Elaboration of Nanostructured Levan-Based Colloid System as a Biological Alternative with Antimicrobial Activity for Applications in the Management of Pathogenic Microorganisms
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, Standards
2.2. Biosynthesis of Bacterial Levan In Vitro Using Streptococcus salivarius K12 and Leuconostoc mesenteroides DSM 20343
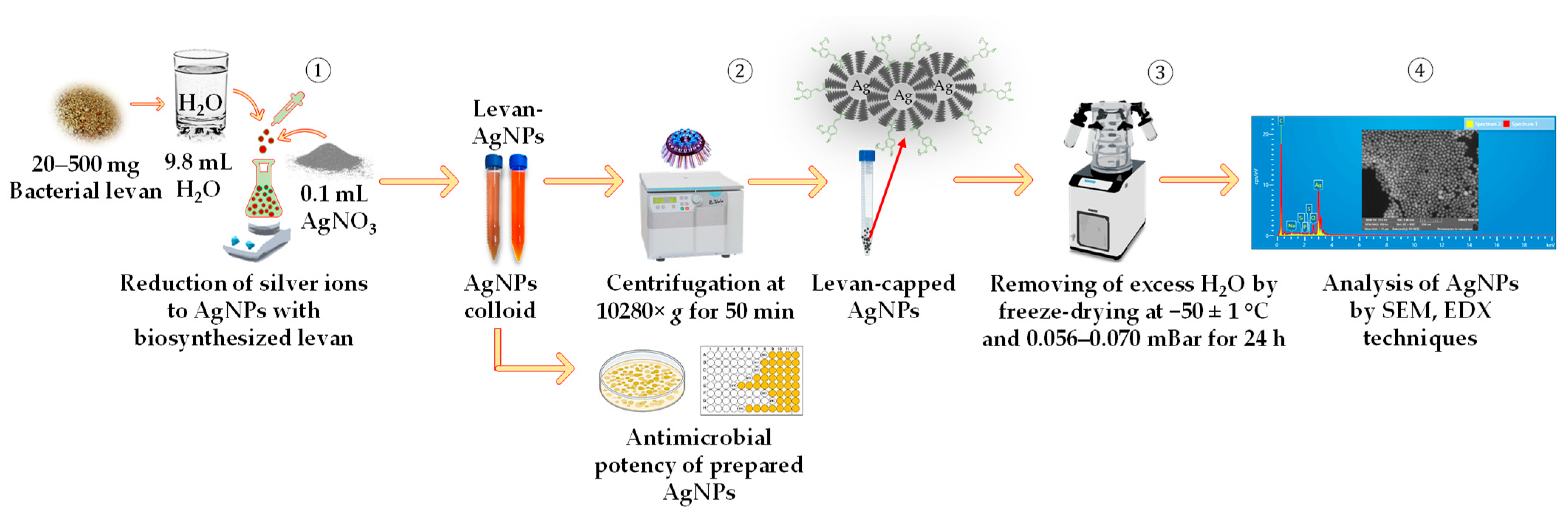
2.3. Preparation of Silver Nanoparticles Using Bacterial Levan Produced by Streptococcus salivarius K12 and Leuconostoc mesenteroides DSM 20343
2.4. Preparation of Bacterial Levan for Free and Total Saccharide Analysis
2.5. The HPLC-RID Conditions for Carbohydrate Analysis
2.6. Preparation of Bacterial Levan for Determination of Molecular Weight and Rheological Features
2.7. The Analytical Multi-Detector Gel Permeation Chromatography/Size-Exclusion Chromatography–Liquid Chromatography (GPC/SEC–HPLC) System Conditions for Bacterial Levan Analysis
2.8. Scanning Electron Microscope (SEM) and Energy-Dispersive X-ray Spectroscope (EDX)
2.9. Fourier-Transform Infrared Spectroscopy (FT-IR)
2.10. In Vitro Susceptibility Tests
2.10.1. Minimum Inhibitory Concentration (MIC)
2.10.2. Minimum Bactericidal Concentration (MBC)
2.10.3. Agar Disc Diffusion Method
2.10.4. Agar Well Diffusion Method
2.11. Statistical Analysis
3. Results and Discussion
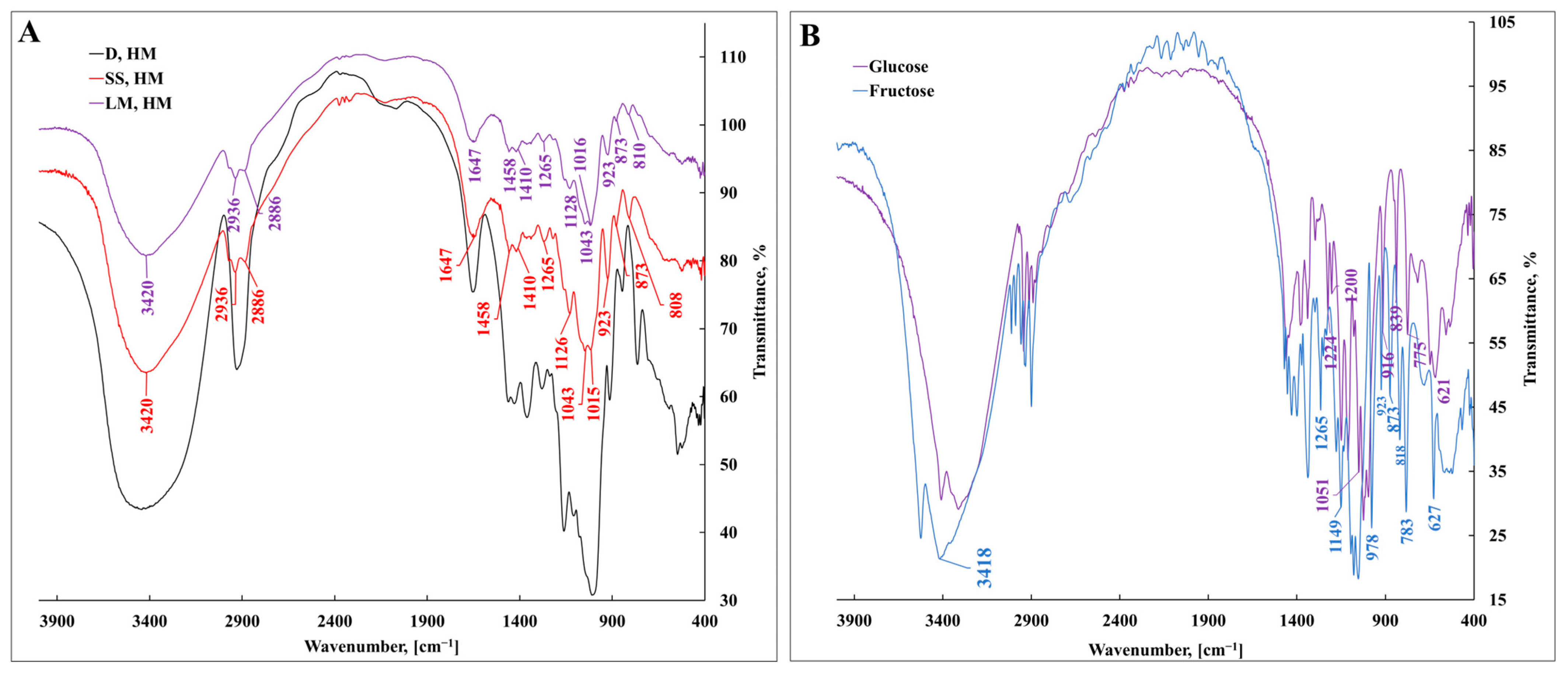
3.1. Characterization of Bacterial Levan Biosynthesized by Streptococcus salivarius K12 and Leuconostoc mesenteroides DSM 20343 Using Fourier-Transform Infrared Spectroscopy
3.2. Surface Morphology and Microstructural Feature Analysis of Bacterial Levan Synthesized by Streptococcus salivarius K12 and Leuconostoc mesenteroides DSM 20343 Using Scanning Electron Microscopy
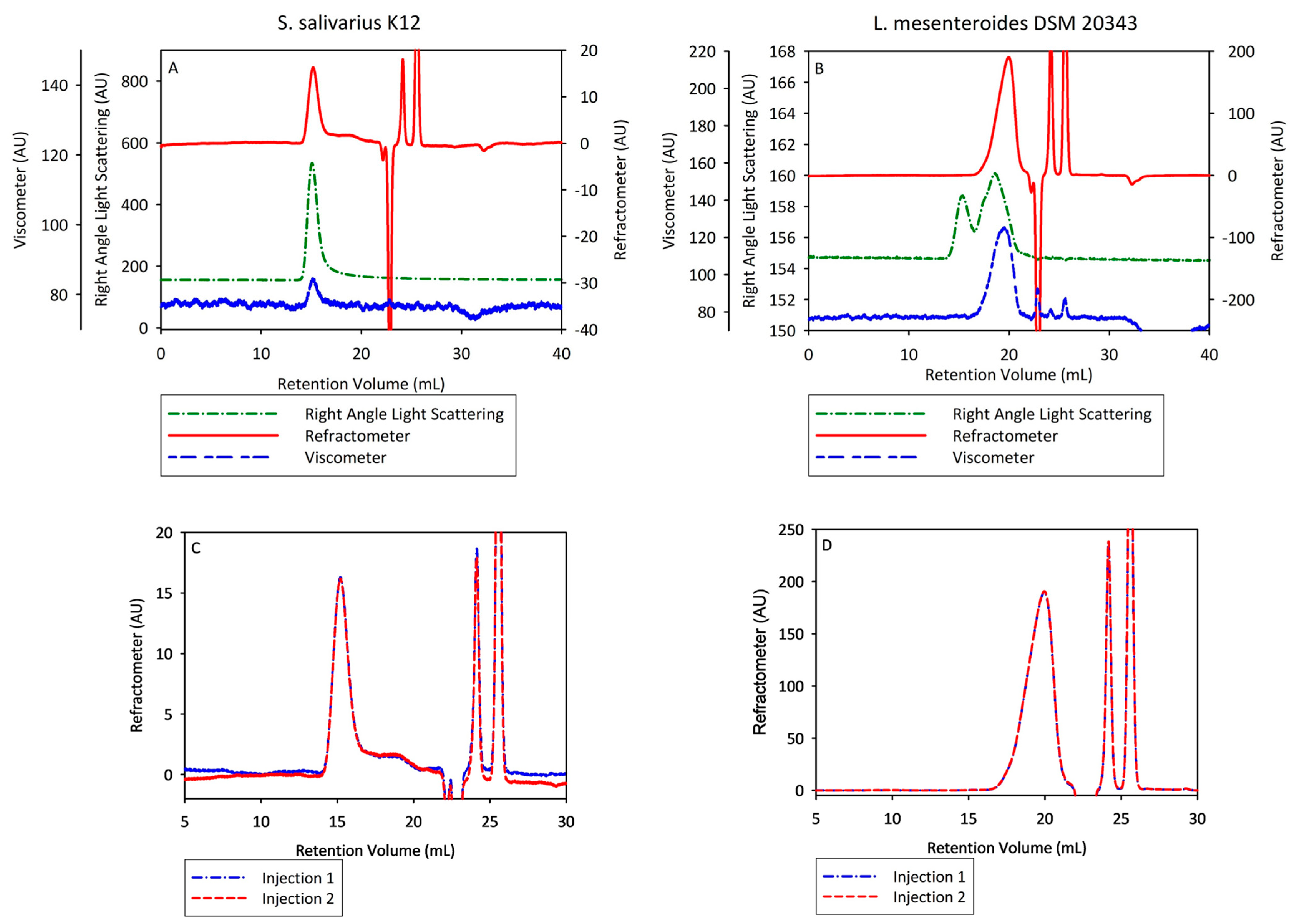
3.3. Characterization of Bacterial Levan Biosynthesized by Streptococcus salivarius K12 and Leuconostoc mesenteroides DSM 20343 by Multi-Detector Gel Permeation Chromatography/Size-Exclusion Chromatography–Liquid Chromatography
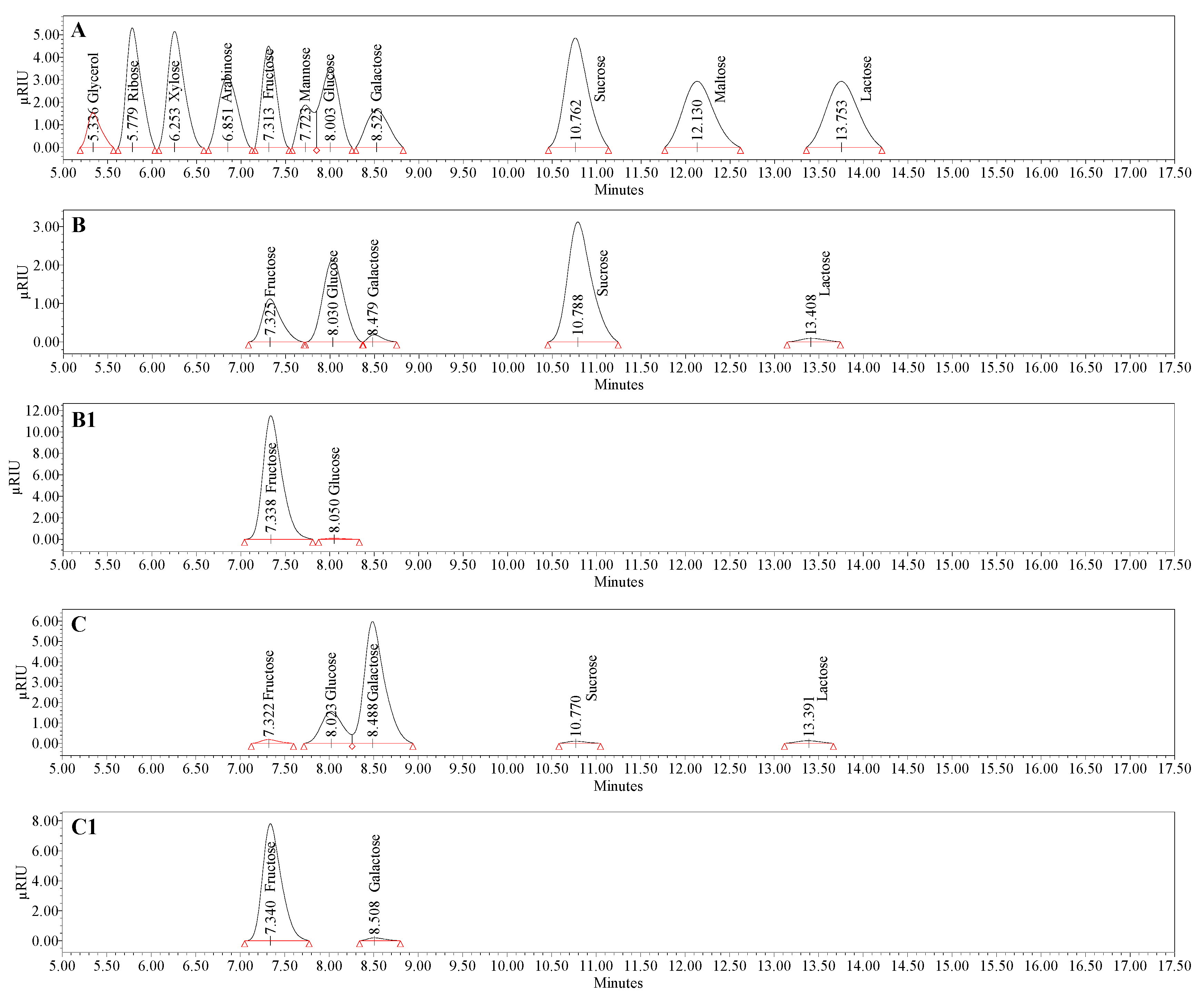
3.4. Compositional Analysis of Bacterial Levan Produced by Streptococcus salivarius K12 and Leuconostoc mesenteroides DSM 20343
3.5. Spectrophotometric Analysis of Elaborated Colloid Systems Based on Bacterial Levan and Silver Nanoparticles by UV–Vis Spectroscopy
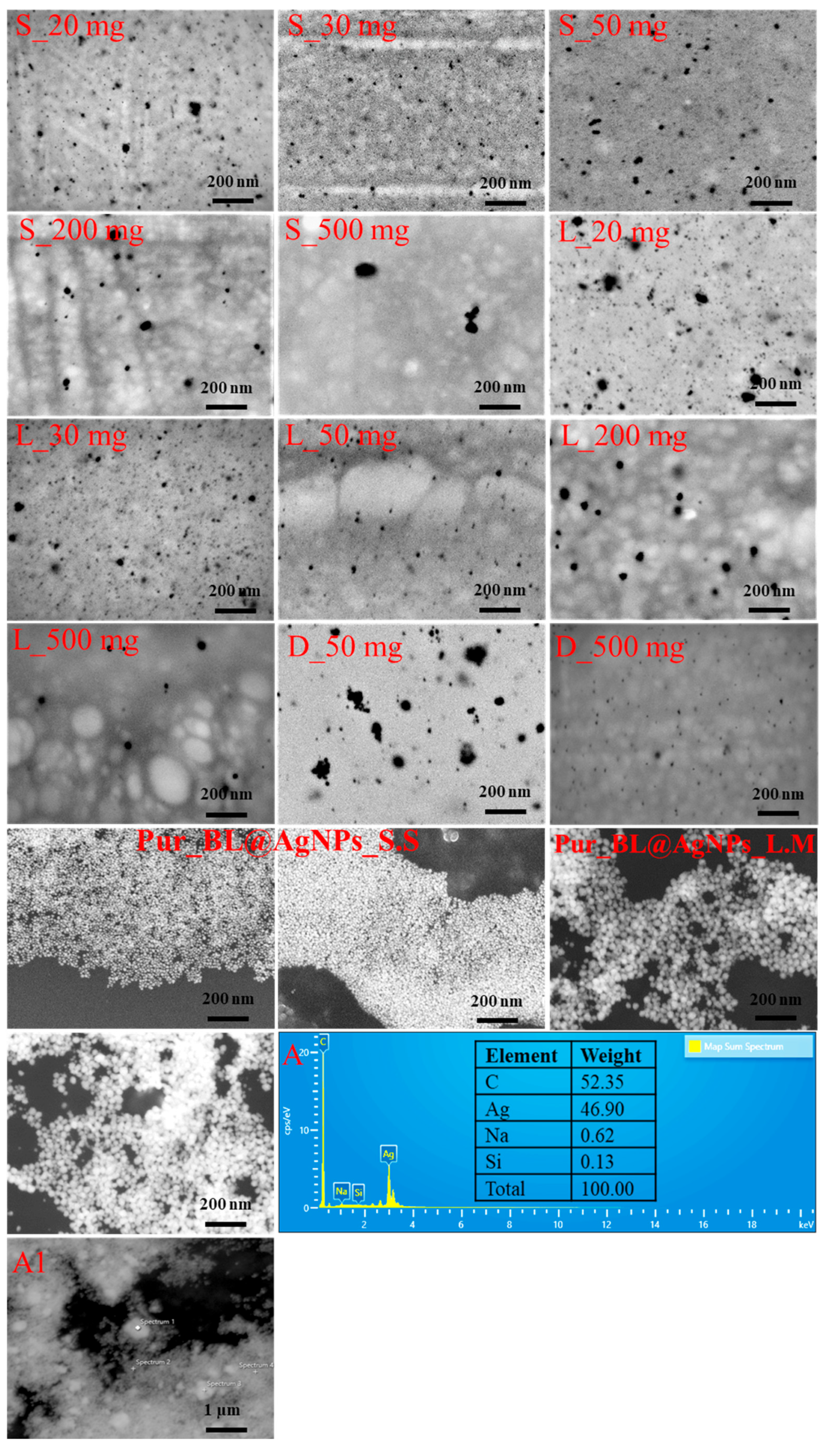
3.6. Structural Analysis of Bacterial Levan-Capped Silver Nanoparticles
3.7. Antimicrobial Activity of Bacterial Levan and Colloid Systems Containing Silver Nanoparticles According to Diameter of Inhibition Zone Values
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Ag | argentum |
| AMR | antimicrobial resistance |
| API | active pharmaceutical ingredient |
| ATCC | American-type culture collection |
| ATF | adenosine triphosphate |
| Au | aurum |
| BL | bacterial levan |
| BLQ | below limit of quantification |
| BSE | back-scattered electron |
| CFU | colony-forming unit |
| CoS | colloid system |
| CoS-BL@AgNPs | colloid system containing silver nanoparticles mediated by bacterial levan |
| Ð | dispersity |
| DeW | deionized water |
| DEX | dextran |
| DEX@AgNPs | dextran-mediated silver nanoparticles |
| dL | deciliter |
| DMF | dimethylformamide |
| DMSO | dimethyl sulfoxide |
| DW | dry weight basis |
| EC | European Commission |
| ECM | extracellular matrix |
| EDX | energy-dispersive X-ray spectroscopy |
| EPS | exopolysaccharide |
| FT-IR | Fourier-transform infrared spectroscopy |
| GPC | gel permeation chromatography |
| H–HW | hard-to-heal wound |
| HPLC | high-performance liquid chromatography |
| IR | infrared radiation |
| IV | intrinsic viscosity |
| kDa | kilodalton |
| LAB | lactic acid bacteria |
| LALS | left-angle light-scattering detector |
| LC–MS | liquid chromatography–mass spectrometry |
| M | mole |
| MBC | minimum bactericidal concentration |
| MDR | multi-drug resistance |
| MeCN | acetonitrile |
| MeOH | methanol |
| MHA | Mueller–Hinton agar |
| MHB | Mueller–Hinton broth |
| MIC | minimum inhibitory concentration |
| mM | millimole |
| Mn | number average molecular weight |
| MRS | De Man, Rogosa, and Sharpe |
| Mw | weight average molecular weight |
| NP | nanoparticle |
| NSAID | nonsteroidal anti-inflammatory drug |
| PCR | polymerase chain reaction |
| PD | polydispersity |
| PTFE | polytetrafluoroethylene |
| PVP | polyvinylpyrrolidone |
| RALS | right-angle light-scattering detector |
| Rh | hydrodynamic radius |
| RID | refractive index detection |
| rpm | revolutions per minute |
| SE | secondary electron |
| SEC | size-exclusion chromatography |
| SEM | scanning electron microscope |
| spp. | species |
| SPR | surface plasmon resonance |
| TCA | trichloroacetic acid |
| UV–Vis | ultraviolet–visible spectroscopy |
References
- Järbrink, K.; Ni, G.; Sönnergren, H.; Schmidtchen, A.; Pang, C.; Bajpai, R.; Car, J. The humanistic and economic burden of chronic wounds: A protocol for a systematic review. Syst. Rev. 2017, 6, 15. [Google Scholar] [CrossRef]
