Choi, Y.S.; Han, Y.W.; Shin, D.W.; Min, K.; Shin, J.; Lee, M.J.; Son, H.; Jeong, D.H.; Kwak, S.-Y. In vivo surface-enhanced Raman scattering nanosensor for the real-time monitoring of multiple stress signalling molecules in plants. Nat. Nanotechnol. 2023, 18, 205–216. [Google Scholar] [CrossRef]
- Weng, S.; Hu, X.; Wang, J.; Tang, L.; Li, P.; Zheng, S.; Zheng, L.; Huang, L.; Xin, Z. Advanced application of Raman spectroscopy and surface-enhanced Raman spectroscopy in plant disease diagnostics: A review. J. Agric. Food Chem. 2021, 69, 2950–2964. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Somborn, A.; Schlehuber, D.; Keuter, V.; Deerberg, G. Raman spectroscopy in crop quality assessment: Focusing on sensing secondary metabolites: A review. Hortic. Res. 2023, 10, uhad074. [Google Scholar] [CrossRef] [PubMed]
- Elli, G.; Hamed, S.; Petrelli, M.; Ibba, P.; Ciocca, M.; Lugli, P.; Petti, L. Field-effect transistor-based biosensors for environmental and agricultural monitoring. Sensors 2022, 22, 4178. [Google Scholar] [CrossRef]
- Dullius, A.; Buffon, G.; Junior, M.F.; Giuliatti, S. Artificial Intelligence in Phycochemicals Recognition. In Value-Added Products from Algae: Phycochemical Production and Applications; Springer: Berlin/Heidelberg, Germany, 2023; pp. 97–122. [Google Scholar]
- Kalita, I.; Bhattacharjee, S.; Saharia, M. Advancements in Medicinal Plant Research: Harnessing Artificial Intelligence, Machine Learning, Deep Learning, and Bioinformatics. In Biotechnology, Multiple Omics, and Precision Breeding in Medicinal Plants; CRC Press: Boca Raton, FL, USA, 2025; pp. 135–145. [Google Scholar]
- Zhong, C.; Li, L.; Wang, Y.-Z. Applications of chemical fingerprints and machine learning in plant ecology: Recent progress and future perspectives. Microchem. J. 2024, 206, 111447. [Google Scholar] [CrossRef]
- Chen, J.; Yang, W.; Tan, G.; Tian, C.; Wang, H.; Zhou, J.; Liao, H. Prediction of the taxonomical classification of the Ranunculaceae family using a machine learning method. New J. Chem. 2022, 46, 5150–5161. [Google Scholar] [CrossRef]
- Abdullah-Zawawi, M.-R.; Govender, N.; Karim, M.B.; Altaf-Ul-Amin, M.; Kanaya, S.; Mohamed-Hussein, Z.-A. Chemoinformatics-driven classification of angiosperms using sulfur-containing compounds and machine learning algorithm. Plant Methods 2022, 18, 118. [Google Scholar] [CrossRef]
- Julier, A.C.M.; Jardine, P.E.; Coe, A.L.; Gosling, W.D.; Lomax, B.H.; Fraser, W.T. Chemotaxonomy as a tool for interpreting the cryptic diversity of Poaceae pollen. Rev. Palaeobot. Palynol. 2016, 235, 140–147. [Google Scholar] [CrossRef]
- Kadam, Y.; Yache, A.; Solunke, P.; Tati, R.; Kachhoria, R.; Buchade, A. Automated Medicinal Plant Identification through Image Processing and Machine Learning. In Proceedings of the 2024 8th International Conference on Computing, Communication, Control and Automation (ICCUBEA), Pune, India, 23–24 August 2024; pp. 1–5. [Google Scholar]
- Ramesh, S.; Hebbar, R.; Niveditha, M.; Pooja, R.; Shashank, N.; Vinod, P.V. Plant disease detection using machine learning. In Proceedings of the 2018 International Conference on Design Innovations for 3Cs Compute Communicate Control (ICDI3C), Bangalore, India, 25–28 April 2018; pp. 41–45. [Google Scholar]
- Zhang, X.; Yang, L.-E.; Hu, Y.; Wu, X.; Wang, Z.; Miao, Y.; Sun, H.; Nie, Z.; Tan, N. Integrating morphology, molecular phylogeny and chemotaxonomy for the most effective authentication in Chinese Rubia with insights into origin and distribution of characteristic Rubiaceae-type cyclopeptides. Ind. Crops Prod. 2023, 191, 115775. [Google Scholar] [CrossRef]
