Nanotechnological Applications Based on Bacterial Encapsulins
Abstract
:1. Introduction
2. Encapsulins
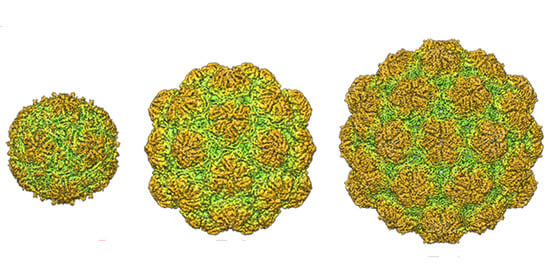
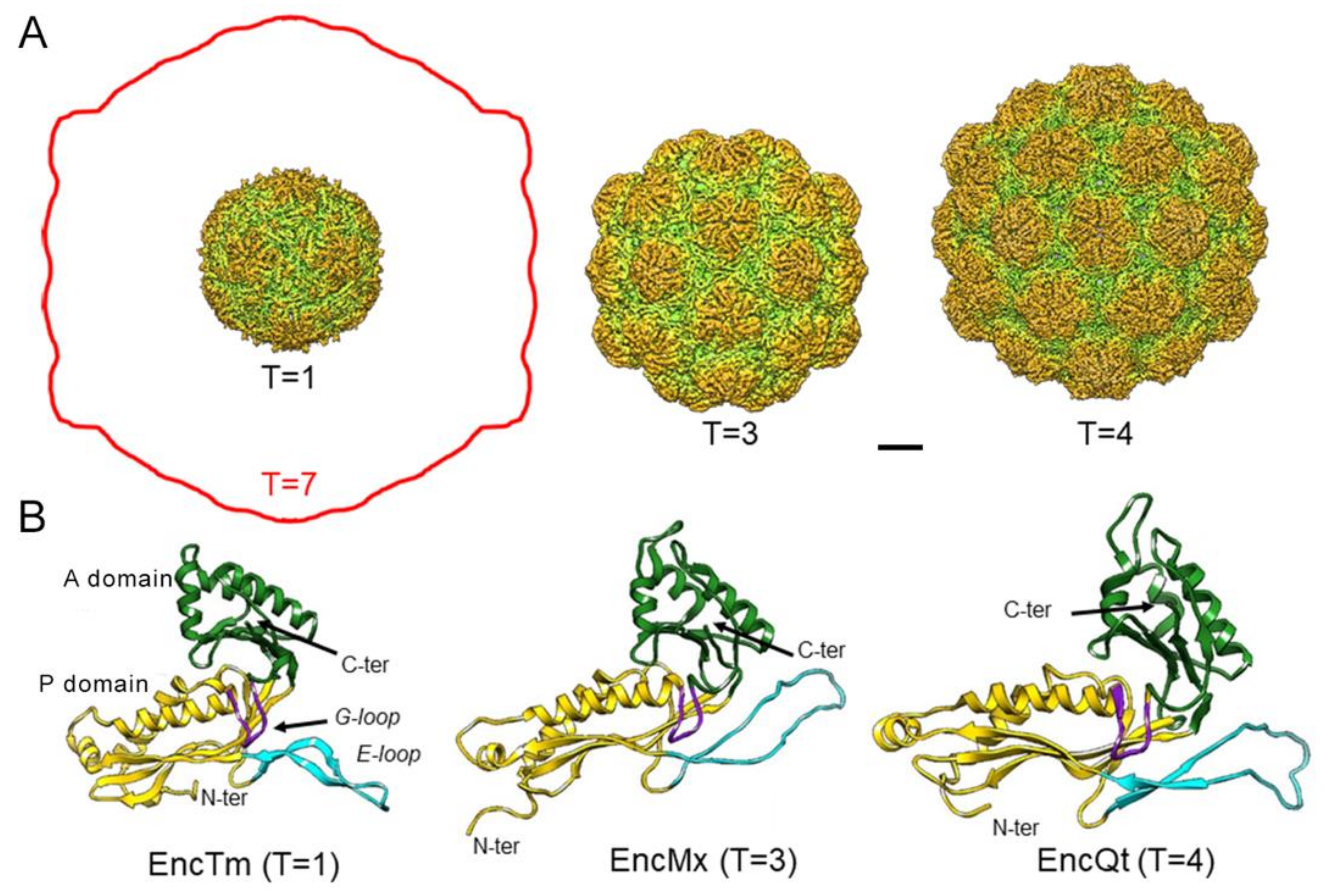
2.1. Nanocompartment Structure
2.2. Encapsulin Systems
2.2.1. Family 1
2.2.2. Family 2
2.2.3. Family 3
2.2.4. Family 4
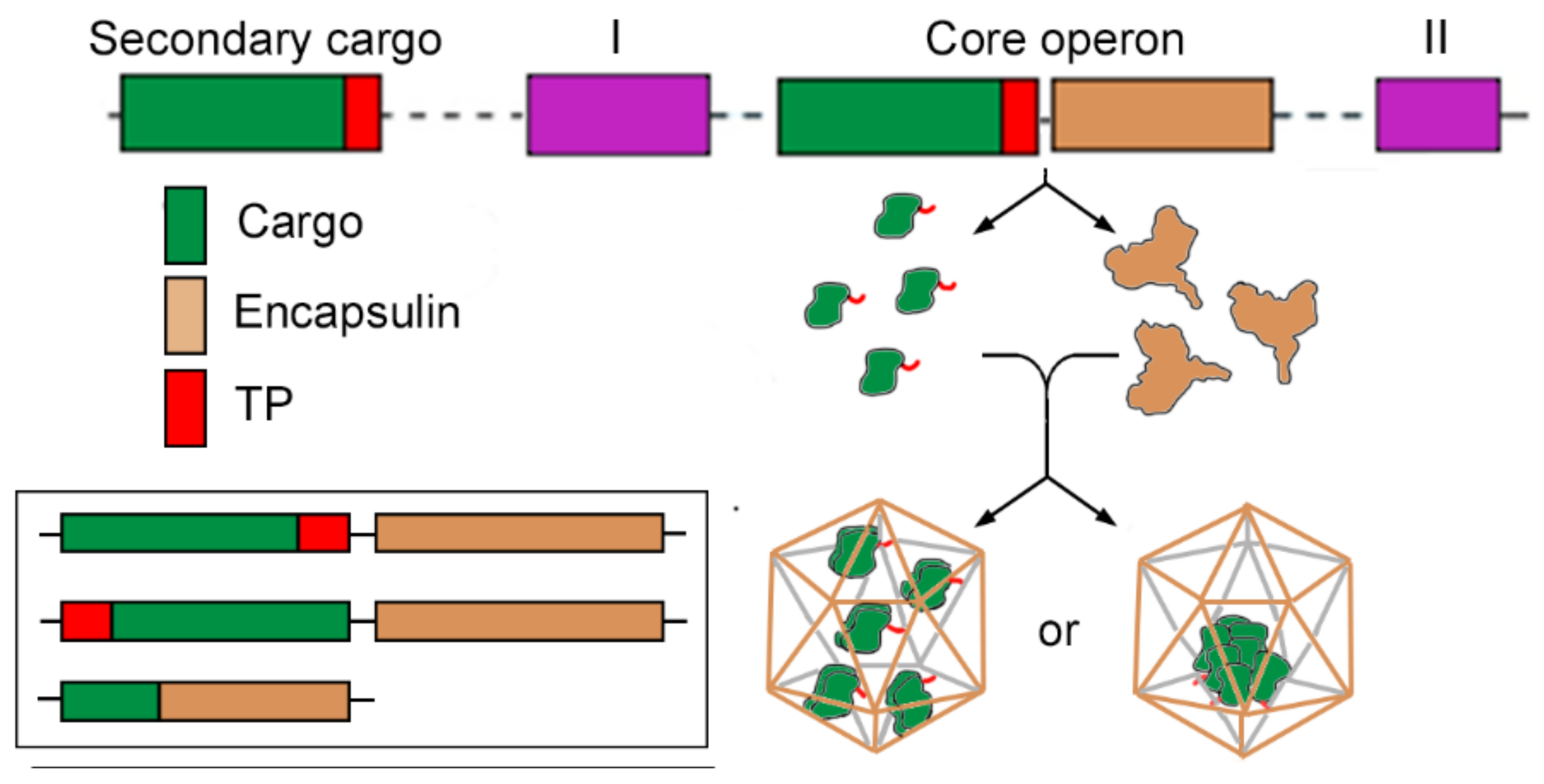
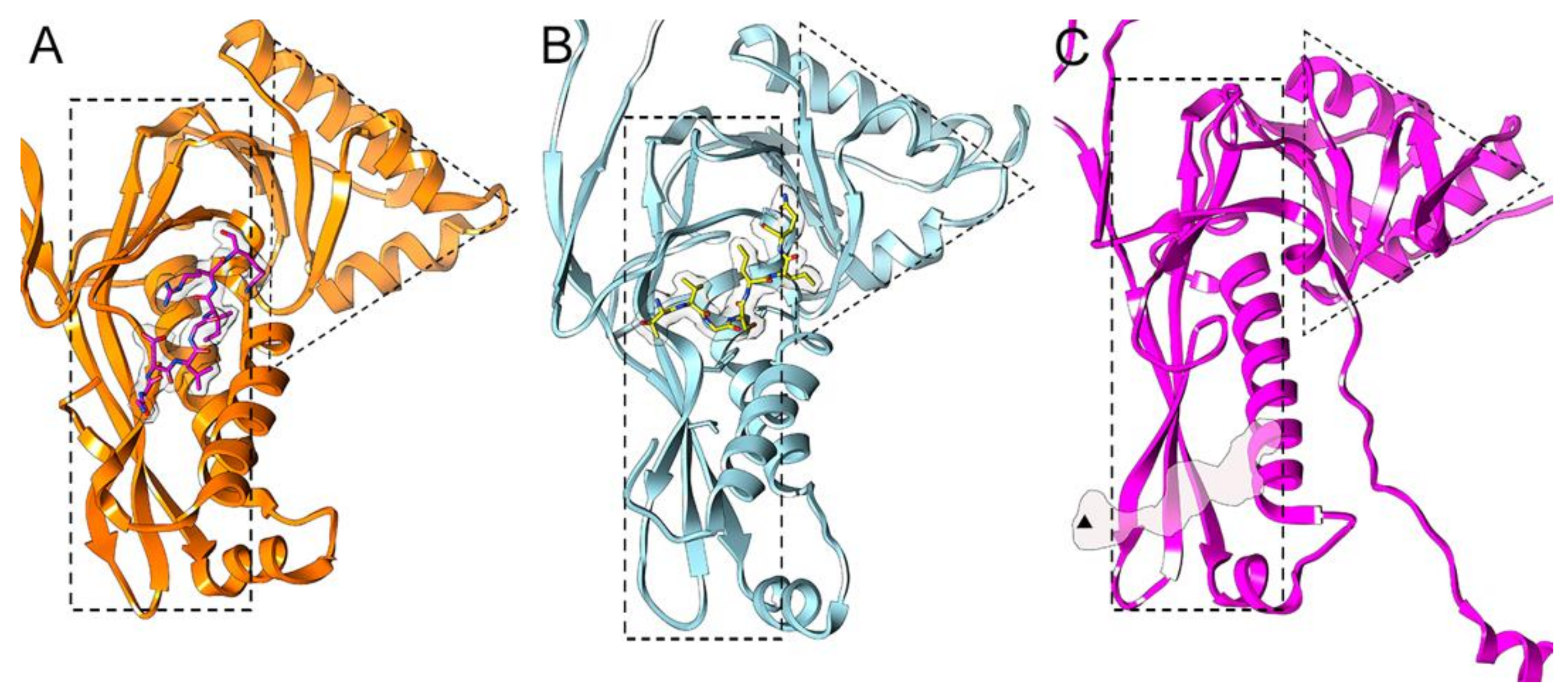
2.3. Cargo Loading
2.4. Effect of Encapsulin on Cargo Protein Function
2.5. Physiological Role of Encapsulins
3. The Encapsulin Toolbox
3.1. Use of Encapsulin Systems for Nanotechnological Applications
3.2. Encapsulin Expression and Purification
3.3. Shell Engineering
4. Encapsulin-Based Nanotechnological Applications
4.1. Encapsulins as Nanoreactors
4.2. Encapsulins as Targeted Delivery Systems and Nanovaccine Platfforms
4.3. Encapsulins as Genetically Encodable Materials for Biological Imaging