- Lindholm, C.; Searle, R. Wound management for the 21st century: Combining effectiveness and efficiency. Int. Wound J. 2016, 13, 5–15. [Google Scholar] [CrossRef]
- Frykberg, R.G.; Banks, J. Challenges in the Treatment of Chronic Wounds. Adv. Wound Care 2015, 4, 560–582. [Google Scholar] [CrossRef]
- Gāliņa, D.; Radenkovs, V.; Kviesis, J.; Valdovska, A. Effect of Essential Oils Supplemented with Caprylic Acid and Sodium Chloride against Faecal ESBL-Producing Escherichia coli Isolated from Pigs. Antibiotics 2022, 11, 461. [Google Scholar] [CrossRef]
- European Commission. European Commission Regulation (EU) 2019/6 of the European Parliament and of the Council of 11 December 2018 on veterinary medicinal products and repealing Directive 2001/82/EC. Off. J. Eur. Union 2019, L4, 43–167. [Google Scholar]
- Olofsson, T.C.; Butler, É.; Lindholm, C.; Nilson, B.; Michanek, P.; Vásquez, A. Fighting Off Wound Pathogens in Horses with Honeybee Lactic Acid Bacteria. Curr. Microbiol. 2016, 73, 463–473. [Google Scholar] [CrossRef]
- Verdú-Soriano, J.; de Cristino-Espinar, M.; Luna-Morales, S.; Dios-Guerra, C.; Caballero-Villarraso, J.; Moreno-Moreno, P.; Casado-Díaz, A.; Berenguer-Pérez, M.; Guler-Caamaño, I.; Laosa-Zafra, O.; et al. Superiority of a Novel Multifunctional Amorphous Hydrogel Containing Olea europaea Leaf Extract (EHO-85) for the Treatment of Skin Ulcers: A Randomized, Active-Controlled Clinical Trial. J. Clin. Med. 2022, 11, 1260. [Google Scholar] [CrossRef]
- O’Dwyer, L. Hard to heal wounds: Dealing with the problematic wound. Vet. Nurse 2015, 6, 316–329. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef]
- Baetke, S.C.; Lammers, T.; Kiessling, F. Applications of nanoparticles for diagnosis and therapy of cancer. Br. J. Radiol. 2015, 88, 20150207. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.-J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Gurung, J.; Khyriem, A.B.; Banik, A.; Lyngdoh, W.V.; Choudhury, B.; Bhattacharyya, P. Association of biofilm production with multidrug resistance among clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa from intensive care unit. Indian J. Crit. Care Med. 2013, 17, 214–218. [Google Scholar] [CrossRef]
- Attinger, C.; Wolcott, R. Clinically Addressing Biofilm in Chronic Wounds. Adv. Wound Care 2012, 1, 127–132. [Google Scholar] [CrossRef]
- Adzitey, F.; Huda, N.; Ali, G.R.R. Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. 3 Biotech 2013, 3, 97–107. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Lux, C.N. Wound healing in animals: A review of physiology and clinical evaluation. Vet. Dermatol. 2021, 33, 91-e27. [Google Scholar] [CrossRef]
- Gallaher, H.M.; Butler, J.R.; Wills, R.W.; Priddy, L.B.; Elder, S.H.; Heller, S.M.; Brinkman, E.; Baumgartner, W. Effects of short- and long-term administration of nonsteroidal anti-inflammatory drugs on osteotomy healing in dogs. Vet. Surg. 2019, 48, 1318–1329. [Google Scholar] [CrossRef]
- Barry, S. Non-steroidal anti-inflammatory drugs inhibit bone healing: A review. Vet. Comp. Orthop. Traumatol. 2010, 23, 385–392. [Google Scholar] [CrossRef]
- Simova, V.; Voslarova, E.; Vecerek, V.; Passantino, A.; Bedanova, I. Effects of travel distance and season of the year on transport-related mortality in cattle. Anim. Sci. J. 2017, 88, 526–532. [Google Scholar] [CrossRef]
- Ghosh, S.; Lahiri, D.; Nag, M.; Dey, A.; Sarkar, T.; Pathak, S.K.; Edinur, H.A.; Pati, S.; Ray, R.R. Bacterial Biopolymer: Its Role in Pathogenesis to Effective Biomaterials. Polymers 2021, 13, 1242. [Google Scholar] [CrossRef]
- Abdalla, A.K.; Ayyash, M.M.; Olaimat, A.N.; Osaili, T.M.; Al-Nabulsi, A.A.; Shah, N.P.; Holley, R. Exopolysaccharides as Antimicrobial Agents: Mechanism and Spectrum of Activity. Front. Microbiol. 2021, 12, 664395. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, J.; Barse, B.; Fais, A.; Delogu, G.L.; Kumar, A. Biopolymer: A Sustainable Material for Food and Medical Applications. Polymers 2022, 14, 983. [Google Scholar] [CrossRef] [PubMed]
- Goy, R.C.; De Britto, D.; Assis, O.B.G. A review of the antimicrobial activity of chitosan. Polímeros 2009, 19, 241–247. [Google Scholar] [CrossRef]
- Aullybux, A.A.; Puchooa, D.; Bahorun, T.; Jeewon, R. Correction to: Phylogenetics and antibacterial properties of exopolysaccharides from marine bacteria isolated from Mauritius seawater. Ann. Microbiol. 2019, 69, 973–974. [Google Scholar] [CrossRef]
- Lahiri, D.; Nag, M.; Dutta, B.; Dey, A.; Sarkar, T.; Pati, S.; Edinur, H.A.; Kari, Z.A.; Noor, N.H.M.; Ray, R.R. Bacterial Cellulose: Production, Characterization, and Application as Antimicrobial Agent. Int. J. Mol. Sci. 2021, 22, 12984. [Google Scholar] [CrossRef]
- Augimeri, R.V.; Varley, A.J.; Strap, J.L. Establishing a Role for Bacterial Cellulose in Environmental Interactions: Lessons Learned from Diverse Biofilm-Producing Proteobacteria. Front. Microbiol. 2015, 6, 1282. [Google Scholar] [CrossRef]
