Surface Architecture Influences the Rigidity of Candida albicans Cells
Abstract
1. Introduction
2. Experimental
2.1. Surface Fabrication
2.2. Scanning Electron Microscopy
2.3. Atomic Force Microscopy
2.4. Surface Wettability
2.5. X-ray Photoelectron Spectrometry
2.6. Microorganism, Culture Conditions, and Sample Preparation
2.7. SEM Analysis
2.8. Confocal Laser Scanning Microscopy (CLSM) Analysis
2.9. Cell Topography
2.10. Cell Rigidity
2.11. Statistical Analysis
3. Results and Discussion
3.1. Surface Characterization
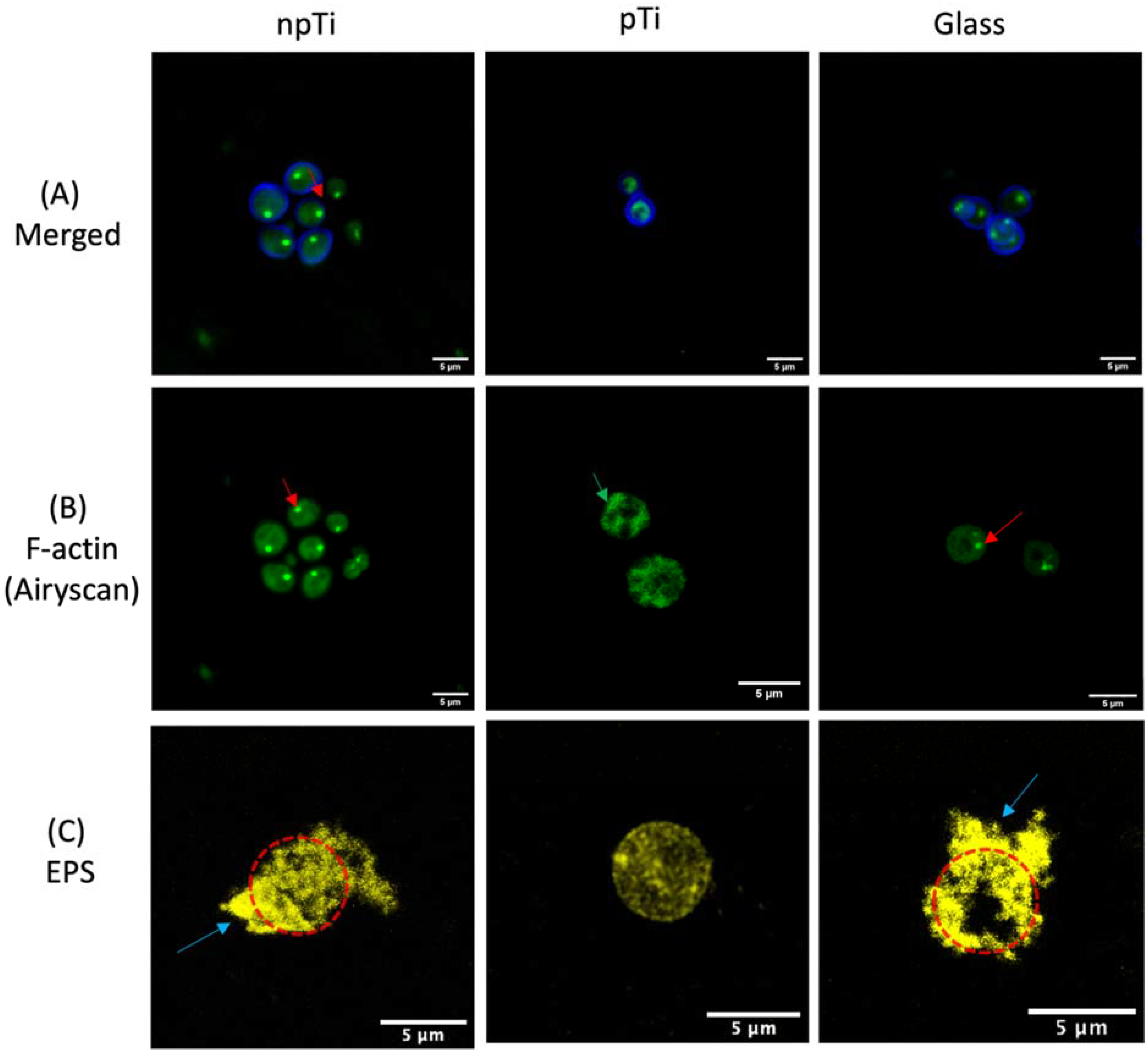
3.2. Cell Morphology of C. Albicans Attached on npTi, pTi, and Glass Surfaces
3.3. Mechanical Properties and Biofilm Formation of Attached C. Albicans Cells on npTi, pTi, and Glass Surfaces
3.4. Physiological Properties of C. Albicans Cells Attached on npTi, pTi, and Glass Surfaces
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kojic, E.M.; Darouiche, R.O. Candida Infections of Medical Devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldavs, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J. Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 2014, 370, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A. Molecular mechanisms of mechanosensing and their roles in fungal contact sensing. Nat. Rev. Microbiol. 2008, 6, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Cavalheiro, M.; Teixeira, M.C. Candida Biofilms: Threats, Challenges, and Promising Strategies. Front. Med. 2018, 5, 28. [Google Scholar] [CrossRef]
- Gow, N.A.; Brown, A.J.; Odds, F.C. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 2002, 5, 366–371. [Google Scholar] [CrossRef]
- Sterzenbach, T.; Helbig, R.; Hannig, C.; Hannig, M. Bioadhesion in the oral cavity and approaches for biofilm management by surface modifications. Clin. Oral Investig. 2020, 24, 4237–4260. [Google Scholar] [CrossRef]
- Liu, Y.S.; Lee, O.K. In Search of the Pivot Point of Mechanotransduction: Mechanosensing of Stem Cells. Cell Transplant. 2014, 23, 1–11. [Google Scholar] [CrossRef]
