Identification of a Novel Osteogenetic Oligodeoxynucleotide (osteoDN) That Promotes Osteoblast Differentiation in a TLR9-Independent Manner
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture
2.3. Alkaline Phosphatase (ALP) Staining
2.4. Alizarin Staining
2.5. Quantitative Real-Time RT-PCR (qPCR)
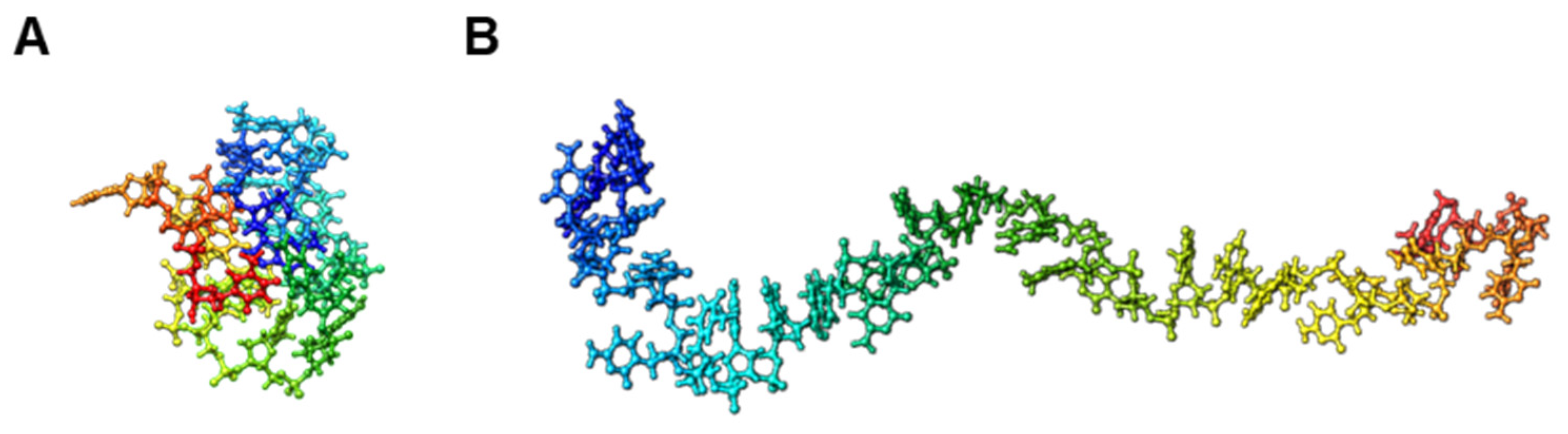
2.6. Trivial Trajectory Parallelization of Multicanonical Molecular Dynamics (TTP-McMD)
2.7. Statistical Analysis
3. Results
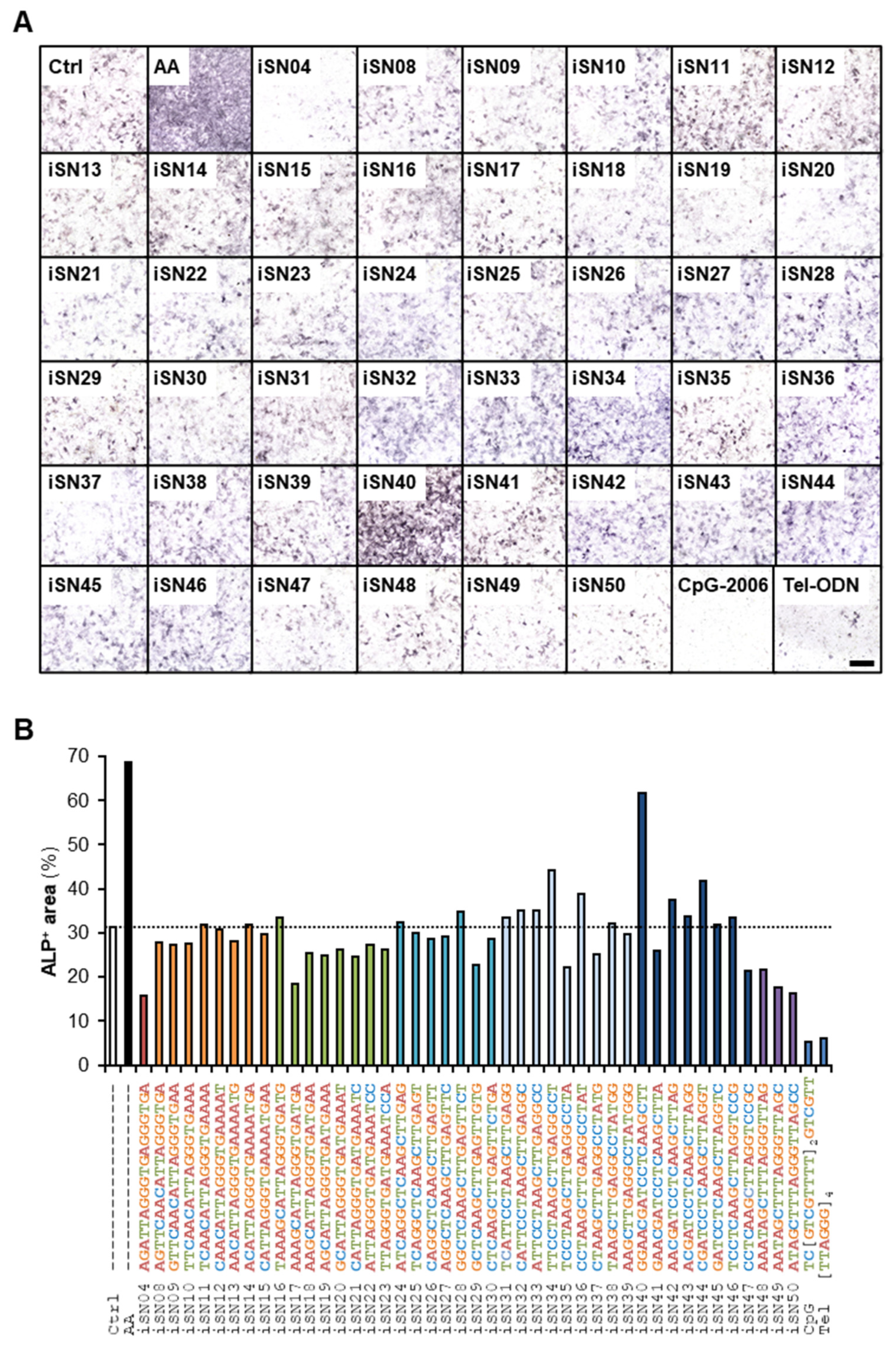
3.1. iSN40 Promotes Osteoblast Differentiation
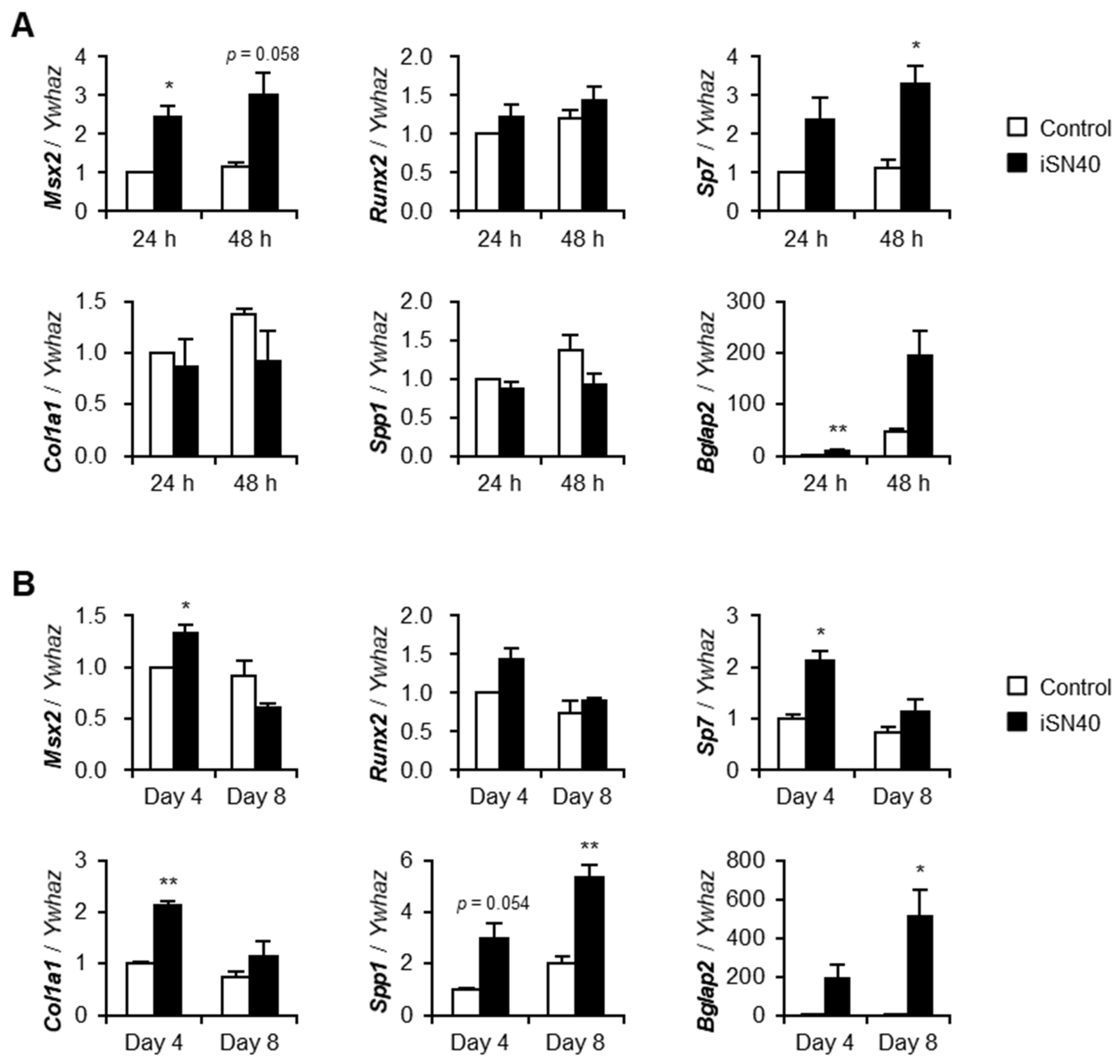
3.2. iSN40 Modulates Osteogenic Gene Expression
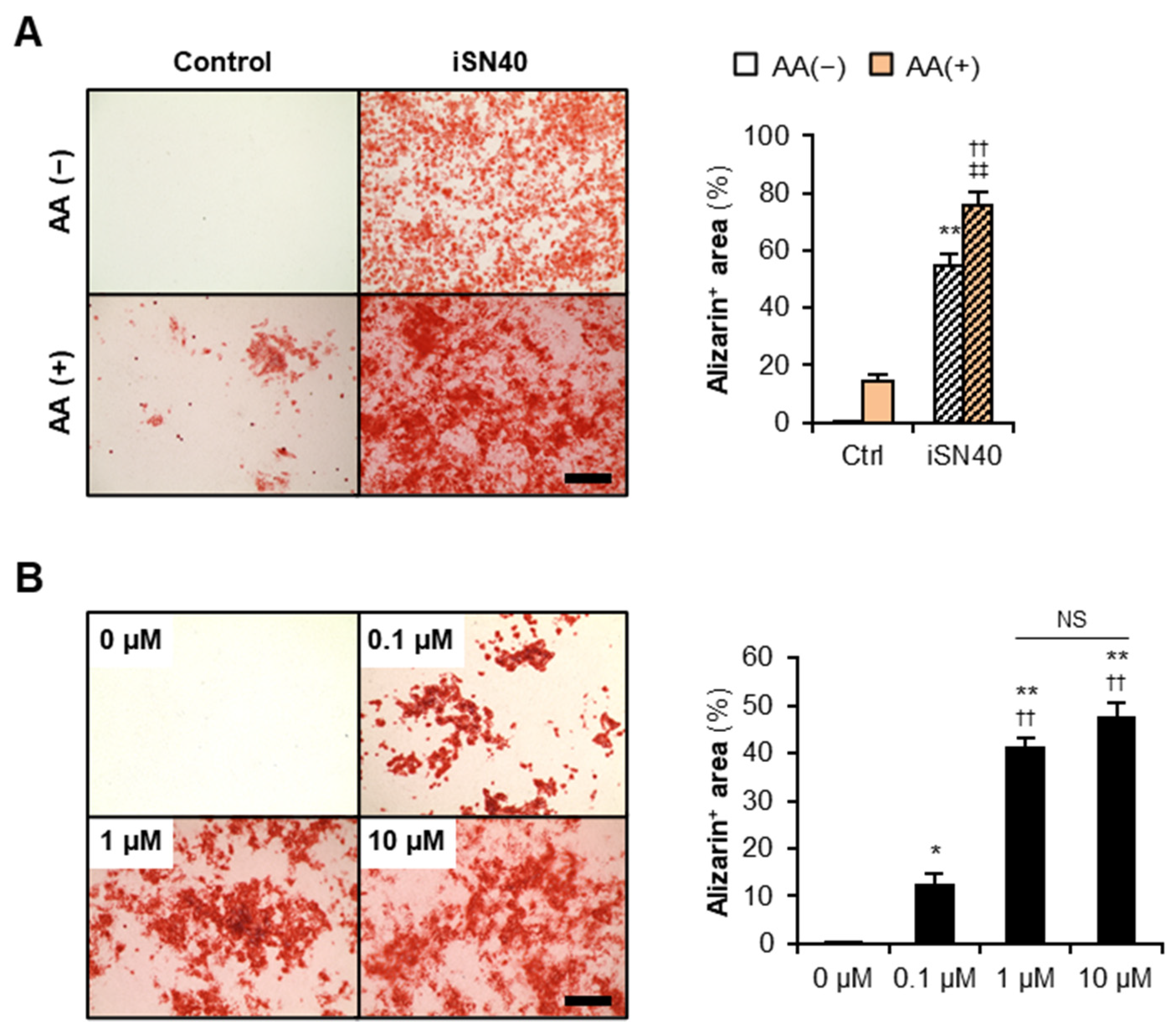
3.3. iSN40 Promotes Osteoblast Calcification
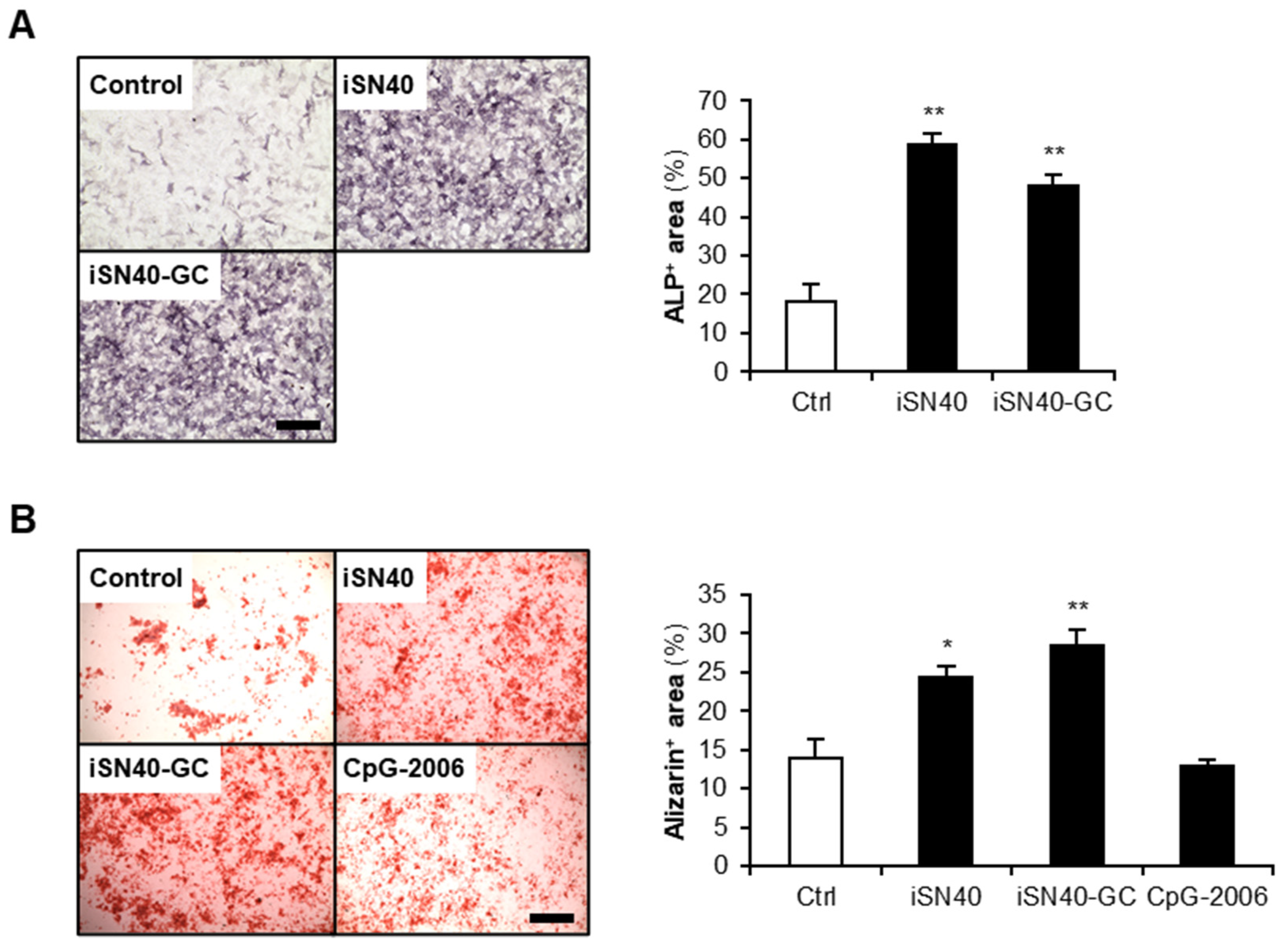
3.4. Osteogenetic Action of iSN40 Is TLR9-Independent
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boros, K.; Freemont, T. Physiology of ageing of the musculoskeletal system. Best Pract. Res. Clin. Rheumatol. 2017, 31, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Pisani, P.; Renna, M.D.; Conversano, F.; Casciaro, E.; Di Paola, M.; Quarta, E.; Muratore, M.; Casciaro, S. Major osteoporotic fragility fractures: Risk factor updates and societal impact. World J. Orthop. 2016, 7, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Corrado, A.; Cici, D.; Rotondo, C.; Maruotti, N.; Cantatore, F.P. Molecular basis of bone aging. Int. J. Mol. Sci. 2020, 21, 3679. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, G.; Wang, Y.; Zhou, J.; Yuan, H.; Li, X.; Liu, Y.; Wang, B. Histone demethylase KDM7A reciprocally regulates adipogenic and osteogenic differentiation via regulation of C/EBPalpha and canonical Wnt signalling. J. Cell. Mol. Med. 2019, 23, 2149–2162. [Google Scholar] [CrossRef]
