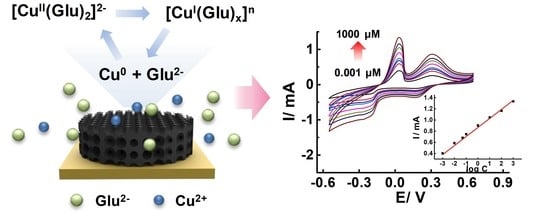
Porous Carbon Boosted Non-Enzymatic Glutamate Detection with Ultra-High Sensitivity in Broad Range Using Cu Ions
Abstract
:1. Introduction
2. Experimental
2.1. Chemicals and Apparatus
2.2. Preparation of Electrode Based on a Chitosan-Derived Carbon Foam
2.3. Characterizations
3. Results and Discussion
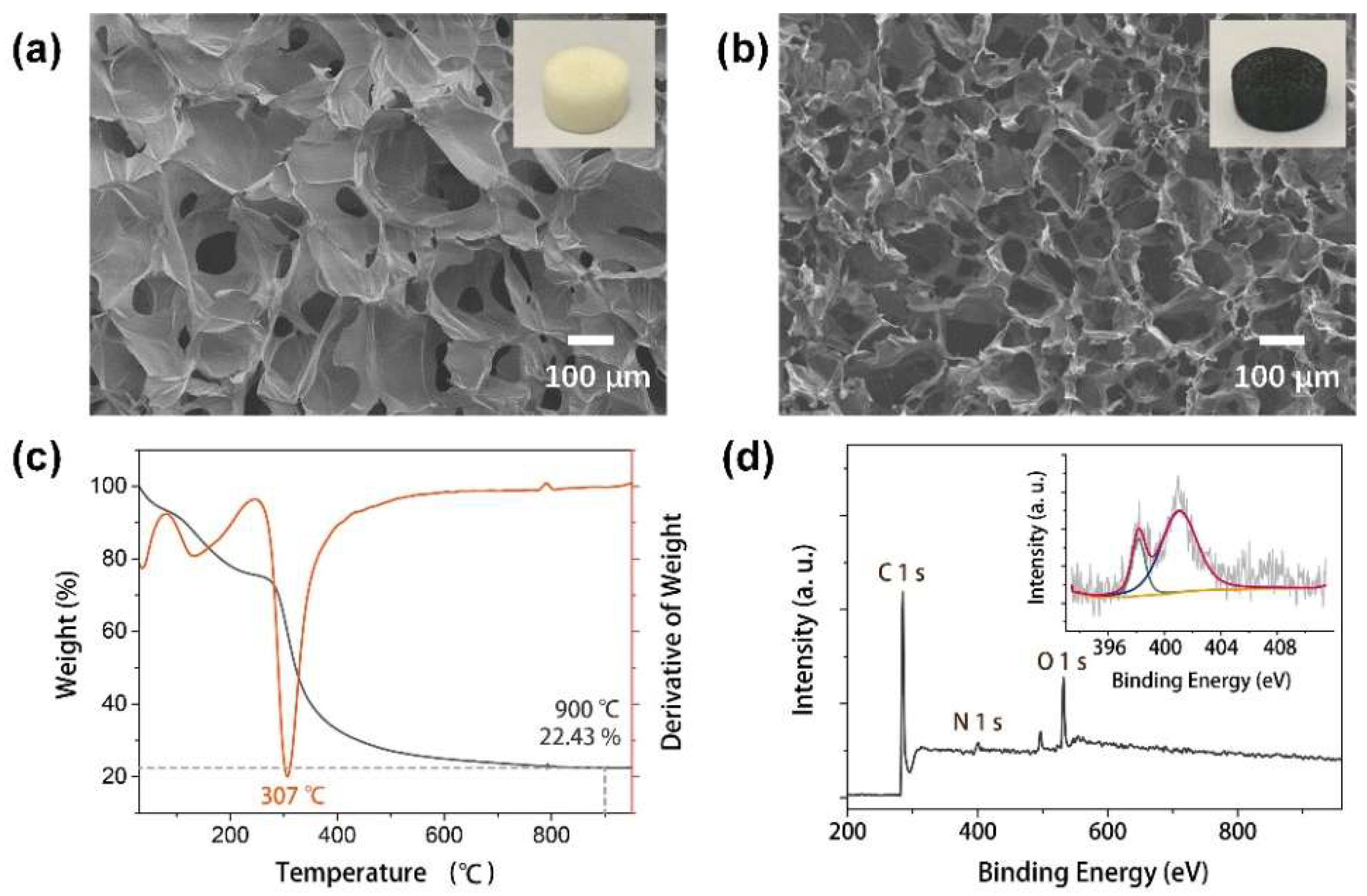
3.1. Chitosan-Derived Carbon Foam Electrode
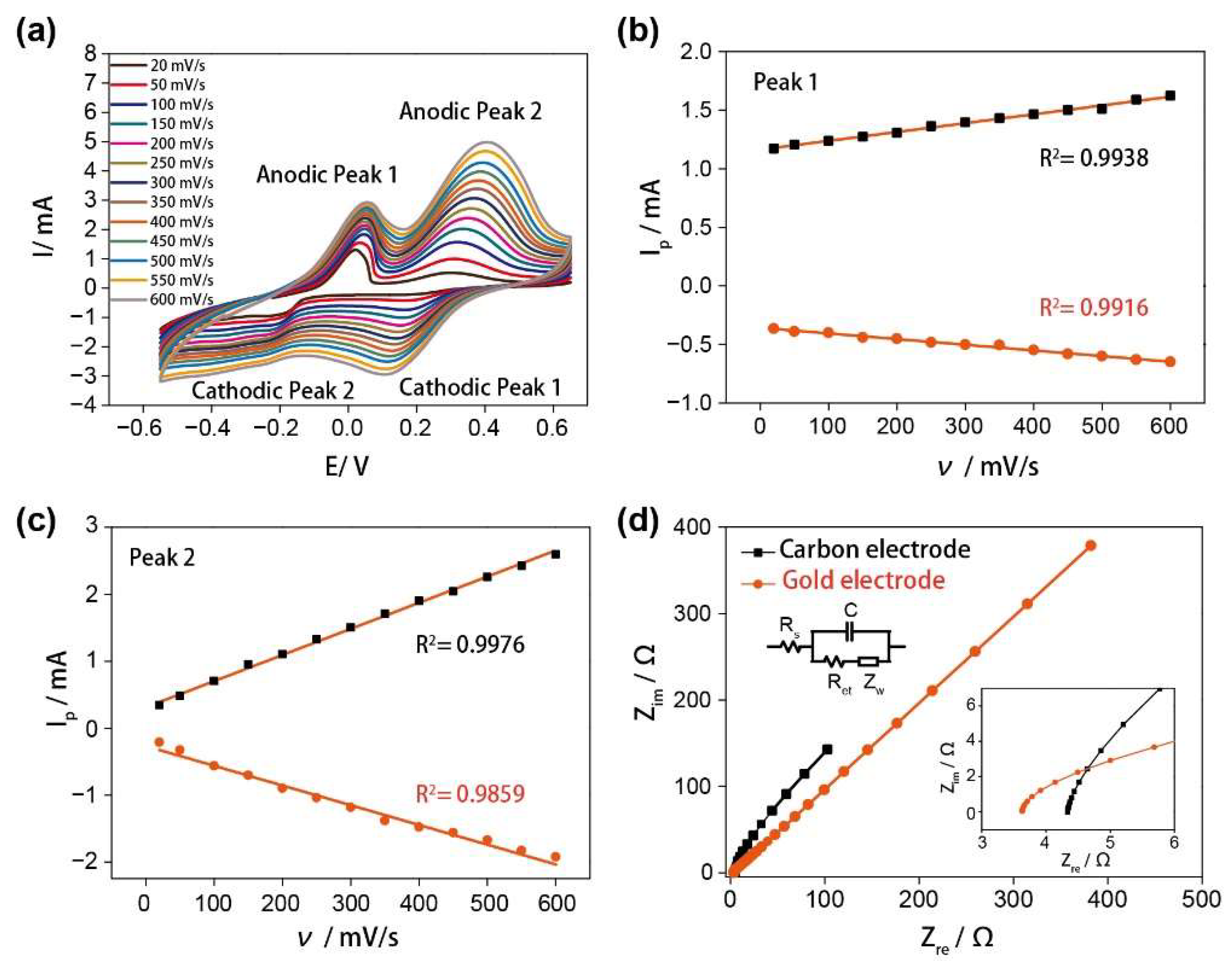
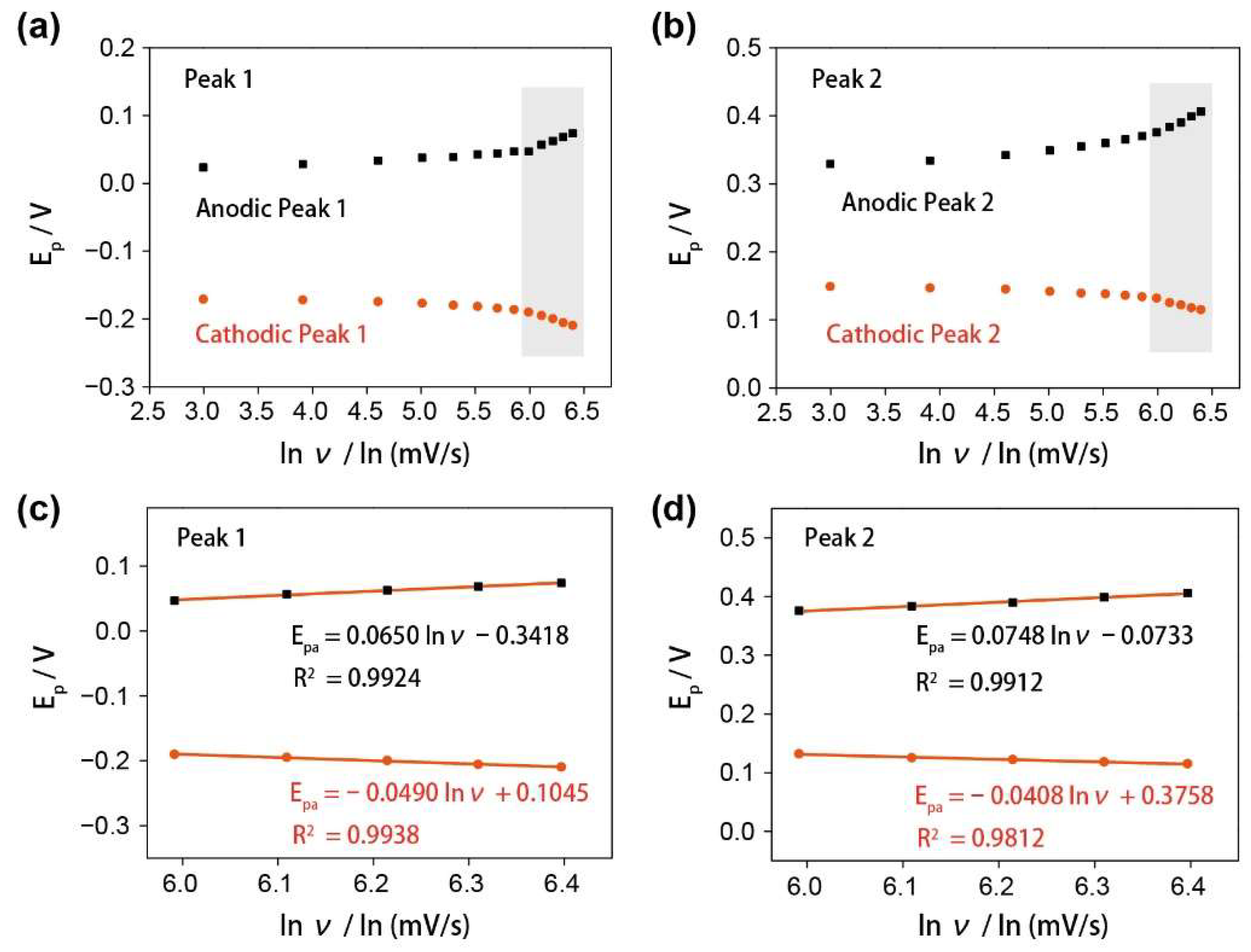
3.2. The Electrochemical Characterization
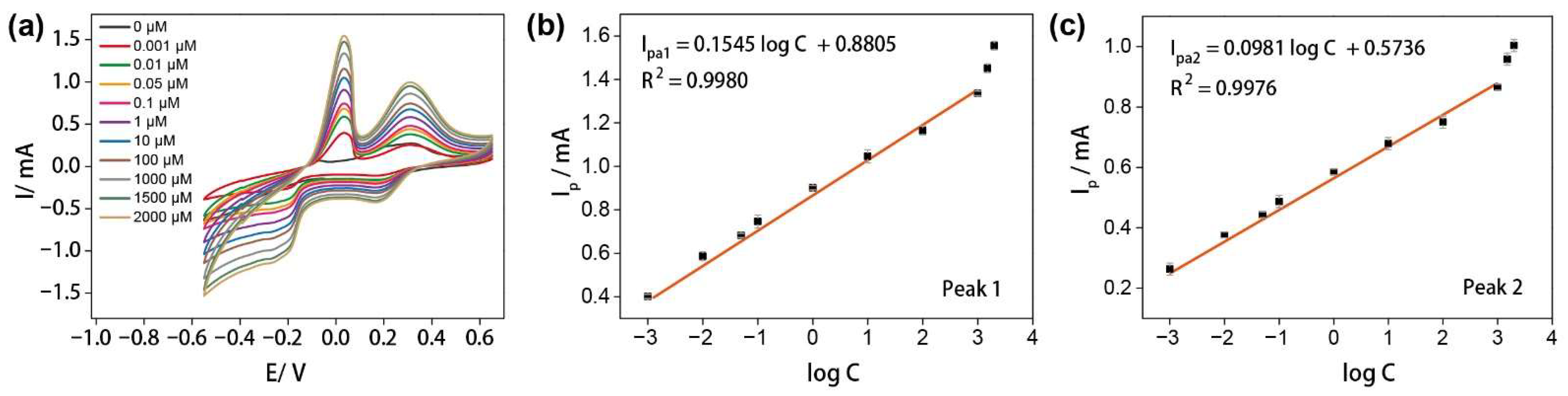
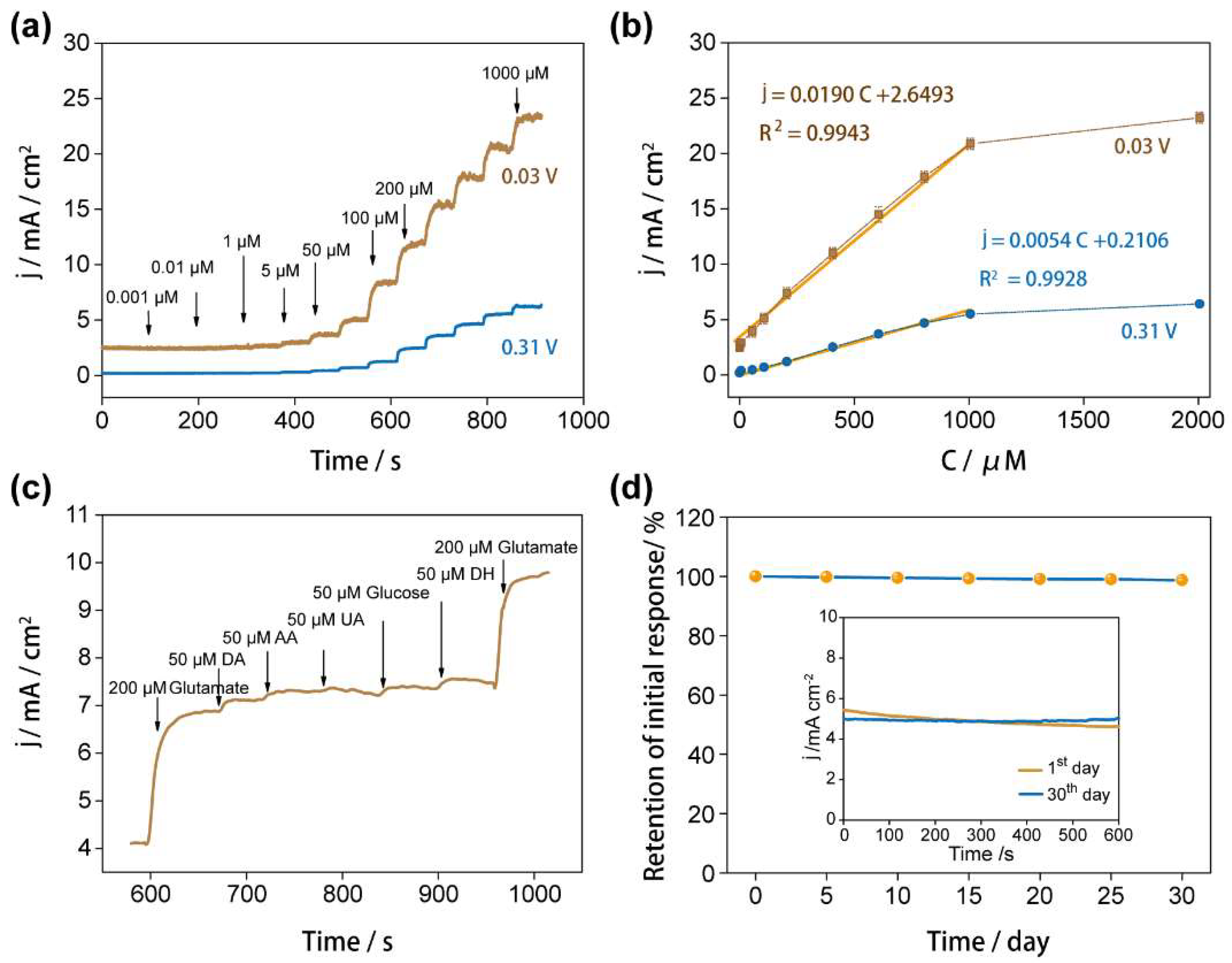
3.3. The Sensing Performances
3.4. The Sensing Mechanisms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Greenamyre, J.T.; Penney, J.B.; Young, A.B.; D’Amato, C.J.; Hicks, S.P.; Shoulson, I. Alterations in L-glutamate binding in Alzheimer’s and Huntington’s diseases. Science 1985, 227, 1496–1499. [Google Scholar] [CrossRef] [PubMed]
- James, C.M.; Houlihan, G.D.; Snell, R.G.; Cheadle, J.P.; Harper, P.S. Late-onset Huntington’s disease: A clinical and molecular study. Age Ageing 1994, 23, 445–448. [Google Scholar] [CrossRef]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Sánchez, A.M.; Montiel, T.; Segovia, J.; Massieu, L. Glutamate toxicity in the striatum of the R6/2 Huntington’s disease transgenic mice is age-dependent and correlates with decreased levels of glutamate transporters. Neurobiol. Dis. 2009, 34, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Pépin, J.; Francelle, L.; Sauvage, M.A.C.d.; Longprez, L.d.; Gipchtein, P.; Cambon, K.; Valette, J.; Brouillet, E.; Flament, J. In vivo imaging of brain glutamate defects in a knock-in mouse model of Huntington’s disease. NeuroImage 2016, 139, 53–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baad-Hansen, L.; Cairns, B.; Ernberg, M.; Svensson, P. Effect of systemic monosodium glutamate (MSG) on headache and pericranial muscle sensitivity. Cephalalgia 2010, 30, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Ray, S.; Nagarajan, K. Glutamic acid as anticancer agent: An overview. Saudi Pharm. J. 2013, 21, 337–343. [Google Scholar] [CrossRef] [Green Version]