- Viela, F.; Granados, D.; Ayuso-Sacido, A.; Rodríguez, I. Biomechanical Cell Regulation by High Aspect Ratio Nanoimprinted Pillars. Adv. Funct. Mater. 2016, 26, 5599–5609. [Google Scholar] [CrossRef]
- Le Saux, G.; Bar-Hanin, N.; Edri, A.; Hadad, U.; Porgador, A.; Schvartzman, M. Nanoscale Mechanosensing of Natural Killer Cells is Revealed by Antigen-Functionalized Nanowires. Adv. Mater. 2019, 31, 1805954. [Google Scholar] [CrossRef]
- Fu, J.; Liu, X.; Tan, L.; Cui, Z.; Liang, Y.; Li, Z.; Zhu, S.; Zheng, Y.; Kwok Yeung, K.W.; Chu, P.K.; et al. Modulation of the Mechanosensing of Mesenchymal Stem Cells by Laser-Induced Patterning for the Acceleration of Tissue Reconstruction through the Wnt/β-Catenin Signaling Pathway Activation. Acta Biomater. 2020, 101, 152–167. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, D.; Du, W.; Zou, J.; Li, X.; Xie, J. Substrate Mechanics Dictate Cell-Cell Communication by Gap Junctions in Stem Cells from Human Apical Papilla. Acta Biomater. 2020, 107, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J. Topographically induced direct cell mechanotransduction. Med. Eng. Phys. 2005, 27, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Salou, L.; Hoornaert, A.; Louarn, G.; Layrolle, P. Enhanced osseointegration of titanium implants with nanostructured surfaces: An experimental study in rabbits. Acta Biomater. 2015, 11, 494–502. [Google Scholar] [CrossRef]
- Wandiyanto, J.V.; Linklater, D.; Tharushi Perera, P.G.; Orlowska, A.; Truong, V.K.; Thissen, H.; Ghanaati, S.; Baulin, V.; Crawford, R.J.; Juodkazis, S.; et al. Pheochromocytoma (PC12) Cell Response on Mechanobactericidal Titanium Surfaces. Materials 2018, 11, 605. [Google Scholar] [CrossRef] [PubMed]
- Senevirathne, S.W.M.A.I.; Hasan, J.; Mathew, A.; Woodruff, M.; Yarlagadda, P.K.D.V. Bactericidal efficiency of micro- and nanostructured surfaces: A critical perspective. RSC Adv. 2021, 11, 1883–1900. [Google Scholar] [CrossRef]