- Mara, M.; Pop, P.; Barata, J. A Comparative Study of Machine Learning Models for Plant Disease Identification. In Proceedings of the 19th International Conference on Soft Computing Models in Industrial and Environmental Applications SOCO, Salamanca, Spain, 9–11 October 2024; pp. 107–116. [Google Scholar]
- Slynko, N.M.; Burmakina, N.V.; Potseluyev, O.M.; Kapustyanchik, S.Y.; Galitsin, G.Y.; Goryachkovskaya, T.N.; Kuybida, L.V.; Shekhovtsov, S.V.; Peltek, S.E.; Shumny, V.K. Gas chromatography-mass spectrometry in the taxonomy of Miscanthus. Вавилoвский Журнал Генетики и Селекции 2019, 23, 1076–1081. [Google Scholar]
- Varshney, D.; Babukhanwala, B.; Khan, J.; Saxena, D.; Singh, A.K. Plant disease detection using machine learning techniques. In Proceedings of the 2022 3rd International Conference for Emerging Technology (INCET), Belgaum, India, 27–29 May 2022; pp. 1–5. [Google Scholar]
- Pargaien, A.V.; Singh, D.; Chauhan, M.; Negi, H.; Chilwal, B.; Pargaien, N. Identification of plant leaves having anti-diabetic property using machine learning. In Proceedings of the 2023 2nd International Conference on Applied Artificial Intelligence and Computing (ICAAIC), Salem, India, 4–6 May 2023; pp. 195–200. [Google Scholar]
- Prajapati, S.; Qureshi, S.; Rao, Y.; Nadkarni, S.; Retharekar, M.; Avhad, A. Plant disease identification using deep learning. In Proceedings of the 2023 4th International Conference for Emerging Technology (INCET), Belgaum, India, 26–28 May 2023; pp. 1–5. [Google Scholar]
- Kalpana, P.; Anandan, R. A Capsule Attention Network for Plant Disease Classification. Trait. Signal 2023, 40, 2051. [Google Scholar] [CrossRef]
- de Azevedo, M.O.; Paucar, V.L. Performance analysis of algorithms based on intelligence of plants. In Proceedings of the IEEE CACIDI 2016-IEEE Conference on Computer Sciences, Buenos Aires, Argentina, 30 November–2 December 2016; pp. 1–6. [Google Scholar]
- Nanda, S.J.; Panda, G.; Majhi, B.; Tah, P. Improved identification of nonlinear MIMO plants using new hybrid FLANN-AIS model. In Proceedings of the 2009 IEEE International Advance Computing Conference, Patiala, India, 6–7 March 2009; pp. 141–146. [Google Scholar]
- Venkateswara Reddy, E.; Naveen Kumar, G.S.; Swathi, B.; Siva Naga Dhipti, G. Deep learning approach for image-based plant species classification. In Proceedings of the International Conference on Soft Computing and Signal Processing, Shanghai, China, 24–26 September 2021; pp. 405–412. [Google Scholar]
- Grapov, D.; Fahrmann, J.; Wanichthanarak, K.; Khoomrung, S. Rise of deep learning for genomic, proteomic, and metabolomic data integration in precision medicine. Omics A J. Integr. Biol. 2018, 22, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Cambiaghi, A.; Ferrario, M.; Masseroli, M. Analysis of metabolomic data: Tools, current strategies and future challenges for omics data integration. Brief. Bioinform. 2017, 18, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Maurya, R.; Mahapatra, S.; Rajput, L. A Lightweight Meta-Ensemble Approach for Plant Disease Detection Suitable for IoT-Based Environments. IEEE Access 2024, 12, 28096–28108. [Google Scholar] [CrossRef]
- Walsh, J.; Mangina, E.; Negrão, S. Advancements in Imaging Sensors and AI for Plant Stress Detection: A Systematic Literature Review. Plant Phenomics 2024, 6, 0153. [Google Scholar] [CrossRef]
- Cardoso, R.; Pereira, T.; Facure, M.; Santos, D.D.; Mercante, L.; Mattoso, L.; Correa, D. Current Progress in Plant Pathogen Detection Enabled by Nanomaterials-based (Bio)Sensors. Sens. Actuators Rep. 2022, 4, 100068. [Google Scholar] [CrossRef]
- Soria, N.C.; Bisson, M.; Atilla-Gokcumen, G.; Aga, D. High-resolution mass spectrometry-based metabolomics reveal the disruption of jasmonic pathway in Arabidopsis thaliana upon copper oxide nanoparticle exposure. Sci. Total Environ. 2019, 693, 133443. [Google Scholar] [CrossRef]