- Zainulabdeen, S.M.S.; Abdul, J.; Salmanand, S.; Khalaf, K. Purification and characterization of levan from Lactobacillus gasseri and its effect against Pseudomonas aeruginosa. Nat. Volatiles Essent. Oils 2021, 8, 5788–5808. [Google Scholar]
- Hamada, M.A.; Hassan, R.A.; Abdou, A.M.; Elsaba, Y.M.; Aloufi, A.S.; Sonbol, H.; Korany, S.M. Bio_fabricated levan polymer from Bacillus subtilis healing properties. Appl. Sci. 2022, 12, 6413. [Google Scholar] [CrossRef]
- Gokmen, G.G.; Silbir, S.; Goksungur, Y.; Kisla, D. The effect of microbial levan on the growth of microorganisms: Inhibitory or stimulatory? Fresenius Environ. Bull. 2020, 29, 1648–1654. [Google Scholar]
- Semjonovs, P.; Shakirova, L.; Treimane, R.; Shvirksts, K.; Auzina, L.; Cleenwerck, I.; Zikmanis, P. Production of extracellular fructans by Gluconobacter nephelii P1464. Lett. Appl. Microbiol. 2016, 62, 145–152. [Google Scholar] [CrossRef]
- Sima, F.; Mutlu, E.C.; Eroglu, M.S.; Sima, L.E.; Serban, N.; Ristoscu, C.; Petrescu, S.M.; Oner, E.T.; Mihailescu, I.N. Levan Nanostructured Thin Films by MAPLE Assembling. Biomacromolecules 2011, 12, 2251–2256. [Google Scholar] [CrossRef]
- González-Garcinuño, Á.; Masa, R.; Hernández, M.; Domínguez, Á.; Tabernero, A.; del Valle, E.M. Levan-Capped Silver Nanoparticles for Bactericidal Formulations: Release and Activity Modelling. Int. J. Mol. Sci. 2019, 20, 1502. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Shahid, S.; Lee, C.-S. Green Synthesis of Gold and Silver Nanoparticles Using Leaf Extract of Clerodendrum inerme; Characterization, Antimicrobial, and Antioxidant Activities. Biomolecules 2020, 10, 835. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, K.B.A.; Kalla, D.; Uppuluri, K.B.; Anbazhagan, V. Green synthesis of silver and gold nanoparticles employing levan, a biopolymer from Acetobacter xylinum NCIM 2526, as a reducing agent and capping agent. Carbohydr. Polym. 2014, 112, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.I.; Hwang, Y.; Sahu, A.; Min, K.; Sung, D.; Tae, G.; Chang, J.H. An injectable and physical levan-based hydrogel as a dermal filler for soft tissue augmentation. Biomater. Sci. 2018, 6, 2627–2638. [Google Scholar] [CrossRef] [PubMed]
- Byun, B.Y.; Lee, S.J.; Mah, J.H. Antipathogenic activity and preservative effect of levan (β-2,6-fructan), a multifunctional polysaccharide. Int. J. Food Sci. Technol. 2014, 49, 238–245. [Google Scholar] [CrossRef]
- Zheng, L.; Li, S.; Luo, J.; Wang, X. Latest Advances on Bacterial Cellulose-Based Antibacterial Materials as Wound Dressings. Front. Bioeng. Biotechnol. 2020, 8, 593768. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.A.; Albetran, H.M.; Alheshibri, M.H.; Timoumi, A.; Algarou, N.A.; Akhtar, S.; Slimani, Y.; Almessiere, M.A.; Alahmari, F.S.; Baykal, A.; et al. Synthesis of Electrospun TiO2 Nanofibers and Characterization of Their Antibacterial and Antibiofilm Potential against Gram-Positive and Gram-Negative Bacteria. Antibiotics 2020, 9, 572. [Google Scholar] [CrossRef] [PubMed]
- Radenkovs, V.; Juhnevica-Radenkova, K.; Jakovlevs, D.; Zikmanis, P.; Galina, D.; Valdovska, A. The Release of Non-Extractable Ferulic Acid from Cereal By-Products by Enzyme-Assisted Hydrolysis for Possible Utilization in Green Synthesis of Silver Nanoparticles. Nanomaterials 2022, 12, 3053. [Google Scholar] [CrossRef]
- Speck, J.C. The Lobry De Bruyn-Alberda Van Ekenstein Transformation. In Advances in Carbohydrate Chemistry; Wolfrom, M.L., Tipson, R.S., Eds.; Academic Press: Cambridge, MA, USA, 1958; Volume 13, pp. 63–103. [Google Scholar]
- Yang, G.; Lin, Q.; Wang, C.; Li, J.; Wang, J.; Zhou, J.; Wang, Y.; Wang, C. Synthesis and characterization of dextran-capped silver nanoparticles with enhanced antibacterial activity. J. Nanosci. Nanotechnol. 2012, 12, 3766–3774. [Google Scholar] [CrossRef]
- Radenkovs, V.; Juhnevica-Radenkova, K.; Kviesis, J.; Lazdina, D.; Valdovska, A.; Vallejo, F.; Lacis, G. Lignocellulose-Degrading Enzymes: A Biotechnology Platform for Ferulic Acid Production from Agro-Industrial Side Streams. Foods 2021, 10, 3056. [Google Scholar] [CrossRef]
- Kourmouli, A.; Valenti, M.; van Rijn, E.; Beaumont, H.J.E.; Kalantzi, O.-I.; Schmidt-Ott, A.; Biskos, G. Can disc diffusion susceptibility tests assess the antimicrobial activity of engineered nanoparticles? J. Nanopart. Res. 2018, 20, 62. [Google Scholar] [CrossRef]
- Jathore, N.R.; Bule, M.V.; Tilay, A.V.; Annapure, U.S. Microbial levan from Pseudomonas fluorescens: Characterization and medium optimization for enhanced production. Food Sci. Biotechnol. 2012, 21, 1045–1053. [Google Scholar] [CrossRef]
- Ibrahim, M.; Alaam, M.; El-Haes, H.; Jalbout, A.F.; de Leon, A. Analysis of the structure and vibrational spectra of glucose and fructose. Eclet. Quim. 2006, 31, 15–21. [Google Scholar] [CrossRef]
- Glišić, S.; Nikolić, G.; Cakić, M.; Trutić, N. Spectroscopic study of copper(II) complexes with carboxymethyl dextran and dextran sulfate. Russ. J. Phys. Chem. A 2015, 89, 1254–1262. [Google Scholar] [CrossRef]