- Takai, E.; Costa, K.D.; Shaheen, A.; Hung, C.T.; Guo, X.E. Osteoblast Elastic Modulus Measured by Atomic Force Microscopy Is Substrate Dependent. Ann. Biomed. Eng. 2005, 33, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Abagnale, G.; Steger, M.; Nguyen, V.H.; Hersch, N.; Sechi, A.; Joussen, S.; Denecke, B.; Merkel, R.; Hoffmann, B.; Dreser, A.; et al. Surface topography enhances differentiation of mesenchymal stem cells towards osteogenic and adipogenic lineages. Biomaterials 2015, 61, 316–326. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Hasan, J.; Webb, H.K.; Gervinskas, G.; Juodkazis, S.; Truong, V.K.; Wu, A.H.F.; Lamb, R.N.; Baulin, V.A.; Watson, G.S.; et al. Bactericidal activity of black silicon. Nat. Commun. 2013, 4, 2838. [Google Scholar] [CrossRef]
- Aburto-Medina, A.; Le, P.H.; MacLaughlin, S.; Ivanova, E. Diversity of experimental designs for the fabrication of antifungal surfaces for the built environment. Appl. Microbiol. Biotechnol. 2021, 105, 2663–2674. [Google Scholar] [CrossRef]
- Razvag, Y.; Neve-Oz, Y.; Sherman, E.; Reches, M. Nanoscale Topography-Rigidity Correlation at the Surface of T Cells. ACS Nano 2019, 13, 346–356. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, Y.; Xu, D.; Alam, M.M.; Shui, L.; Chen, H. Cell Elasticity Measurement Using a Microfluidic Device with Real-Time Pressure Feedback. Lab Chip 2020, 20, 2343–2353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Guo, Y.; Liu, X.; Chen, X.G.; Wu, Q.; Chen, G.Q. Engineering Cell Wall Synthesis Mechanism for Enhanced PHB Accumulation in E. coli. Metab. Eng. 2018, 45, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Asgari, M.; Brulé, V.; Western, T.L.; Pasini, D. Nano-Indentation Reveals a Potential Role for Gradients of Cell Wall Stiffness in Directional Movement of The Resurrection Plant Selaginella lepidophylla. Sci. Rep. 2020, 10, 506. [Google Scholar] [CrossRef] [PubMed]