- Maity, D.; Kumar, R.T.R. Highly sensitive amperometric detection of glutamate by glutamic oxidase immobilized Pt nanoparticle decorated multiwalled carbon nanotubes(MWCNTs)/polypyrrole composite. Biosens. Bioelectron. 2019, 130, 307–314. [Google Scholar] [CrossRef]
- Vaquero, J.; Butterworth, R.F. The brain glutamate system in liver failure. J. Neurochem. 2006, 98, 661–669. [Google Scholar] [CrossRef]
- Grace, A.A. Phasic versus tonic dopamine release and the modulation of dopamine system responsivity: A hypothesis for the etiology of schizophrenia. Neuroscience 1991, 41, 1–24. [Google Scholar] [CrossRef]
- Schultz, J.; Uddin, Z.; Singh, G.; Howlader, M.M.R. Glutamate sensing in biofluids: Recent advances and research challenges of electrochemical sensors. Analyst 2020, 145, 321–347. [Google Scholar] [CrossRef] [PubMed]
- Isoaho, N.; Peltola, E.; Sainio, S.; Wester, N.; Protopopova, V.; Wilson, B.P.; Koskinen, J.; Laurila, T. Carbon nanostructure based platform for enzymatic glutamate biosensors. J. Phys. Chem. C. 2017, 121, 4618–4626. [Google Scholar] [CrossRef]
- Scoggin, J.L.; Tan, C.; Nguyen, N.H.; Kansakar, U.; Madadi, M.; Siddiqui, S.; Arumugam, P.U.; DeCoster, M.A.; Murray, T.A. An enzyme-based electrochemical biosensor probe with sensitivity to detect astrocytic versus glioma uptake of glutamate in real time in vitro. Biosens. Bioelectron. 2019, 126, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Claussen, J.C.; Artiles, M.S.; McLamore, E.S.; Mohanty, S.; Shi, J.; Rickus, J.L.; Fisher, T.S.; Porterfield, D.M. Electrochemical glutamate biosensing with nanocube and nanosphere augmented single-walled carbon nanotube networks: A comparative study. J. Mater. Chem. 2011, 21, 11224. [Google Scholar] [CrossRef]