- Rutkovskiy, A.; Stenslokken, K.O.; Vaage, I.J. Osteoblast differentiation at a glance. Med. Sci. Monit. Basic Res. 2016, 22, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, G.; Zhang, Y.; Wang, J.; Wang, Y.; Cheng, L.; Sun, M.; Rui, Y. Activation of JNK signaling in osteoblasts is inversely correlated with collagen synthesis in age-related osteoporosis. Biochem. Biophys. Res. Commun. 2018, 504, 771–776. [Google Scholar] [CrossRef]
- Eriksen, C.G.; Olsen, H.; Husted, L.B.; Sorensen, L.; Carstens, M.; Soballe, K.; Langdahl, B.L. The expression of IL-6 by osteoblasts is increased in healthy elderly individuals: Stimulated proliferation and differentiation are unaffected by age. Calcif. Tissue Int. 2010, 87, 414–423. [Google Scholar] [CrossRef]
- Becerikli, M.; Jaurich, H.; Schira, J.; Schulte, M.; Dobele, C.; Wallner, C.; Abraham, S.; Wagner, J.M.; Dadras, M.; Kneser, U.; et al. Age-dependent alterations in osteoblast and osteoclast activity in human cancellous bone. J. Cell. Mol. Med. 2017, 21, 2773–2781. [Google Scholar] [CrossRef] [Green Version]
- Awasthi, H.; Mani, D.; Singh, D.; Gupta, A. The underlying pathophysiology and therapeutic approaches for osteoporosis. Med. Res. Rev. 2018, 38, 2024–2057. [Google Scholar] [CrossRef]
- Corrado, A.; Sanpaolo, E.R.; Di Bello, S.; Cantatore, F.P. Osteoblast as a target of anti-osteoporotic treatment. Postgrad. Med. 2017, 129, 858–865. [Google Scholar] [CrossRef]