- Heinisch, J.J.; Rodicio, R. Physical and Chemical Stress Factors in Yeast. In Biology of Microorganisms on Grapes, in Must and in Wine; König, H., Unden, G., Fröhlich, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 275–291. [Google Scholar]
- Nguyen, D.H.K.; Loebbe, C.; Linklater, D.P.; Xu, X.; Vrancken, N.; Katkus, T.; Juodkazis, S.; Maclaughlin, S.; Baulin, V.; Crawford, R.J.; et al. The idiosyncratic self-cleaning cycle of bacteria on regularly arrayed mechano-bactericidal nanostructures. Nanoscale 2019, 11, 16455–16462. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Cheng, S.; Tanaka, Y.; Hosokawa, Y.; Yalikun, Y.; Li, M. Mechanical properties of single cells: Measurement methods and applications. Biotechnol. Adv. 2020, 45, 107648. [Google Scholar] [CrossRef] [PubMed]
- Le, P.H.; Nguyen, D.H.K.; Aburto-Medina, A.; Linklater, D.P.; Crawford, R.J.; MacLaughlin, S.; Ivanova, E.P. Nanoscale Surface Roughness Influences Candida albicans Biofilm Formation. ACS Appl. Bio Mater. 2020, 3, 8581–8591. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Truong, V.K.; Wang, J.Y.; Berndt, C.C.; Jones, R.T.; Yusuf, I.I.; Peake, I.; Schmidt, H.W.; Fluke, C.; Barnes, D.; et al. Impact of nanoscale roughness of titanium thin film surfaces on bacterial retention. Langmuir 2010, 26, 1973–1982. [Google Scholar] [CrossRef]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Open Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophoton. Int. 2004, 11, 36–42. [Google Scholar]
- Galdiero, E.; de Alteriis, E.; De Natale, A.; D’Alterio, A.; Siciliano, A.; Guida, M.; Lombardi, L.; Falanga, A.; Galdiero, S. Eradication of Candida albicans persister cell biofilm by the membranotropic peptide gH625. Sci. Rep. 2020, 10, 5780. [Google Scholar] [CrossRef]
- Hassan, A.N.; Frank, J.F.; Qvist, K.B. Direct observation of bacterial exopolysaccharides in dairy products using confocal scanning laser microscopy. J. Dairy Sci. 2002, 85, 1705–1708. [Google Scholar] [CrossRef]