- Patabadige, D.; Millet, L.; Aufrecht, J.; Shankles, P.; Standaert, R.; Retterer, S.; Doktycz, M. Label-free time- and space-resolved exometabolite sampling of growing plant roots through nanoporous interfaces. Sci. Rep. 2019, 9, 10272. [Google Scholar] [CrossRef]
- Jalili, A.; Bagherifar, R.; Nokhodchi, A.; Conway, B.; Javadzadeh, Y. Current Advances in Nanotechnology-Mediated Delivery of Herbal and Plant-Derived Medicines. Adv. Pharm. Bull. 2023, 13, 712–722. [Google Scholar] [CrossRef]
- Dewi, M.; Chaerunisaa, A.; Muhaimin, M.; Joni, I. Improved Activity of Herbal Medicines through Nanotechnology. Nanomaterials 2022, 12, 4073. [Google Scholar] [CrossRef]
- Prabhakar, P.; Anand, K.; Bala, I.; Shakya, R.; Massaon, H.K.; Suwalka, A.; Cp, B. Revolutionizing Herbal Medicine: Exploring Nano Drug Delivery Systems. Sumat. Med. J. 2023, 6, 143–154. [Google Scholar] [CrossRef]
- Bayer, I.S. Controlled drug release from nanoengineered polysaccharides. Pharmaceutics 2023, 15, 1364. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.; Singh, S.; Ramamurthy, P.C.; Dhanjal, D.S.; Subramanian, J.; Singh, J.; Singh, A. Plant Secondary Metabolites: A Biosensing Approach. In Advances in Agricultural and Industrial Microbiology: Volume 1: Microbial Diversity and Application in Agroindustry; Springer: Berlin/Heidelberg, Germany, 2022; pp. 249–268. [Google Scholar]
- Geetha, P.; Sudha, K.; Praveena, H.D. Nano Engineering Concepts, Principles and Applications in Food Technology. In Nanoelectronics Devices: Design, Materials, and Applications-Part II; Bentham Science Publishers: Sharjah, United Arab Emirates, 2023; pp. 246–279. [Google Scholar] [CrossRef]
- Pandey, Y.; Ambwani, S. Nano Metal based Herbal theranostics for Cancer management: Coalescing nature’s boon with nanotechnological advancement. Curr. Pharm. Biotechnol. 2021, 23, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Hezekiah, O. Business Modelling for the Quality Control and Commercialisation of Engineered Nano-Materials. Ph.D. Thesis, Durban University of Technology, Durban, South Africa, 2021. [Google Scholar]
- Munir, S.; Ahmed, S.; Ibrahim, M.; Khalid, M.; Ojha, S. A Spellbinding Interplay Between Biological Barcoding and Nanotechnology. Front. Bioeng. Biotechnol. 2020, 8, 556291. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, M.; Kim, J.-T.; Kim, W.-R.; Kim, J.-Y.; Jung, B.-S.; Choi, H.-J.; Chi, H.-Y.; Govindasamy, R.; Kim, S.-H. Safeguarding public health: Advanced detection of food adulteration using nanoparticle-based sensors. Crit. Rev. Anal. Chem. 2024, 1–21. [Google Scholar] [CrossRef]
- Kumar, R. Nanotechnology in herbal medicine: Challenges and future perspectives. In Nanotechnology in Herbal Medicine; Elsevier: Amsterdam, The Netherlands, 2023; pp. 515–548. [Google Scholar]
- Singh, N.; Yadav, S.S. Nanotechnological advancement in spices adulteration detection and authenticity validation. Food Control 2024, 167, 110806. [Google Scholar] [CrossRef]
- Gasmi, A.; Shanaida, M.; Oleshchuk, O.; Semenova, Y.; Mujawdiya, P.K.; Ivankiv, Y.; Pokryshko, O.; Noor, S.; Piscopo, S.; Adamiv, S. Natural ingredients to improve immunity. Pharmaceuticals 2023, 16, 528. [Google Scholar] [CrossRef]
- Benedetti, R.; Bajardi, F.; Capozziello, S.; Carafa, V.; Conte, M.; Del Sorbo, M.R.; Nebbioso, A.; Singh, M.; Stunnenberg, H.G.; Valadan, M. Different approaches to unveil biomolecule configurations and their mutual interactions. Anal. Lett. 2021, 54, 40–56. [Google Scholar] [CrossRef]