- McClung, M.R.; Lewiecki, E.M.; Cohen, S.B.; Bolognese, M.A.; Woodson, G.C.; Moffett, A.H.; Peacock, M.; Miller, P.D.; Lederman, S.N.; Chesnut, C.H.; et al. Bone Loss Study Group. N. Engl. J. Med. 2006, 354, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Diedhiou, D.; Cuny, T.; Sarr, A.; Norou Diop, S.; Klein, M.; Weryha, G. Efficacy and safety of denosumab for the treatment of osteoporosis: A systematic review. Ann. Endocrinol. 2015, 76, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Canalis, E. Wnt signalling in osteoporosis: Mechanisms and novel therapeutic approaches. Nat. Rev. Endocrinol. 2013, 10, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; St Onge, E.L.; Whalen, K.L. Romosozumab: A novel agent in the treatment for postmenopausal osteoporosis. J. Pharm. Technol. 2021, 37, 45–52. [Google Scholar] [CrossRef]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip. Rev. RNA 2020, 11, e1594. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Nigar, S.; Shimosato, T. Cooperation of oligodeoxynucleotides and synthetic molecules as enhanced immune modulators. Front. Nutr. 2019, 6, 140. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Wan, M.; Zhang, Y.; Sun, L.; Sun, R.; Hu, D.; Zhou, X.; Wang, L.; Wu, X.; Wang, L.; et al. Inhibition of a C-rich oligodeoxynucleotide on activation of immune cells in vitro and enhancement of antibody response in mice. Immunology 2010, 131, 501–512. [Google Scholar] [CrossRef]
- Feng, Z.; Shen, Y.; Wang, L.; Cheng, L.; Wang, J.; Li, Q.; Shi, W.; Sun, X. An oligodeoxynucleotide with promising modulation activity for the proliferation and activation of osteoblast. Int. J. Mol. Sci. 2011, 12, 2543–2555. [Google Scholar] [CrossRef]