- Adya, A.K.; Canetta, E.; Walker, G.M. Atomic force microscopic study of the influence of physical stresses on Saccharomyces cerevisiae and Schizosaccharomyces pombe. FEMS Yeast Res. 2006, 6, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.C.; Dimitriadis, E.K.; Horkay, F. Robust Strategies for Automated AFM Force Curve Analysis—II: Adhesion-Influenced Indentation of Soft, Elastic Materials. J. Biomech. Eng. 2007, 129, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, X.; Ren, B.; Cheng, L. The regulation of hyphae growth in Candida albicans. Virulence 2020, 11, 337–348. [Google Scholar] [CrossRef]
- Nguyen, D.H.K.; Wang, J.; Sbarski, I.; Juodkazis, S.; Crawford, R.J.; Ivanova, E.P. Influence of Amorphous, Carbon-Derived Wrinkled Surface Topologies on the Colonization of Pseudomonas aeruginosa Bacteria. Adv. Mater. Interfaces 2019, 6, 1801890. [Google Scholar] [CrossRef]
- Formosa, C.; Schiavone, M.; Martin-Yken, H.; François, J.M.; Duval, R.E.; Dague, E. Nanoscale effects of caspofungin against two yeast species, Saccharomyces cerevisiae and Candida albicans. Antimicrob. Agents Chemother. 2013, 57, 3498–3506. [Google Scholar] [CrossRef]
- Kheur, S.; Singh, N.; Bodas, D.; Rauch, J.-Y.; Jambhekar, S.; Kheur, M.; Rajwade, J. Nanoscale silver depositions inhibit microbial colonization and improve biocompatibility of titanium abutments. Colloids Surf. B Biointerfaces 2017, 159, 151–158. [Google Scholar] [CrossRef]
- Bhat, S.V.; Sultana, T.; Körnig, A.; McGrath, S.; Shahina, Z.; Dahms, T.E.S. Correlative Atomic Force Microscopy Quantitative Imaging-Laser Scanning Confocal Microscopy Quantifies the Impact of Stressors on Live Cells in Real-Time. Sci. Rep. 2018, 8, 8305. [Google Scholar] [CrossRef]
- Khoury, Z.H.; Vila, T.; Puthran, T.R.; Sultan, A.S.; Montelongo-Jauregui, D.; Melo, M.A.S.; Jabra-Rizk, M.A. The Role of Candida albicans Secreted Polysaccharides in Augmenting Streptococcus mutans Adherence and Mixed Biofilm Formation: In vitro and in vivo Studies. Front. Microbiol. 2020, 11, 307. [Google Scholar] [CrossRef]