- Lu, R.; Gan, W.; Wu, B.-H.; Zhang, Z.; Guo, Y.; Wang, H.-F. C−H Stretching Vibrations of Methyl, Methylene and Methine Groups at the Vapor/Alcohol (n = 1−8) Interfaces. J. Phys. Chem. B 2005, 109, 14118–14129. [Google Scholar] [CrossRef] [PubMed]
- Al-Qaysi, S.A.S.; Al-Haideri, H.; Al-Shimmary, S.M.; Abdulhameed, J.M.; Alajrawy, O.I.; Al-Halbosiy, M.M.; Moussa, T.A.A.; Farahat, M.G. Bioactive Levan-Type Exopolysaccharide Produced by Pantoea agglomerans ZMR7: Characterization and Optimization for Enhanced Production. J. Microbiol. Biotechnol. 2021, 31, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Herrera, A.; Rodríguez, H.; Torres-Lugo, M.; Rinaldi, C. Functionalization of magnetite (Fe 3O 4) nanoparticles for cancer treatment. In Proceedings of the 2006 NSTI Nanotechnology Conference Trade Show—NSTI Nanotech 2006 Technical Proceedings, Boston, MA, USA, 7–11 May 2006; Volume 2, pp. 75–78. [Google Scholar]
- Nikolić, G.S.; Cakić, M.D.; Glišić, S.; Cvetković, D.J.; Mitić, Ž.J.; Marković, D.Z. Study of green nanoparticles and biocom-plexes based on exopolysaccharide by modern Fourier transform spectroscopy. In Fourier Transforms—High-Tech Application and Current Trends; Nikolić, G., Cvetković, D., Cakić, M., Eds.; IntechOpen Limited: London, UK, 2017; pp. 149–174. ISBN 978-953-51-2894-6. [Google Scholar]
- Mummaleti, G.; Sarma, C.; Kalakandan, S.; Sivanandham, V.; Rawson, A.; Anandharaj, A. Optimization and extraction of edible microbial polysaccharide from fresh coconut inflorescence sap: An alternative substrate. LWT 2021, 138, 110619. [Google Scholar] [CrossRef]
- Nastase, R.; Tatibouët, J.-M.; Fourré, E. Depolymerization of inulin in the highly reactive gas phase of a non thermal plasma at atmospheric pressure. Plasma Process. Polym. 2018, 15, 1800067. [Google Scholar] [CrossRef]
- Mamay; Wahyuningrum, D.; Hertadi, R. Isolation and Characterization of Levan from Moderate Halophilic Bacteria Bacillus licheniformis BK AG21. Procedia Chem. 2015, 16, 292–298. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Yi, H. Potential antitumor and anti-inflammatory activities of an extracellular polymeric substance (EPS) from Bacillus subtilis isolated from a housefly. Sci. Rep. 2022, 12, 1383. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, M.; Amoozegar, M.A.; Ventosa, A. Screening and comparative assay of poly-hydroxyalkanoates produced by bacteria isolated from the Gavkhooni Wetland in Iran and evaluation of poly-β-hydroxybutyrate production by halotolerant bacterium Oceanimonas sp. GK1. Ann. Microbiol. 2015, 65, 517–526. [Google Scholar] [CrossRef]
- Liu, C.; Lu, J.; Lu, L.; Liu, Y.; Wang, F.; Xiao, M. Isolation, structural characterization and immunological activity of an exopolysaccharide produced by Bacillus licheniformis 8-37-0-1. Bioresour. Technol. 2010, 101, 5528–5533. [Google Scholar] [CrossRef] [PubMed]
- Hertadi, R.; Amari, M.M.S.; Ratnaningsih, E. Enhancement of antioxidant activity of levan through the formation of nanoparticle systems with metal ions. Heliyon 2020, 6, e04111. [Google Scholar] [CrossRef] [PubMed]
- Thakham, N.; Thaweesak, S.; Teerakulkittipong, N.; Traiosot, N.; Kaikaew, A.; Lirio, G.A.; Jangiam, W. Structural Characterization of Functional Ingredient Levan Synthesized by Bacillus siamensis Isolated from Traditional Fermented Food in Thailand. Int. J. Food Sci. 2020, 2020, 7352484. [Google Scholar] [CrossRef]
- Permatasari, N.U.; Ratnaningsih, E.; Hertadi, R. The Use of Response Surface Method in Optimization of Levan Production by Heterologous Expressed Levansucrase from Halophilic Bacteria Bacillus licheniformis BK2. IOP Conf. Ser. Earth Environ. Sci. 2018, 209, 012015. [Google Scholar] [CrossRef]
- Qin, Q.Q.; Xia, B.S.; Xiong, Y.; Zhang, S.X.; Luo, Y.B.; Hao, Y.L. Structural Characterization of the Exopolysaccharide Produced by Streptococcus thermophilus 05-34 and Its In Situ Application in Yogurt. J. Food Sci. 2011, 76, C1226–C1230. [Google Scholar] [CrossRef]
- Omar, M.M.; Awda, J.M. Production of levansucrase from local isolate Bacillus lichniformans MJ8 and enzymic synthesis and characterization of levan. Appl. Microbiol. Open Access 2022, 8, 1–9. [Google Scholar]
- Demirci, T.; Hasköylü, M.E.; Eroğlu, M.S.; Hemberger, J.; Öner, E.T. Levan-based hydrogels for controlled release of Amphotericin B for dermal local antifungal therapy of Candidiasis. Eur. J. Pharm. Sci. 2020, 145, 105255. [Google Scholar] [CrossRef]
- de Siqueira, E.C.; Rebouças, J.d.S.; Pinheiro, I.O.; Formiga, F.R. Levan-based nanostructured systems: An overview. Int. J. Pharm. 2020, 580, 119242. [Google Scholar] [CrossRef]
- Shimizu, N.; Abea, A.; Ushiyama, T.; Öner, E.T. Effect of temperature on the hydrolysis of levan treated with compressed hot water fluids. Food Sci. Nutr. 2020, 8, 2004–2014. [Google Scholar] [CrossRef]