- Batra, B.; Kumari, S.; Pundir, C.S. Construction of glutamate biosensor based on covalent immobilization of glutmate oxidase on polypyrrole nanoparticles/polyaniline modified gold electrode. Enzyme Microb. Tech. 2014, 57, 69–77. [Google Scholar] [CrossRef]
- Jamal, M.; Hasan, M.; Mathewson, A.; Razeeb, K.M. Disposable sensor based on enzyme-free Ni nanowire array electrode to detect glutamate. Biosens. Bioelectron. 2013, 40, 213–218. [Google Scholar] [CrossRef]
- Batra, B.; Pundir, C.S. An amperometric glutamate biosensor based on immobilization of glutamate oxidase onto carboxylated multiwalled carbon nanotubes/gold nanoparticles/chitosan composite film modified Au electrode. Biosens. Bioelectron. 2013, 47, 496–501. [Google Scholar] [CrossRef]
- Jamal, M.; Chakrabarty, S.; Shao, H.; McNulty, D.; Yousuf, M.A.; Furukawa, H.; Khosla, A.; Razeeb, K.M. A non-enzymatic glutamate sensor based on nickel oxide nanoparticle. Microsyst. Technol. 2018, 24, 4217–4223. [Google Scholar] [CrossRef]
- Baciu, D.D.; Bîrjega, R.; Mărăscu, V.; Zăvoianu, R.; Matei, A.; Vlad, A.; Cojocaru, A.; Visan, T. Enhanced voltammetric response of monosodium glutamate on screen-printed electrodes modified with NiAl layered double hydroxide films. Surf. Interfaces 2021, 24, 101055. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, T.; Niu, Y.; Ye, B.C. Electrochemical detection of glutamate by metal–organic frameworks-derived Ni@NC electrocatalysts. Microchem. J. 2022, 175, 107229. [Google Scholar] [CrossRef]
- Zeynaloo, E.; Yang, Y.P.; Dikici, E.; Landgraf, R.; Bachas, L.G.; Daunert, S. Design of a mediator-free, non-enzymatic electrochemical biosensor for glutamate detection. Nanomed. Nanotech. Biol. Med. 2021, 31, 102305. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Uddin, M.T.; Asiri, A.M.; Awual, M.R.; Fazal, M.A.; Rahman, M.M.; Islam, M.A. Fabrication of selective l-glutamic acid sensor in electrochemical technique from wet-chemically prepared RuO2 doped ZnO nanoparticles. Mater. Chem. Phys. 2020, 251, 123029. [Google Scholar] [CrossRef]