- Akashi, T.; Kanbe, T.; Tanaka, K. The role of the cytoskeleton in the polarized growth of the germ tube in Candida albicans. Microbiology 1994, 140 Pt 2, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Walther, A.; Wendland, J. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich Syndrome protein homolog Wal1p. Eukaryot. Cell 2004, 3, 471–482. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pruyne, D.; Legesse-Miller, A.; Gao, L.; Dong, Y.; Bretscher, A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004, 20, 559–591. [Google Scholar] [CrossRef] [PubMed]
- Sheikh-Hamad, D.; Gustin, M.C. MAP kinases and the adaptive response to hypertonicity: Functional preservation from yeast to mammals. Am. J. Physiol. Ren. Physiol. 2004, 287, F1102–F1110. [Google Scholar] [CrossRef]
- Oberholzer, U.; Nantel, A.; Berman, J.; Whiteway, M. Transcript profiles of Candida albicans cortical actin patch mutants reflect their cellular defects: Contribution of the Hog1p and Mkc1p signaling pathways. Eukaryot. Cell 2006, 5, 1252–1265. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Schiavone, M.; Formosa-Dague, C.; Elsztein, C.; Teste, M.A.; Martin-Yken, H.; De Morais, M.A., Jr.; Dague, E.; François, J.M. Evidence for a Role for the Plasma Membrane in the Nanomechanical Properties of the Cell Wall as Revealed by an Atomic Force Microscopy Study of the Response of Saccharomyces cerevisiae to Ethanol Stress. Appl. Environ. Microbiol. 2016, 82, 4789–4801. [Google Scholar] [CrossRef]
- Quilès, F.; Accoceberry, I.; Couzigou, C.; Francius, G.; Noël, T.; El-Kirat-Chatel, S. AFM Combined to ATR-FTIR Reveals Candida Cell Wall Changes under Caspofungin Treatment. Nanoscale 2017, 9, 13731–13738. [Google Scholar] [CrossRef]
- Gonçalves, S.; Silva, P.M.; Felício, M.R.; de Medeiros, L.N.; Kurtenbach, E.; Santos, N.C. Psd1 Effects on Candida albicans Planktonic Cells and Biofilms. Front. Cell. Infect. Microbiol. 2017, 7, 249. [Google Scholar] [CrossRef]