- Shen, Y.; Feng, Z.; Lin, C.; Hou, X.; Wang, X.; Wang, J.; Yu, Y.; Wang, L.; Sun, X. An oligodeoxynucleotide that induces differentiation of bone marrow mesenchymal stem cells to osteoblasts in vitro and reduces alveolar bone loss in rats with periodontitis. Int. J. Mol. Sci. 2012, 13, 2877–2892. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Zheng, Y.; Li, H.; Lin, H.; Chen, Z.; Tian, Y.; Chen, H.; Zhang, P.; Xu, X.; Shen, Y. The Toll-like receptor ligand, CpG oligodeoxynucleotides, regulate proliferation and osteogenic differentiation of osteoblast. J. Orthop. Surg. Res. 2020, 15, 327. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Shen, Y.; Zhang, C.; Zhang, L.; Qin, Y.; Yu, Y.; Wang, L.; Sun, X. A specific oligodeoxynucleotide promotes the differentiation of osteoblasts via ERK and p38 MAPK pathways. Int. J. Mol. Sci. 2012, 13, 7902–7914. [Google Scholar] [CrossRef] [PubMed]
- Nigar, S.; Yamamoto, Y.; Okajima, T.; Shigemori, S.; Sato, T.; Ogita, T.; Shimosato, T. Synergistic oligodeoxynucleotide strongly promotes CpG-induced interleukin-6 production. BMC Immunol. 2017, 18, 44. [Google Scholar] [CrossRef] [Green Version]
- Shinji, S.; Umezawa, K.; Nihashi, Y.; Nakamura, S.; Shimosato, T.; Takaya, T. Identification of the myogenetic oligodeoxynucleotides (myoDNs) that promote differentiation of skeletal muscle myoblasts by targeting nucleolin. Front. Cell Dev. Biol. 2021, 8, 616706. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Yonekura, S.; Shimosato, T.; Takaya, T. Myogenetic oligodeoxynucleotide (myoDN) recovers the differentiation of skeletal muscle myoblasts deteriorated by diabetes mellitus. Front. Physiol. 2021, 12, 679152. [Google Scholar] [CrossRef] [PubMed]
- Nihashi, Y.; Shinji, S.; Umezawa, K.; Shimosato, T.; Ono, T.; Kagami, H.; Takaya, T. Myogenetic oligodeoxynucleotide complexed with berberine promotes differentiation of chicken myoblasts. Anim. Sci. J. 2021, 92, e13597. [Google Scholar] [CrossRef]