- Pitiphattharabun, S.; Meesombad, K.; Panomsuwan, G.; Jongprateep, O. MWCNT/Ti-doped ZnO nanocomposite as electrochemical sensor for detecting glutamate and ascorbic acid. Int. J. Appl. Ceram. Technol. 2022, 19, 467–479. [Google Scholar] [CrossRef]
- Pary, P.; Bengoa, L.N.; Egli, W.A. Electrochemical characterization of a Cu(II)-glutamate alkaline solution for copper electrodeposition. J. Electrochem. Soc. 2015, 162, D275–D282. [Google Scholar] [CrossRef]
- Wang, M.; Ma, Y.; Sun, Y.; Hong, S.Y.; Lee, S.K.; Yoon, B.; Chen, L.; Ci, L.; Nam, J.D.; Chen, X.; et al. Hierarchical porous chitosan sponges as robust and recyclable adsorbents for anionic dye adsorption. Sci. Rep. 2017, 7, 18054. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Fu, L.; Liu, N.; Liu, M.; Wang, Y.; Liu, Z. Synthesis of nitrogen-doped graphene using embedded carbon and nitrogen sources. Adv. Mater. 2011, 23, 1020–1024. [Google Scholar] [CrossRef]
- Tang, L.; Ji, R.; Li, X.; Teng, K.S.; Lau, S.P. Energy-level structure of nitrogen-doped graphene quantum dots. J. Mater. Chem. C. 2013, 1, 4908–4915. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, Z.; Dai, L. Carbon-based electrocatalysts for advanced energy conversion and storage. Sci. Adv. 2015, 1, 1500564. [Google Scholar] [CrossRef] [Green Version]
- Hughes, G.; Pemberton, R.M.; Fielden, P.R.; Hart, J.P. Development of a novel reagentless, screen-printed amperometric biosensor based on glutamate dehydrogenase and NAD+, integrated with multi-walled carbon nanotubes for the determination of glutamate in food and clinical applications. Sens. Actuators B Chem. 2015, 216, 614–621. [Google Scholar] [CrossRef]
- Shahdeo, D.; Roberts, A.; Archana, G.J.; Shrikrishna, N.S.; Mahari, S.; Nagamani, K.; Gandhi, S. Label free detection of SARS-CoV-2 Receptor Binding Domain (RBD) protein by fabrication of gold nanorods deposited on electrochemical immunosensor (GDEI). Biosens. Bioelectron. 2022, 212, 114406. [Google Scholar] [CrossRef]