- Pei, F.; Ma, Y.; Chen, X.; Liu, H. Purification and structural characterization and antioxidant activity of levan from Bacillus megaterium PFY-147. Int. J. Biol. Macromol. 2020, 161, 1181–1188. [Google Scholar] [CrossRef]
- Xu, W.; Liu, Q.; Bai, Y.; Yu, S.; Zhang, T.; Jiang, B.; Mu, W. Physicochemical properties of a high molecular weight levan from Brenneria sp. EniD312. Int. J. Biol. Macromol. 2018, 109, 810–818. [Google Scholar] [CrossRef]
- Chu, B. Laser Light Scattering Basic Principles and Practice, 2nd ed.; Academic Press: Cambridge, MA, USA, 1991; ISBN 9780323161886. [Google Scholar]
- Santos-Moriano, P.; Fernandez-Arrojo, L.; Poveda, A.; Jimenez-Barbero, J.; Ballesteros, A.O.; Plou, F.J. Levan versus fructooligosaccharide synthesis using the levansucrase from Zymomonas mobilis: Effect of reaction conditions. J. Mol. Catal. B Enzym. 2015, 119, 18–25. [Google Scholar] [CrossRef]
- Hou, Y.; Huang, F.; Yang, H.; Cong, H.; Zhang, X.; Xie, X.; Yang, H.; Tong, Q.; Luo, N.; Zhu, P.; et al. Factors affecting the production and molecular weight of levan in enzymatic synthesis by recombinant Bacillus subtilis levansucrase SacB-T305A. Polym. Int. 2021, 70, 185–192. [Google Scholar] [CrossRef]
- Newbrun, E.; Baker, S. Physico-chemical characteristics of the levan produced by Streptococcus salivarius. Carbohydr. Res. 1968, 6, 165–170. [Google Scholar] [CrossRef]
- Osman, A.; Lin, E.; Hwang, D.S. A sticky carbohydrate meets a mussel adhesive: Catechol-conjugated levan for hemostatic and wound healing applications. Carbohydr. Polym. 2023, 299, 120172. [Google Scholar] [CrossRef]
- Ua-Arak, T.; Jakob, F.; Vogel, R.F. Fermentation pH Modulates the Size Distributions and Functional Properties of Gluconobacter albidus TMW 2.1191 Levan. Front. Microbiol. 2017, 8, 807. [Google Scholar] [CrossRef]
- Zhang, H.; Ren, W.; Guo, Q.; Xiong, Z.; Wang, G.; Xia, Y.; Lai, P.; Yin, B.; Ai, L. Characterization of a yogurt-quality improving exopolysaccharide from Streptococcus thermophilus AR333. Food Hydrocoll. 2018, 81, 220–228. [Google Scholar] [CrossRef]
- Hundschell, C.S.; Jakob, F.; Wagemans, A.M. Molecular Weight Dependent Structure and Polymer Density of the Exopoly-Saccharide Levan. arXiv 2019, arXiv:1909.07737. [Google Scholar]
- Arvidson, S.A.; Rinehart, B.T.; Gadala-Maria, F. Concentration regimes of solutions of levan polysaccharide from Bacillus sp. Carbohydr. Polym. 2006, 65, 144–149. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, X.; Cai, L.; Chen, R.; Zhang, Q.; Wang, X. Determination of Levan from Bacillus licheniformis by Ultraviolet Spectrophotometry. Trop. J. Pharm. Res. 2015, 14, 679–685. [Google Scholar] [CrossRef]
- Simms, P.J.; Boyko, W.J.; Edwards, J.R. The structural analysis of a levan produced by Streptococcus salivarius SS2. Carbohydr. Res. 1990, 208, 193–198. [Google Scholar] [CrossRef]
- Joaquim, E.O.; Hayashi, A.H.; Torres, L.M.B.; Figueiredo-Ribeiro, R.C.L.; Shiomi, N.; de Sousa, F.S.; Lago, J.H.G.; Carvalho, M.A.M. Chemical Structure and Localization of Levan, the Predominant Fructan Type in Underground Systems of Gomphrena marginata (Amaranthaceae). Front. Plant Sci. 2018, 9, 1745. [Google Scholar] [CrossRef]
- Murphy, D. Structure of a levan produced by Bacillus polymyxa. Can. J. Chem. 1952, 30, 872–878. [Google Scholar] [CrossRef]
- Lima, J.P.V.; Campos, P.T.A.; Paiva, M.F.; Linares, J.J.; Dias, S.C.L.; Dias, J.A. Dehydration of Fructose to 5-Hydroxymethylfurfural: Effects of Acidity and Porosity of Different Catalysts in the Conversion, Selectivity, and Yield. Chemistry 2021, 3, 1189–1202. [Google Scholar] [CrossRef]
- Bekers, M.; Upite, D.; Kaminska, E.; Laukevics, J.; Grube, M.; Vigants, A.; Linde, R. Stability of levan produced by Zymomonas mobilis. Process. Biochem. 2005, 40, 1535–1539. [Google Scholar] [CrossRef]
- Kennedy, J.F.; Stevenson, D.L.; White, C.A.; Viikari, L. The chromatographic behaviour of a series of fructo-oligosaccharides derived from levan produced by the fermentation of sucrose by Zymomonas mobilis. Carbohydr. Polym. 1989, 10, 103–113. [Google Scholar] [CrossRef]
- Matulová, M.; Husárová, S.; Capek, P.; Sancelme, M.; Delort, A.-M. NMR structural study of fructans produced by Bacillus sp. 3B6, bacterium isolated in cloud water. Carbohydr. Res. 2011, 346, 501–507. [Google Scholar] [CrossRef]
- Xu, Y.; Coda, R.; Shi, Q.; Tuomainen, P.; Katina, K.; Tenkanen, M. Exopolysaccharides Production during the Fermentation of Soybean and Fava Bean Flours by Leuconostoc mesenteroides DSM 20343. J. Agric. Food Chem. 2017, 65, 2805–2815. [Google Scholar] [CrossRef]
- Kemala, P.; Idroes, R.; Khairan, K.; Ramli, M.; Jalil, Z.; Idroes, G.M.; Tallei, T.E.; Helwani, Z.; Safitri, E.; Iqhrammullah, M.; et al. Green Synthesis and Antimicrobial Activities of Silver Nanoparticles Using Calotropis gigantea from Ie Seu-Um Geothermal Area, Aceh Province, Indonesia. Molecules 2022, 27, 5310. [Google Scholar] [CrossRef]
- Li, J.; Tian, X.; Habasi, M.; Chen, K.; Pang, N.; Hu, P.; Aisa, H.A. A phytoreduction route for selective synthesis of highly stable Ag and Ag:AgCl hybrid nanocolloids. CrystEngComm 2012, 14, 7621–7625. [Google Scholar] [CrossRef]