- Ma, S.; Ge, W.; Yan, Y.; Huang, X.; Ma, L.; Li, C.; Yu, S.; Chen, C. Effects of Streptococcus sanguinis Bacteriocin on Deformation, Adhesion Ability, and Young’s Modulus of Candida albicans. Biomed. Res. Int. 2017, 2017, 5291486. [Google Scholar] [CrossRef]
- Çolak, A.; Ikeh, M.A.C.; Nobile, C.J.; Baykara, M.Z. In Situ Imaging of Candida albicans Hyphal Growth via Atomic Force Microscopy. mSphere 2020, 5, e00946-20. [Google Scholar] [CrossRef] [PubMed]
- Ene, I.V.; Adya, A.K.; Wehmeier, S.; Brand, A.C.; Maccallum, D.M.; Gow, N.A.R.; Brown, A.J.P. Host Carbon Sources Modulate Cell Wall Architecture, Drug Resistance and Virulence in A Fungal Pathogen. Cell. Microbiol. 2012, 14, 1319–1335. [Google Scholar] [CrossRef] [PubMed]
- Ene, I.V.; Walker, L.A.; Schiavone, M.; Lee, K.K.; Martin-Yken, H.; Dague, E.; Gow, N.A.R.; Munro, C.A.; Brown, A.J.P. Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance. mBio 2015, 6, e00986-15. [Google Scholar] [CrossRef]
- Yang, L.; Gao, Q.; Ge, L.; Zhou, Q.; Warszawik, E.M.; Bron, R.; Lai, K.W.C.; Van Rijn, P. Topography Induced Stiffness Alteration of Stem Cells Influences Osteogenic Differentiation. Biomater. Sci. 2020, 8, 2638–2652. [Google Scholar] [CrossRef] [PubMed]
- Alalwan, H.; Nile, C.J.; Rajendran, R.; McKerlie, R.; Reynolds, P.; Gadegaard, N.; Ramage, G. Nanoimprinting of biomedical polymers reduces candidal physical adhesion. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1045–1049. [Google Scholar] [CrossRef]
- Pessoa, R.S.; dos Santos, V.P.; Cardoso, S.B.; Doria, A.C.O.C.; Figueira, F.R.; Rodrigues, B.V.M.; Testoni, G.E.; Fraga, M.A.; Marciano, F.R.; Lobo, A.O.; et al. TiO2 coatings via atomic layer deposition on polyurethane and polydimethylsiloxane substrates: Properties and effects on C. albicans growth and inactivation process. Appl. Surf. Sci. 2017, 422, 73–84. [Google Scholar] [CrossRef]
- Nowlin, K.; Boseman, A.; Covell, A.; LaJeunesse, D. Adhesion-dependent rupturing of Saccharomyces cerevisiae on biological antimicrobial nanostructured surfaces. J. R. Soc. Interface 2015, 12, 20140999. [Google Scholar] [CrossRef]