- Yang, J.; Cho, M.; Lee, Y. Synthesis of hierarchical NiCo2O4 hollow nanorods via sacrificial-template accelerate hydrolysis for electrochemical glucose oxidation. Biosens. Bioelectron. 2016, 75, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, Y.; Su, L.; Zhang, H.; Lei, Y. Preparation and characterization of NiO–Ag nanofibers, NiO nanofibers, and porous Ag: Towards the development of a highly sensitive and selective non-enzymatic glucose sensor. J. Mater. Chem. 2010, 20, 9918–9926. [Google Scholar] [CrossRef]
- Lee, S.H.; Yang, J.; Han, Y.J.; Cho, M.; Lee, Y. Rapid and highly sensitive MnOx nanorods array platform for a glucose analysis. Sens. Actuators B Chem. 2015, 218, 137–144. [Google Scholar] [CrossRef]
- Jung, H.; Lee, S.H.; Yang, J.; Cho, M.; Lee, Y. Ni(OH)2@Cu dendrite structure for highly sensitive glucose determination. RSC Adv. 2014, 4, 47714–47720. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.H.; Cho, M.; Lee, Y. Solvent-assisted morphology confinement of a nickel sulfide nanostructure and its application for non-enzymatic glucose sensor. Biosens. Bioelectron. 2016, 85, 587–595. [Google Scholar] [CrossRef]
- Chou, S.; Wang, J.; Liu, H.; Dou, S. Electrochemical deposition of porous Co3O4 nanostructured thin film for lithium-ion battery. J. Power Sources 2008, 182, 359–364. [Google Scholar] [CrossRef]
- Bukharov, M.S.; Shtyrlin, V.G.; Mukhtarov, A.S.; Mamin, G.V.; Stapf, S.; Mattea, C.; Krutikov, A.A.; Il’in, A.N.; Serov, N.Y. Study of structural and dynamic characteristics of copper(II) amino acid complexes in solutions by combined EPR and NMR relaxation methods. Phys. Chem. Chem. Phys. 2014, 16, 9411–9421. [Google Scholar] [CrossRef]
- Primo, A.; Atienzar, P.; Sanchez, E.; Delgado, J.M.; García, H. From biomass wastes to large-area, high-quality, N-doped graphene: Catalyst-free carbonization of chitosan coatings on arbitrary substrates. Chem. Comm. 2012, 48, 9254–9256. [Google Scholar] [CrossRef]