- Hussein, E.A.M.; Mohammad, A.A.-H.; Harraz, F.A.; Ahsan, M.F. Biologically Synthesized Silver Nanoparticles for Enhancing Tetracycline Activity Against Staphylococcus aureus and Klebsiella pneumoniae. Braz. Arch. Biol. Technol. 2019, 62, e19180266. [Google Scholar] [CrossRef]
- Jeong, S.-H.; Choi, H.; Kim, J.Y.; Lee, T.-W. Silver-Based Nanoparticles for Surface Plasmon Resonance in Organic Optoelectronics. Part. Part. Syst. Charact. 2015, 32, 164–175. [Google Scholar] [CrossRef]
- Rosa, R.M.; Silva, J.C.; Sanches, I.S.; Henriques, C. Simultaneous photo-induced cross-linking and silver nanoparticle formation in a PVP electrospun wound dressing. Mater. Lett. 2017, 207, 145–148. [Google Scholar] [CrossRef]
- Alex, K.V.; Pavai, P.T.; Rugmini, R.; Prasad, M.S.; Kamakshi, K.; Sekhar, K.C. Green Synthesized Ag Nanoparticles for Bio-Sensing and Photocatalytic Applications. ACS Omega 2020, 5, 13123–13129. [Google Scholar] [CrossRef]
- Carré-Rangel, L.; Alonso-Nuñez, G.; Espinoza-Gómez, H.; Flores-López, L.Z. Green Synthesis of Silver Nanoparticles: Effect of Dextran Molecular Weight Used as Stabilizing-Reducing Agent. J. Nanosci. Nanotechnol. 2015, 15, 9849–9855. [Google Scholar] [CrossRef]
- Saravanan, L.; Diwakar, S.; Mohankumar, R.; Pandurangan, A.; Jayavel, R. Synthesis, Structural and Optical Properties of PVP Encapsulated CdS Nanoparticles. Nanomater. Nanotechnol. 2011, 1, 1. [Google Scholar] [CrossRef]
- Kutsevol, N.; Chumachenko, V.; Rawiso, M.; Shyichuk, A. Green synthesis of silver nanoparticles using dextran-graft-polyacrylamide as template. Micro Nano Lett. 2016, 11, 256–259. [Google Scholar] [CrossRef]
- Cakić, M.; Glišić, S.; Nikolić, G.; Nikolić, G.M.; Cakić, K.; Cvetinov, M. Synthesis, characterization and antimicrobial activity of dextran sulphate stabilized silver nanoparticles. J. Mol. Struct. 2016, 1110, 156–161. [Google Scholar] [CrossRef]
- Bubonja-Šonje, M.; Knežević, S.; Abram, M. Challenges to antimicrobial susceptibility testing of plant-derived polyphenolic compounds. Arh. Hig. Rada Toksikol. 2020, 71, 300–311. [Google Scholar] [CrossRef]
- Biliuta, G.; Bostănaru-Iliescu, A.-C.; Mareș, M.; Pavlov-Enescu, C.; Năstasă, V.; Burduniuc, O.; Coseri, S. Antibacterial and Antifungal Silver Nanoparticles with Tunable Size Embedded in Various Cellulose-Based Matrices. Molecules 2022, 27, 6680. [Google Scholar] [CrossRef]
- Malik, M.; Iqbal, M.A.; Malik, M.; Raza, M.A.; Shahid, W.; Choi, J.R.; Pham, P.V. Biosynthesis and Characterizations of Silver Nanoparticles from Annona squamosa Leaf and Fruit Extracts for Size-Dependent Biomedical Applications. Nanomaterials 2022, 12, 616. [Google Scholar] [CrossRef] [PubMed]
- Sahu, N.; Soni, D.; Chandrashekhar, B.; Satpute, D.B.; Saravanadevi, S.; Sarangi, B.K.; Pandey, R.A. Synthesis of silver nanoparticles using flavonoids: Hesperidin, naringin and diosmin, and their antibacterial effects and cytotoxicity. Int. Nano Lett. 2016, 6, 173–181. [Google Scholar] [CrossRef]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa Biofilms: Host Response and Clinical Implications in Lung Infections. Am. J. Respir. Cell Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef] [PubMed]
- El Din, S.N.; El-Tayeb, T.A.; Abou-Aisha, K.; El-Azizi, M. In vitro and in vivo antimicrobial activity of combined therapy of silver nanoparticles and visible blue light against Pseudomonas aeruginosa. Int. J. Nanomed. 2016, 11, 1749–1758. [Google Scholar] [CrossRef]
- Abeer Mohammed, A.B.; Abd Elhamid, M.M.; Khalil, M.K.M.; Ali, A.S.; Abbas, R.N. The potential activity of biosynthesized silver nanoparticles of Pseudomonas aeruginosa as an antibacterial agent against multidrug-resistant isolates from intensive care unit and anticancer agent. Environ. Sci. Eur. 2022, 34, 109. [Google Scholar] [CrossRef]
- Edwards-Jones, V. Silver nanoparticles: An overview of scientific toxicity and safety data and introduction of a new dressing, Venus Ag. Wounds UK 2022, 18, 22–29. [Google Scholar]
- Vinicius de Oliveira Brisola Maciel, M.; da Rosa Almeida, A.; Machado, M.H.; Elias, W.C.; Gonçalves da Rosa, C.; Teixeira, G.L.; Noronha, C.M.; Bertoldi, F.C.; Nunes, M.R.; Dutra de Armas, R.; et al. Green synthesis, characteristics and antimicrobial activity of silver nanoparticles mediated by essential oils as reducing agents. Biocatal. Agric. Biotechnol. 2020, 28, 101746. [Google Scholar] [CrossRef]
- Ansar, S.; Tabassum, H.; Aladwan, N.S.M.; Ali, M.N.; Almaarik, B.; AlMahrouqi, S.; Abudawood, M.; Banu, N.; Alsubki, R. Eco friendly silver nanoparticles synthesis by Brassica oleracea and its antibacterial, anticancer and antioxidant properties. Sci. Rep. 2020, 10, 18564. [Google Scholar] [CrossRef]
- Ashour, A.A.; Raafat, D.; El-Gowelli, H.M.; El-Kamel, A.H. Green synthesis of silver nanoparticles using cranberry powder aqueous extract: Characterization and antimicrobial properties. Int. J. Nanomed. 2015, 10, 7207–7221. [Google Scholar] [CrossRef]