- Sun, L.; Wang, S.; Zhu, T. 1, 3, 5-Triethynylbenzene and melamine as monomers to synthesize three-dimensional network porous aromatic frameworks based silica/florisil for determination of carbendazim and thiabendazole in spinach. J. Sep. Sci. 2020, 43, 2842–2849. [Google Scholar] [CrossRef]
- Tavakoli, Z.; Ranjbar, F.; Tackallou, S.H.; Ranjbar, B. Nanostructures for the Prevention, Diagnosis, and Treatment of COVID-19: A Review. Part. Part. Syst. Charact. 2025, 42, 2400083. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, Y.; Huang, W.; Wang, Y.; Hu, X. A novel AuNPs-doped COFs composite as electrochemical probe for chlorogenic acid detection with enhanced sensitivity and stability. Sens. Actuators B Chem. 2018, 276, 362–369. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, D.; Zhou, S.-K.; Yang, L.; Yao, W.-F.; Cheng, F.-F.; Zhu, J.-J.; Zhang, L. Analytical and biomedical applications of nanomaterials in Chinese herbal medicines research. TrAC Trends Anal. Chem. 2022, 156, 116690. [Google Scholar] [CrossRef]
- Bernela, M.; Seth, M.; Kaur, N.; Sharma, S.; Pati, P. Harnessing the potential of nanobiotechnology in medicinal plants. Ind. Crops Prod. 2023, 194, 116266. [Google Scholar] [CrossRef]
- Sharma, B.; Yadav, D. Metabolomics and Network Pharmacology in the Exploration of the Multi-Targeted Therapeutic Approach of Traditional Medicinal Plants. Plants 2022, 11, 3243. [Google Scholar] [CrossRef] [PubMed]
- Shivalkar, S.; Chowdhary, P.; Afshan, T.; Chaudhary, S.; Roy, A.; Samanta, S.; Sahoo, A. Nanoengineering of biohybrid micro/nanobots for programmed biomedical applications. Colloids Surf. B Biointerfaces 2022, 222, 113054. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.; Wang, Y.; Xu, D.; Liang, C.; Guo, J.; Ma, X. Biocompatibility of artificial micro/nanomotors for use in biomedicine. Nanoscale 2019, 11, 14099–14112. [Google Scholar] [CrossRef]
- Li, J.; Kang, L.; Yu, Y.; Long, Y.; Jeffery, J.; Cai, W.; Wang, X. Study of Long-Term Biocompatibility and Bio-Safety of Implantable Nanogenerators. Nano Energy 2018, 51, 728–735. [Google Scholar] [CrossRef]
- Omanović-Mikličanin, E.; Stambolić, A. 5.1 Application of nanospectroscopy in food science and agriculture. Opt. Nanospectroscopy 2022, 311. [Google Scholar] [CrossRef]
- Fu, L.; Zheng, Y.; Zhang, P.; Zhu, J.; Zhang, H.; Zhang, L.; Su, W. Embedding leaf tissue in graphene ink to improve signals in electrochemistry-based chemotaxonomy. Electrochem. Commun. 2018, 92, 39–42. [Google Scholar] [CrossRef]
- Yien, R.M.K.; Matos, A.P.d.S.; Gomes, A.C.C.; Garófalo, D.d.A.; Santos-Oliveira, R.; Simas, N.K.; Ricci-Júnior, E. Nanotechnology promoting the development of products from the biodiversity of the Asteraceae family. Nutrients 2023, 15, 1610. [Google Scholar] [CrossRef]
- Pandey, K.; Chudasama, D. Hybrid Authentication Scheme Based on Cyber-Physical System use in Agriculture. J. Artif. Intell. Res. Adv. 2022, 9, 24–35. [Google Scholar]
- Ali, F.; Neha, K.; Parveen, S. Current regulatory landscape of nanomaterials and nanomedicines: A global perspective. J. Drug Deliv. Sci. Technol. 2022, 80, 104118. [Google Scholar] [CrossRef]
- Rodríguez-Gómez, F.D.; Penon, O.; Monferrer, D.; Rivera-Gil, P. Classification system for nanotechnology-enabled health products with both scientific and regulatory application. Front. Med. 2023, 10, 1212949. [Google Scholar] [CrossRef]
- Azahar, N.I.; Arifin, M.A.; Mahmood, S. Natural Product-Based Nanomedicine. In Nanoengineered Materials for Medical and Healthcare Applications; Wiley: Hoboken, NJ, USA, 2025; pp. 53–91. [Google Scholar]
- Ghosh, M.; Kumar, R. Regulatory issues in nanotechnology. In Nanotechnology Theranostics in Livestock Diseases and Management; Springer: Berlin/Heidelberg, Germany, 2024; pp. 765–788. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).