- Primo, A.; Sánchez, E.; Delgado, J.M.; García, H. High-yield production of N-doped graphitic platelets by aqueous exfoliation of pyrolyzed chitosan. Carbon. 2014, 68, 777–783. [Google Scholar] [CrossRef]
- Hao, P.; Zhao, Z.; Leng, Y.; Tian, J.; Sang, Y.; Boughton, R.I.; Wong, C.P.; Liu, H.; Yang, B. Graphene-based nitrogen self-doped hierarchical porous carbon aerogels derived from chitosan for high performance supercapacitors. Nano Energy 2015, 15, 9–23. [Google Scholar] [CrossRef]
- Beitollahi, H.; Ardakani, M.M.; Ganjipour, B.; Naeimi, H. Novel 2,2′-[1,2-ethanediylbis(nitriloethylidyne)]-bis-hydroquinone double-wall carbon nanotube paste electrode for simultaneous determination of epinephrine, uric acid and folic acid. Biosens. Bioelectron. 2008, 24, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Laviron, E. General expression of the linear potential sweep voltammogram in the case of diffusionless electrochemical systems. J. Electronal. Chem. 1979, 101, 19–28. [Google Scholar] [CrossRef]
- Pufahl, R.A.; Singer, C.P.; Peariso, K.L.; Lin, S.J.; Schmidt, P.J.; Fahrni, C.J.; Culotta, V.C.; Penner-Hahn, J.E.; O’Halloran, V.T. Metal ion chaperone function of the soluble Cu(I) receptor atx1. Science 1997, 278, 853–856. [Google Scholar] [CrossRef]
- Fitts, J.P.; Persson, P.; Brown, G.E.; Parks, G.A. Structure and bonding of Cu(II)–glutamate complexes at the γ-Al2O3–water interface. J. Colloid Interface Sci. 1999, 220, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Valora, G.; Bonomo, R.P.; Tabbì, G. An EPR and voltammetric study of simple and mixed copper(II) complexes with l- or d-glutamate and l-arginate in aqueous solution. Inorg. Chim. Acta 2016, 453, 62–68. [Google Scholar] [CrossRef]
- Balland, V.; Hureau, C.; Savéant, J.M. Electrochemical and homogeneous electron transfers to the Alzheimer amyloid-β copper complex follow a preorganization mechanism. Proc. Natl. Acad. Sci. USA 2010, 107, 17113–17118. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Cho, M.; Lee, Y. Iridium oxide dendrite as a highly efficient dual electro-catalyst for water splitting and sensing of H2O2. J. Electrochem. Soc. 2017, 164, 3029–3035. [Google Scholar] [CrossRef]