- Latimer, J.; Forbes, S.; McBain, A.J. Attenuated Virulence and Biofilm Formation in Staphylococcus aureus following Sublethal Exposure to Triclosan. Antimicrob. Agents Chemother. 2012, 56, 3092–3100. [Google Scholar] [CrossRef]
- Najafi, M.; Moghaddam, M.N.; Yousefi, E. The Effect of Silver Nanoparticles on Pyocyanin Production of Pseudomonas aeruginosa Isolated from Clinical Specimens. Avicenna J. Med. Biotechnol. 2021, 13, 98–103. [Google Scholar] [CrossRef]
- Qureshi, R.; Qamar, M.U.; Shafique, M.; Muzammil, S.; Rasool, M.H.; Ahmad, I.; Ejaz, H. Antibacterial efficacy of silver nanoparticles against metallo-beta-lactamase (bla(NDM), bla(VIM), bla(OXA)) producing clinically isolated Pseudomonas aeruginosa. Pak. J. Pharm. Sci. 2021, 34, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.D.; Olson, M.E. Current concepts in biofilm formation of Staphylococcus epidermidis. Futur. Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Swolana, D.; Kępa, M.; Idzik, D.; Dziedzic, A.; Kabała-Dzik, A.; Wąsik, T.J.; Wojtyczka, R.D. The Antibacterial Effect of Silver Nanoparticles on Staphylococcus epidermidis Strains with Different Biofilm-Forming Ability. Nanomaterials 2020, 10, 1010. [Google Scholar] [CrossRef]
- Dove, A.S.; Dzurny, D.I.; Dees, W.R.; Qin, N.; Nunez Rodriguez, C.C.; Alt, L.A.; Ellward, G.L.; Best, J.A.; Rudawski, N.G.; Fujii, K.; et al. Silver nanoparticles enhance the efficacy of aminoglycosides against antibiotic-resistant bacteria. Front. Microbiol. 2023, 13, 1064095. [Google Scholar] [CrossRef]




| Component | Chemical Formula | Amount, g L−1 |
|---|---|---|
| Bacteriological peptone | – | 10.0 |
| Beef extract | – | 10.0 |
| Yeast extract | – | 5.0 |
| Sucrose | C12H22O11 | 50.0 |
| Dipotassium hydrogen phosphate | K2HPO4 | 2.0 |
| Sodium acetate | C2H3NaO2 | 5.0 |
| Diammonium citrate | C6H14N2O7 | 2.0 |
| Magnesium sulfate | MgSO4 | 0.2 |
| Manganous sulfate | MnSO4 | 0.05 |
| Tween® 80 | C64H124O26 | 1.0 |
| Wavenumber, cm−1 | Assignment |
|---|---|
| IR | |
| 3524–3314 | νOH |
| 3015–2870 | νCH symmetric/asymmetric |
| 1647 | νCH |
| 1637 | δC=O |
| 1458 | δCH2 + δOCH + δCCH |
| 1410 | δCH3 |
| 1380–1430 | δOCH + δCOH + δCCH |
| 1340 | δCCH + δOCH |
| 1265 | Fructose δOH in plane, δCCO |
| 1224, 1200 | Glucose δCH + δOH in plane |
| 1149–995 | νCO + νCC + δCCC |
| 977 | νCO + δCCO |
| 923–916 | νCO + νCCH + νs ring of pyranose |
| 873 | νCC + δCCH + δCH fructose |
| 818–839 | δCH |
| 783,775 | δCCH + δCCO |
| 627 | CH2 + CH |
| 621 | CH2 |
| Replicate | Mw, g moL−1 | Mn, g moL−1 | PD | Rh, nm | IV, dL g−1 | Recovery, % |
|---|---|---|---|---|---|---|
| Bacterial levan biosynthesized by Streptococcus salivarius K12 | ||||||
| 1 | 15.146000 | 14.127000 | 1.07 | 36.3 | 0.21 | 54.3 |
| 2 | 15.724000 | 14.508000 | 1.08 | 35.3 | 0.19 | 55.0 |
| Average | 15.435000 | 14.317500 | 1.08 | 35.8 | 0.20 | 54.7 |
| STDEV | 0.409 | 0.269 | 0.007 | 0.707 | 0.014 | 0.495 |
| Bacterial levan biosynthesized by Leuconostoc mesenteroides DSM 20343 | ||||||
| 1 | 26.700 | 12.500 | 2.14 | 3.5 | 0.14 | 81.8 |
| 2 | 26.400 | 12.000 | 2.20 | 3.5 | 0.14 | 80.6 |
| Average | 26.600 | 12.300 | 2.17 | 3.5 | 0.14 | 81.2 |
| STDEV | 0.212 | 0.354 | 0.042 | 0.000 | 0.000 | 0.849 |
| Carb | BL_S. salivarius K12 | BL_L. mesenteroides DSM 20343 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Free | Bound, 0.1 M HCl 3 h | Bound, 0.1 M HCl 6 h | Bound, 0.2 M HCl 6 h | Bound, 0.5 M HCl 6 h | Bound, 1.0 M TCA 6 h | Bound, 0.05 M oxalic acid 3 h | Free | Bound, 0.1 M HCl 3 h | Bound, 0.1 M HCl 6 h | Bound, 0.2 M HCl 6 h | Bound, 0.5 M HCl 6 h | Bound, 1.0 M TCA 6 h | Bound, 0.05 M Oxalic Acid 3 h | |
| Gly | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Xyl | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Ara | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Fru | 0.4 ± 0.0 f | 85.7 ± 0.1 b | 84.9 ± 0.1 b | 77.1 ± 0.1 d | 56.9 ± 0.1 e | 80.2 ± 0.1 b | 92.0 ± 0.0 a | 0.1 ± 0.0 g | 58.1 ± 0.0 c | 60.6 ± 0.00 b | 53.1 ± 0.1 e | 38.4 ± 0.0 f | 55.0 ± 0.0 d | 62.3 ± 0.0 a |
| Glu | 0.7 ± 0.0 e | 2.5 ± 0.0 d | 4.7 ± 0.1 b | 4.8 ± 0.0 b | 5.8 ± 0.0 a | 4.9 ± 0.1 b | 4.2 ± 0.1 c | 0.4 ± 0.0 c | BLQ | BLQ | 5.3 ± 0.0 b | 10.3 ± 0.0 a | 5.0 ± 0.1 b | BLQ |
| Gala | 0.3 ± 0.0 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 2.5 ± 0.0 d | 7.3 ± 0.1 c | 12.7 ± 0.1 a | 12.2 ± 0.1 ab | 11.9 ± 0.0 b | 12.2 ± 0.0 a | 12.6 ± 0.0 a |
| Suc | 1.2 ± 0.0 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 0.05 ± 0.0 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Mal | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Lac | 0.1 ± 0.0 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 0.05 ± 0.0 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Tot | 2.7 ± 0.0 f | 88.2 ± 0.2 b | 89.6 ± 0.2 b | 81.9 ± 0.1 d | 62.7 ± 0.1 e | 85.1 ± 0.2 c | 96.2 ± 0.1 a | 3.1 ± 0.0 f | 65.4 ± 0.1 e | 73.3 ± 0.1 a | 70.6 ± 0.2 b | 60.6 ± 0.0 d | 72.2 ± 0.1 c | 74.9 ± 0.1 a |