- Nihashi, Y.; Yamamoto, M.; Shimosato, T.; Takaya, T. Myogenetic oligodeoxynucleotide restores differentiation and reverses inflammation of myoblasts aggravated by cancer-conditioned medium. bioRxiv 2021, 469038. [Google Scholar] [CrossRef]
- Nohira, N.; Shinji, S.; Nakamura, S.; Nihashi, Y.; Shimosato, T.; Takaya, T. Myogenetic oligodeoxynucleotides as anti-nucleolin aptamers inhibit the growth of embryonal rhabdomyosarcoma cells. bioRxiv 2021, 464889. [Google Scholar] [CrossRef]
- Macke, T.J.; Case, D.A. Modeling unusual nucleic acid structures. In Molecular Modeling of Nucleic Acids; Leontis, N.B., SantaLucia, J., Eds.; American Chemical Society: Washington, DC, USA, 1998; pp. 379–393. [Google Scholar]
- Ikebe, J.; Umezawa, K.; Kamiya, N.; Sugihara, T.; Yonezawa, Y.; Takano, Y.; Nakamura, H.; Higo, J. Theory for trivial trajectory parallelization of multicanonical molecular dynamics and application to a polypeptide in water. J. Comput. Chem. 2011, 32, 1286–1297. [Google Scholar] [CrossRef]
- Maier, J.A.; Matinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Tsui, V.; Case, D.A. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers 2000, 56, 275–291. [Google Scholar] [CrossRef]
- Pohar, J.; Lainscek, D.; Fukui, R.; Yamamoto, C.; Miyake, K.; Jerala, R.; Bencina, M. Species-specific minimal sequence motif for oligodeoxyribonucleotides activating mouse TLR9. J. Immunol. 