- Ammam, M.; Fransaer, J. Highly sensitive and selective glutamate microbiosensor based on cast polyurethane/AC-electrophoresis deposited multiwalled carbon nanotubes and then glutamate oxidase/electrosynthesized polypyrrole/Pt electrode. Biosens. Bioelectron. 2010, 25, 1597–1602. [Google Scholar] [CrossRef]
- Kaivosoja, E.; Tujunen, N.; Jokinen, V.; Protopopova, V.; Heinilehto, S.; Koskinen, J.; Laurila, T. Glutamate detection by amino functionalized tetrahedral amorphous carbon surfaces. Talanta 2015, 141, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Wisetsuwannaphum, S.; Foord, J.S. Glutamate biosensors based on diamond and graphene platforms. Faraday Discuss. 2014, 172, 457–472. [Google Scholar] [CrossRef]
- Chakraborty, S.; Raj, C.R. Amperometric biosensing of glutamate using carbon nanotube based electrode. Electrochem. Commun. 2007, 9, 1323–1330. [Google Scholar] [CrossRef]
- Batra, B.; Yadav, M.; Pundir, C.S. l-Glutamate biosensor based on l-glutamate oxidase immobilized onto ZnO nanorods/polypyrrole modified pencil graphite electrode. Biochem. Eng. J. 2016, 105, 428–436. [Google Scholar] [CrossRef]
- Özel, R.E.; Ispas, C.; Ganesana, M.; Leiter, J.C.; Andreescu, S. Glutamate oxidase biosensor based on mixed ceria and titania nanoparticles for the detection of glutamate in hypoxic environments. Biosens. Bioelectron. 2014, 52, 397–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Electrodes | Enzyme | Linear Range (μM) | Limit of Detection (μM) | Sensitivities (μA/mM∙cm2) | Reference |
|---|---|---|---|---|---|
| GlutOx/cMWCNT-AuNPs-CHIT/Au | GlutOx | 5–500 | 1.6 | 155 | [17] |
| GlutOx/Pt-SWCNT/PAA | GlutOx | 0.05–1600 | 0.0046 | 27.4 | [31] |
| PU/GlutOx/MWCNT/PPy/Pt | GlutOx | 0.3–500 | 0.3 | 0.384 | [48] |
| GlutOx/APTES/ta-C/Pt | GlutOx | 10–500 | 10 | 2.9 | [49] |
| GlutOx/BDD/Pt | GlutOx | 0.5–50 | 0.35 | 24 | [50] |