| Test Culture | Average Zone of Inhibition, mm | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BLss@AgNPs 20 | BLlm@AgNPs 20 | BLss@AgNPs 50 | BLss@AgNPs 500 | BLdex@AgNPs 500 | ||||||
| D | W | D | W | D | W | D | W | D | W | |
| Pseudomonas aeruginosa ATCC 27853 | 10.1 ± 0.0 a | 14.1 ± 0.2 c | 10.1 ± 0.1 a | 14.3 ± 0.1 c | 9.2 ± 0.0 ab | 17.4 ± 0.2 b | 8.1 ± 0.0 b | 21.2 ± 0.2 a | 9.3 ± 0.1 ab | 10.1 ± 0.1 d |
| Staphylococcus aureus ATCC 6538P | 10.2 ± 0.0 ab | 11.2 ± 0.1 b | 10.3 ± 0.1 ab | 11.4 ± 0.1 b | 11.1 ± 0.1 a | 14.2 ± 0.1 a | 10.1 ± 0.1 ab | 15.5 ± 0.3 a | 9.1 ± 0.0 b | 12.2 ± 0.2 b |
| Escherichia coli ATCC 25922 | 9.3 ± 0.0 bc | 13.1 ± 0.1 c | 10.2 ± 0.0 ab | 13.4 ± 0.1 c | 10.3 ± 0.1 ab | 15.4 ± 0.1 b | 11.1 ± 0.1 a | 19.3 ± 0.1 a | 8.1 ± 0.0 c | 10.3 ± 0.2 d |
| Enterococcus aerogenes ATCC 13048 | 8.1 ± 0.0 ab | 12.4 ± 0.0 b | 9.1 ± 0.0 a | 12.4 ± 0.0 b | 8.2 ± 0.0 ab | 12.4 ± 0.0 b | 9.2 ± 0.0 a | 15.1 ± 0.2 a | 7.2 ± 0.0 b | 6.1 ± 0.0 c |
| Pseudomonas aeruginosa * | 9.4 ± 0.0 a | 6.1 ± 0.0 a | 9.1 ± 0.0 a | 6.2 ± 0.0 a | 9.3 ± 0.0 a | 6.1 ± 0.0 a | 9.4 ± 0.0 a | 6.2 ± 0.0 a | 9.4 ± 0.1 a | 6.2 ± 0.0 a |
| Staphylococcus epidermidis * | 9.1 ± 0.1 b | 11 ± 0.0 c | 10.2 ± 0.2 ab | 13.3 ± 0.1 b | 11.1 ± 0.1 a | 15.1 ± 0.1 a | 10.2 ± 0.0 ab | 16.2 ± 0.1 a | 9.3 ± 0.1 b | 11.1 ± 0.1 c |
| Escherichia coli * | 10.2 ± 0.1 a | 6.2 ± 0.0 d | 10.1 ± 0.2 a | 9.2 ± 0.2 c | 11.1 ± 0.0 a | 11.3 ± 0.1 b | 11.2 ± 0.1 a | 14.4 ± 0.2 a | 10.1 ± 0.1 a | 6.1 ± 0.0 d |
| Enterococcus faecium * | 9.3 ± 0 b | 8.2 ± 0.0 c | 9.2 ± 0.0 b | 11.2 ± 0.0 b | 11.3 ± 0.1 a | 13.1 ± 0.0 a | 8.1 ± 0.0 b | 14.3 ± 0.1 a | 9.4 ± 0.0 b | 8.1 ± 0.0 c |
| Test Culture | Average MBC and MIC, µL mL−1 AgNPs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BLss@AgNPs 20 | BLlm@AgNPs 20 | BLss@AgNPs 50 | BLss@AgNPs 500 | BLdex@AgNPs 500 | ||||||
| MBC (95%) | MIC | MBC (95%) | MIC | MBC (95%) | MIC | MBC (95%) | MIC | MBC (95%) | MIC | |
| Pseudomonas aeruginosa ATCC 27853 | 5.2 ± 0.1 a | 2.6 ± 0.0 b | 5.2 ± 0.1 a | 5.2 ± 0.0 a | 5.2 ± 0.0 a | 5.2 ± 0.0 a | 5.2 ± 0.0 a | 5.2 ± 0.4 a | 2.6 ± 0.1 b | 2.6 ± 0.0 b |
| Staphylococcus aureus ATCC 6538P | 41.7 ± 0.3 b | 20.8 ± 0.3 b | 41.7 ± 0.7 b | 20.8 ± 0.1 b | 20.8 ± 0.3 c | 20.8 ± 0.4 b | 83.3 ± 2.4 a | 83.3 ± 2.1 a | 41.7 ± 0.6 b | 20.8 ± 0.8 b |
| Escherichia coli ATCC 25922 | 5.2 ± 0.0 b | 5.2 ± 0.0 a | 10.4 ± 0.1 a | 2.6 ± 0.0 b | 2.6 ± 0.0 c | 2.6 ± 0.0 b | 10.4 ± 0.4 a | 5.2 ± 0.3 a | 2.6 ± 0.0 c | 2.6 ± 0.0 b |
| Enterococcus aerogenes ATCC 13048 | 20.8 ± 0.4 a | 10.4 ± 0.1 b | 10.4 ± 0.0 b | 10.4 ± 0.1 b | 10.4 ± 0.0 b | 10.4 ± 0.1 b | 10.4 ± 0.3 b | 10.4 ± 0.4 b | 20.8 ± 0.8 a | 20.8 ± 0.7 a |
| Pseudomonas aeruginosa * | 5.2 ± 0.0 a | 5.2 ± 0.1 a | 5.2 ± 0.0 a | 5.2 ± 0.0 a | 5.2 ± 0.0 a | 5.2 ± 0.0 a | 5.2 ± 0.1 a | 5.2 ± 0.1 a | 5.2 ± 0.1 a | 5.2 ± 0.1 a |
| Staphylococcus epidermidis * | 20.8 ± 0.2 a | 10.4 ± 0.2 b | 20.8 ± 0.2 a | 10.4 ± 0.1 b | 20.8 ± 0.4 a | 10.4 ± 0.2 b | 20.8 ± 0.9 a | 10.4 ± 0.5 b | 20.8 ± 0.9 a | 10.4 ± 0.3 b |
| Escherichia coli * | 5.2 ± 0.0 b | 5.2 ± 0.0 b | 5.2 ± 0.0 b | 5.2 ± 0.0 b | 5.2 ± 0.1 b | 5.2 ± 0.0 b | 10.4 ± 0.4 a | 10.4 ± 0.4 a | 5.2 ± 0.1 b | 5.2 ± 0.1 b |
| Enterococcus faecium * | 20.8 ± 0.1 a | 5.2 ± 0.0 b | 10.4 ± 0.1 b | 10.4 ± 0.1 a | 10.4 ± 0.2 b | 5.2 ± 0.0 b | 10.4 ± 0.6 b | 10.4 ± 0.4 a | 20.8 ± 1.4 a | 5.2 ± 0.1 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radenkovs, V.; Valdovska, A.; Galina, D.; Cairns, S.; Jakovlevs, D.; Gaidukovs, S.; Cinkmanis, I.; Juhnevica-Radenkova, K. Elaboration of Nanostructured Levan-Based Colloid System as a Biological Alternative with Antimicrobial Activity for Applications in the Management of Pathogenic Microorganisms. Nanomaterials 2023, 13, 2969. https://doi.org/10.3390/nano13222969
Radenkovs V, Valdovska A, Galina D, Cairns S, Jakovlevs D, Gaidukovs S, Cinkmanis I, Juhnevica-Radenkova K. Elaboration of Nanostructured Levan-Based Colloid System as a Biological Alternative with Antimicrobial Activity for Applications in the Management of Pathogenic Microorganisms. Nanomaterials. 2023; 13(22):2969. https://doi.org/10.3390/nano13222969
Chicago/Turabian StyleRadenkovs, Vitalijs, Anda Valdovska, Daiga Galina, Stefan Cairns, Dmitrijs Jakovlevs, Sergejs Gaidukovs, Ingmars Cinkmanis, and Karina Juhnevica-Radenkova. 2023. "Elaboration of Nanostructured Levan-Based Colloid System as a Biological Alternative with Antimicrobial Activity for Applications in the Management of Pathogenic Microorganisms" Nanomaterials 13, no. 22: 2969. https://doi.org/10.3390/nano13222969
APA StyleRadenkovs, V., Valdovska, A., Galina, D., Cairns, S., Jakovlevs, D., Gaidukovs, S., Cinkmanis, I., & Juhnevica-Radenkova, K. (2023). Elaboration of Nanostructured Levan-Based Colloid System as a Biological Alternative with Antimicrobial Activity for Applications in the Management of Pathogenic Microorganisms. Nanomaterials, 13(22), 2969. https://doi.org/10.3390/nano13222969