2015, 195, 4396–4405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sackesen, C.; van de Veen, W.; Akdis, M.; Soyer, O.; Zumkehr, J.; Ruckert, B.; Stanic, B.; Kalayci, O.; Alkan, S.S.; Gursel, I.; et al. Suppression of B-cell activation and IgE, IgA, IgG1 and IgG4 production by mammalian telomeric oligonucleotides. Allergy 2013, 68, 593–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komori, T. Regulation of osteoblast differentiation by Runx2. Adv. Exp. Med. Biol. 2010, 658, 43–49. [Google Scholar]
- Moser, S.C.; van der Eerden, B.C.J. Osteocalcin: A versatile bone-derived hormone. Front. Endocrinol. 2018, 9, 794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.M.; Lee, E.H. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng. Part B Rev. 2013, 19, 254–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vollmer, J.; Krieg, A.M. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv. Drug Deliv. Rev. 2009, 61, 195–2004. [Google Scholar] [CrossRef]
- Nemoto, E.; Honda, T.; Kanaya, S.; Takada, H.; Shimauchi, H. Expression of functional Toll-like receptors and nucleotide-binding oligomerization domain proteins in murine cementoblasts and their upregulation during cell differentiation. J. Periodontal Res. 2008, 43, 585–593. [Google Scholar] [CrossRef]
- Charles, J.F.; Nakamura, M.C. Bone and the innate immune system. Curr. Osteoporos. Rep. 2014, 12, 1–8. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed, K.M.F.; Boeckler, J.; Dorfer, C.E. TLR expression profile of human alveolar bone proper-derived stem/progenitor cells and osteoblasts. J. Craniomaxillofac. Surg. 2017, 45, 2054–2060. [Google Scholar] [CrossRef]