- Kollu, N.V.; LaJeunesse, D.R. Cell Rupture and Morphogenesis Control of the Dimorphic Yeast Candida albicans by Nanostructured Surfaces. ACS Omega 2021, 6, 1361–1369. [Google Scholar] [CrossRef]
- Chaffin, W.L.; Lopez-Ribot, J.L.; Casanova, M.; Gozalbo, D.; Martinez, J.P. Cell wall and secreted proteins of Candida albicans: Identification, function, and expression. Microbiol. Mol. Biol. Rev. 1998, 62, 130–180. [Google Scholar] [CrossRef]
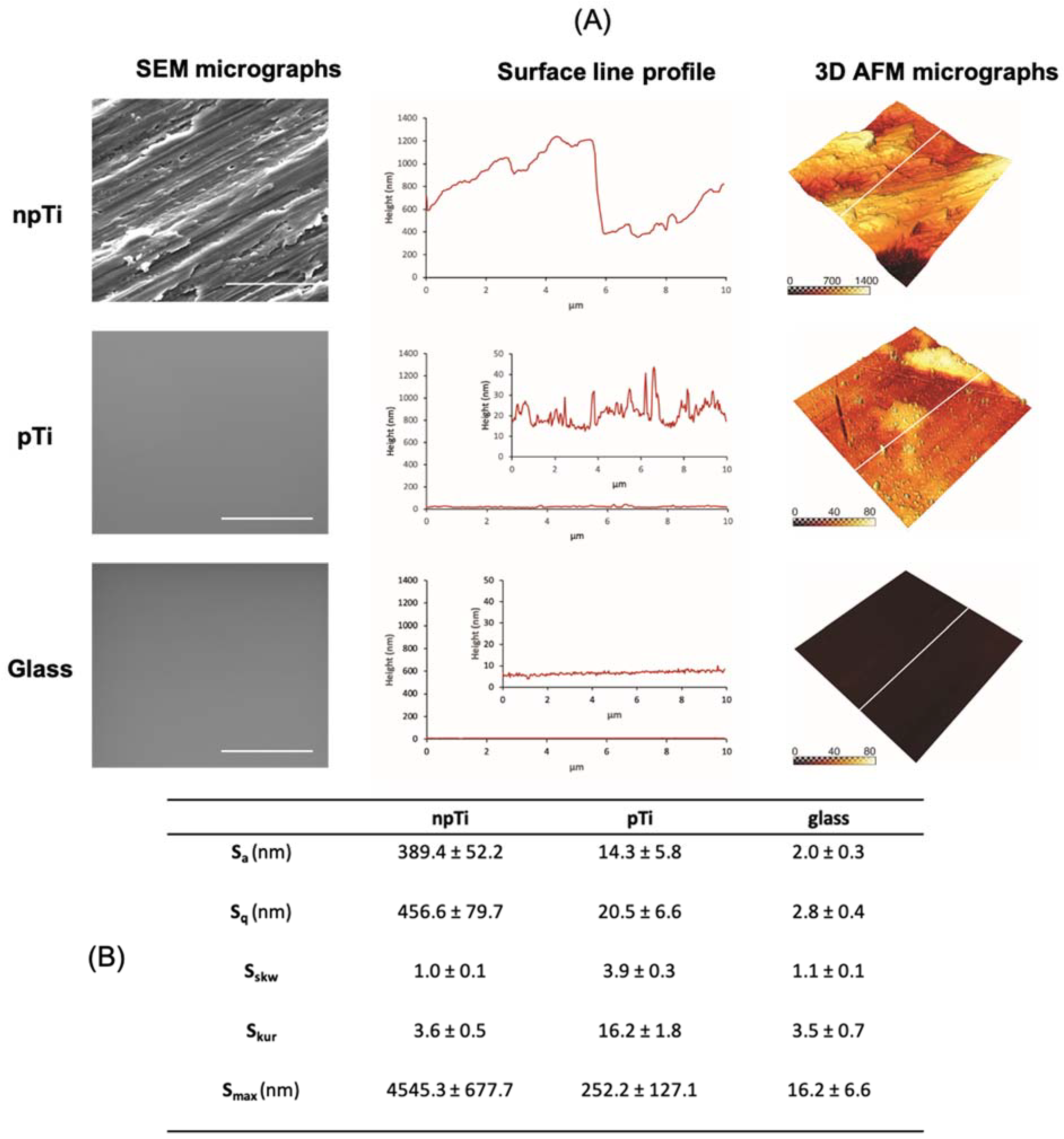

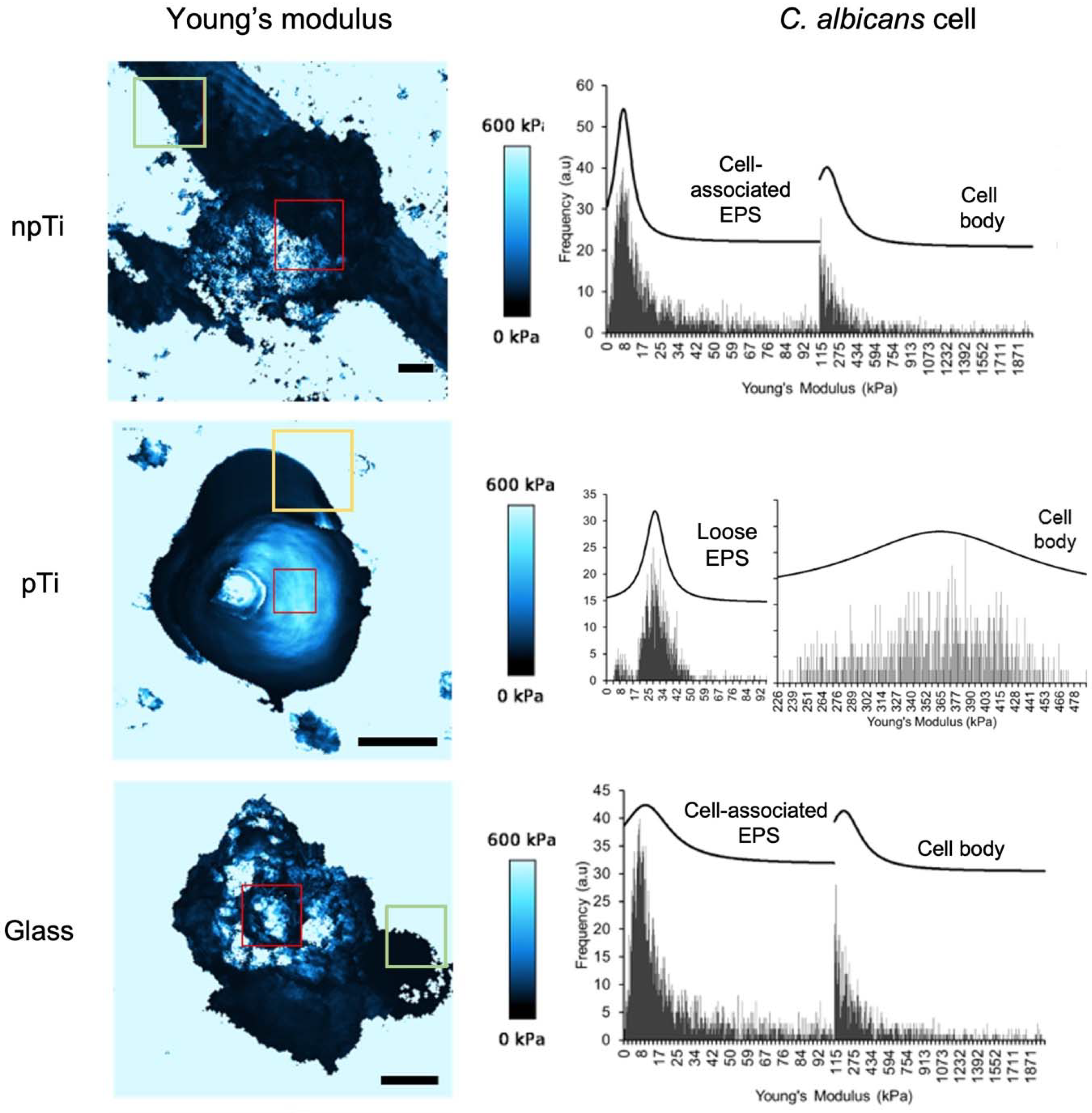
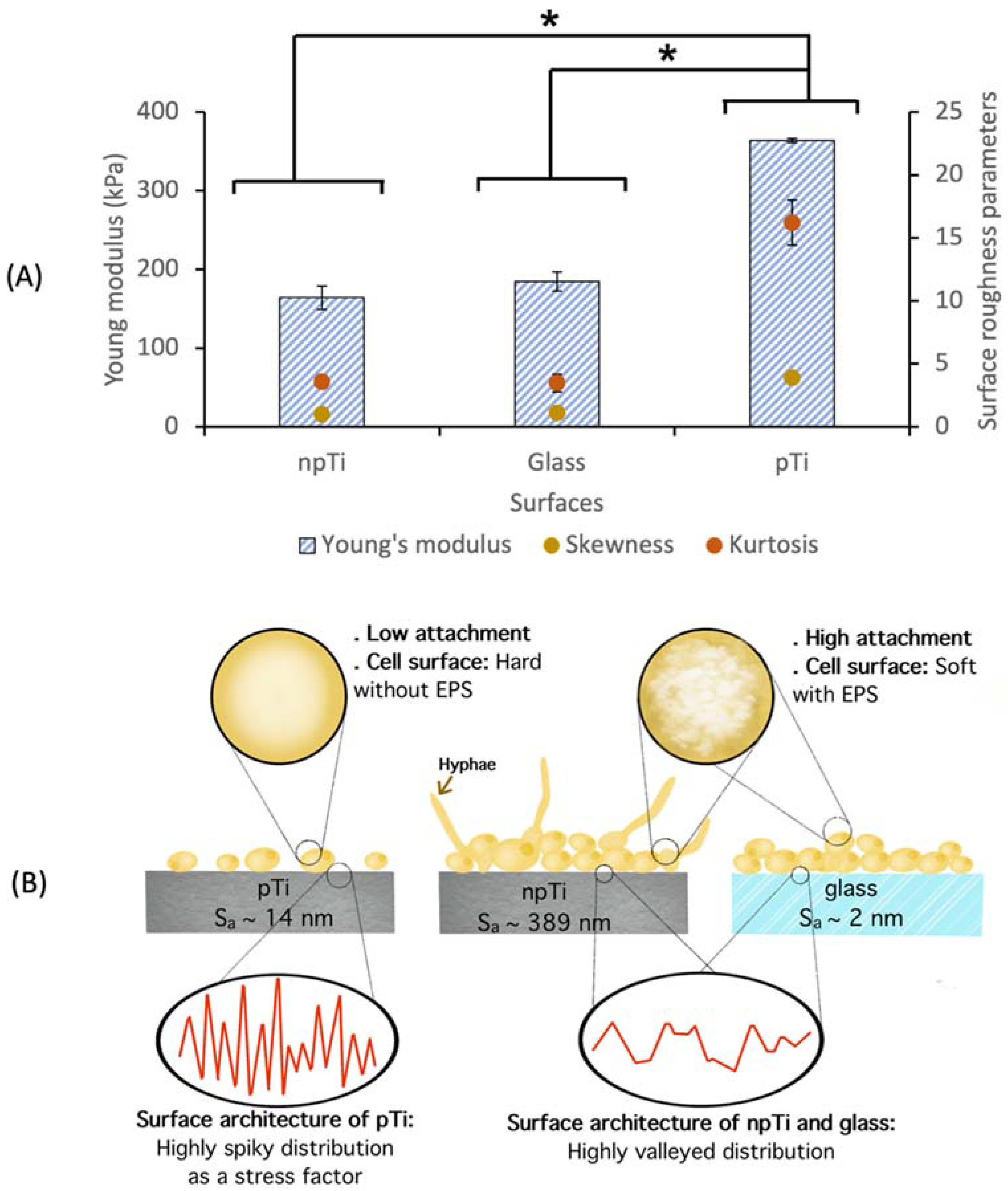
| npTi | pTi | Glass | ||
|---|---|---|---|---|
| C. albicans Cell Surface | Sa (µm) | 3.3 ± 0.2 | 2.6 ± 0.1 * | 3.3 ± 0.2 |
| Cell body (kPa) | 164.2 ± 15.0 | 363.8 ± 2.5 * | 184.6 ± 12.0 | |
| Cell-associated EPS (kPa) | 7.8 ± 0.1 | Not detected | 10.3 ± 0.7 | |
| Loose EPS (kPa) b | 9.1 ± 0.1 | 29.0 ± 0.2 | 2.2 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, P.H.; Nguyen, D.H.K.; Medina, A.A.; Linklater, D.P.; Loebbe, C.; Crawford, R.J.; MacLaughlin, S.; Ivanova, E.P. Surface Architecture Influences the Rigidity of Candida albicans Cells. Nanomaterials 2022, 12, 567. https://doi.org/10.3390/nano12030567
Le PH, Nguyen DHK, Medina AA, Linklater DP, Loebbe C, Crawford RJ, MacLaughlin S, Ivanova EP. Surface Architecture Influences the Rigidity of Candida albicans Cells. Nanomaterials. 2022; 12(3):567. https://doi.org/10.3390/nano12030567
Chicago/Turabian StyleLe, Phuc H., Duy H. K. Nguyen, Arturo Aburto Medina, Denver P. Linklater, Christian Loebbe, Russell J. Crawford, Shane MacLaughlin, and Elena P. Ivanova. 2022. "Surface Architecture Influences the Rigidity of Candida albicans Cells" Nanomaterials 12, no. 3: 567. https://doi.org/10.3390/nano12030567
APA StyleLe, P. H., Nguyen, D. H. K., Medina, A. A., Linklater, D. P., Loebbe, C., Crawford, R. J., MacLaughlin, S., & Ivanova, E. P. (2022). Surface Architecture Influences the Rigidity of Candida albicans Cells. Nanomaterials, 12(3), 567. https://doi.org/10.3390/nano12030567