| Glutamate dehydrogenase/CNT/GCE | Glutamate dehydrogenase | 2–225 | 2 | 0.71 | [51] |
| GlutOx/ta-C/CNFs | GlutOx | 20–500 | 0.000767 | 18.8 | [12] |
| GlutOx/ZnO nanorods/PPy/PGE | GlutOx | 0.02–500 | 0.18 | N/A | [52] |
| GlutOx/CeO2/TiO2/CHIT/o-PD/Pt | GlutOx | 5–90 | 0.594 | 793 pA/µM | [53] |
| Pt/Ni nanowire array | No enzyme | 500–8000 | 135 | 65 | [16] |
| NiO/chit/GCE | No enzyme | 1000–8000 | 272 | 11 | [18] |
| Ni@NC/GCE | No enzyme | 0.005–500 | 135 | - | [20] |
| GluBP/Au NP/SPCE | No enzyme | 0.1–0.8 | 0.15 | - | [21] |
| ZnO/RuO2 NPs/GCE | No enzyme | 0.0001–10 | 9.6 × 10−5 | 5.42 × 103 | [22] |
| MWCNT/Ti-doped ZnO/GCE | No enzyme | 100–1000 1000–10,000 | 11.59 | 25 4.7 | [23] |
| Cu2+ assisted carbon foam | No enzyme | 0.001–1000 | 0.001 | 1.9 × 104 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Han, J.; Tong, Z.; Qin, J.; Wang, M.; Suhr, J.; Nam, J.; Xiao, L.; Jia, S.; Chen, X. Porous Carbon Boosted Non-Enzymatic Glutamate Detection with Ultra-High Sensitivity in Broad Range Using Cu Ions. Nanomaterials 2022, 12, 1987. https://doi.org/10.3390/nano12121987
Ma Y, Han J, Tong Z, Qin J, Wang M, Suhr J, Nam J, Xiao L, Jia S, Chen X. Porous Carbon Boosted Non-Enzymatic Glutamate Detection with Ultra-High Sensitivity in Broad Range Using Cu Ions. Nanomaterials. 2022; 12(12):1987. https://doi.org/10.3390/nano12121987
Chicago/Turabian StyleMa, Yifei, Jiemin Han, Zhaomin Tong, Jieling Qin, Mei Wang, Jonghwan Suhr, Jaedo Nam, Liantuan Xiao, Suotang Jia, and Xuyuan Chen. 2022. "Porous Carbon Boosted Non-Enzymatic Glutamate Detection with Ultra-High Sensitivity in Broad Range Using Cu Ions" Nanomaterials 12, no. 12: 1987. https://doi.org/10.3390/nano12121987
APA StyleMa, Y., Han, J., Tong, Z., Qin, J., Wang, M., Suhr, J., Nam, J., Xiao, L., Jia, S., & Chen, X. (2022). Porous Carbon Boosted Non-Enzymatic Glutamate Detection with Ultra-High Sensitivity in Broad Range Using Cu Ions. Nanomaterials, 12(12), 1987. https://doi.org/10.3390/nano12121987