- Amcheslavsky, A.; Bar-Shavit, Z. Toll-like receptor 9 ligand blocks osteoclast differentiation through induction of phosphatase. J. Bone Miner. Res. 2007, 22, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Vimalraj, S.; Arumugam, B.; Miranda, P.J.; Selvamurugan, N. Runx2: Structure, function, and phosphorylation in osteoblast differentiation. Int. J. Biol. Macromol. 2015, 78, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Artigas, N.; Urena, C.; Rodriguez-Carballo, E.; Rosa, J.L.; Ventura, F. Mitogen-activated protein kinase (MAPK)-regulated interactions between Osterix and Runx2 are critical for the transcriptional osteogenic program. J. Biol. Chem. 2014, 289, 27105–27117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, H.; Kuang, P.; Cui, H.; Luo, Q.; Liu, H.; Lu, Y.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; et al. Sodium fluoride induces apoptosis in mouse splenocytes by activating ROS-dependent NF-κB signaling. Oncotarget 2017, 8, 114428–114441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Xu, Y.; Fu, Q.; Dong, Y. Osterix is required for Sonic hedgehog-induced osteoblastic MC3T3-E1 cell differentiation. Cell Biochem. Biophys. 2012, 64, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Takayama, T.; Dai, J.; Tachi, K.; Shohara, R.; Kasai, H.; Imamura, K.; Yamano, S. The potential of stromal cell-derived factor-1 delivery using a collagen membrane for bone regeneration. J. Biomater. Appl. 2017, 31, 1049–1061. [Google Scholar] [CrossRef]
- Gao, J.; Feng, Z.; Wang, X.; Zeng, M.; Liu, J.; Han, S.; Xu, J.; Chen, L.; Cao, K.; Long, J.; et al. SIRT3/SOD2 maintains osteoblast differentiation and bone formation by regulating mitochondrial stress. Cell Death Differ. 2018, 25, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.J.; Tseng, H.C.; Wong, S.W.; Wang, Z.; Deng, M.; Ko, C.C. Dopaminergic effects on in vitro osteogenesis. Bone Res. 2015, 3, 15020. [Google Scholar] [CrossRef] [Green Version]
- Veazey, K.J.; Colding, M.C. Selection of stable reference genes for quantitative rt-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLoS ONE 2011, 6, e27592. [Google Scholar] [CrossRef] [Green Version]
- Nihashi, Y.; Miyoshi, M.; Umezawa, K.; Shimosato, T.; Takaya, T. Identification of a novel osteogenetic oligodeoxynucleotide (osteoDN) that promotes osteoblast differentiation in a TLR9-independent manner. bioRxiv 2022, 485101. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nihashi, Y.; Miyoshi, M.; Umezawa, K.; Shimosato, T.; Takaya, T. Identification of a Novel Osteogenetic Oligodeoxynucleotide (osteoDN) That Promotes Osteoblast Differentiation in a TLR9-Independent Manner. Nanomaterials 2022, 12, 1680. https://doi.org/10.3390/nano12101680
Nihashi Y, Miyoshi M, Umezawa K, Shimosato T, Takaya T. Identification of a Novel Osteogenetic Oligodeoxynucleotide (osteoDN) That Promotes Osteoblast Differentiation in a TLR9-Independent Manner. Nanomaterials. 2022; 12(10):1680. https://doi.org/10.3390/nano12101680
Chicago/Turabian StyleNihashi, Yuma, Mana Miyoshi, Koji Umezawa, Takeshi Shimosato, and Tomohide Takaya. 2022. "Identification of a Novel Osteogenetic Oligodeoxynucleotide (osteoDN) That Promotes Osteoblast Differentiation in a TLR9-Independent Manner" Nanomaterials 12, no. 10: 1680. https://doi.org/10.3390/nano12101680
APA StyleNihashi, Y., Miyoshi, M., Umezawa, K., Shimosato, T., & Takaya, T. (2022). Identification of a Novel Osteogenetic Oligodeoxynucleotide (osteoDN) That Promotes Osteoblast Differentiation in a TLR9-Independent Manner. Nanomaterials, 12(10), 1680. https://doi.org/10.3390/nano12101680