4.4. Encapsulin-Based Metallic Nanoparticles
5. Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greening, C.; Lithgow, T. Formation and function of bacterial organelles. Nat. Rev. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, J.; Lowe, J. Prokaryotic cytoskeletons: Protein filaments organizing small cells. Nat. Rev. Microbiol. 2018, 16, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Surovtsev, I.V.; Jacobs-Wagner, C. Subcellular Organization: A Critical Feature of Bacterial Cell Replication. Cell 2018, 172, 1271–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, C.R.; Wan, J.; Komeili, A. Organelle Formation in Bacteria and Archaea. Annu. Rev. Cell Dev. Biol. 2018, 34, 217–238. [Google Scholar] [CrossRef]
- Jones, J.A.; Giessen, T.W. Advances in encapsulin nanocompartment biology and engineering. Biotechnol. Bioeng. 2021, 118, 491–505. [Google Scholar] [CrossRef]
- Sutter, M.; Boehringer, D.; Gutmann, S.; Gunther, S.; Prangishvili, D.; Loessner, M.J.; Stetter, K.O.; Weber-Ban, E.; Ban, N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 2008, 15, 939–947. [Google Scholar] [CrossRef]
- Uebe, R.; Schuler, D. Magnetosome biogenesis in magnetotactic bacteria. Nat. Rev. Microbiol. 2016, 14, 621–637. [Google Scholar] [CrossRef]
- Fuerst, J.A. Intracellular compartmentation in planctomycetes. Annu. Rev. Microbiol. 2005, 59, 299–328. [Google Scholar] [CrossRef]
- Azaldegui, C.A.; Vecchiarelli, A.G.; Biteen, J.S. The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 2021, 120, 1123–1138. [Google Scholar] [CrossRef]
- Kerfeld, C.A.; Aussignargues, C.; Zarzycki, J.; Cai, F.; Sutter, M. Bacterial microcompartments. Nat. Rev. Microbiol. 2018, 16, 277–290. [Google Scholar] [CrossRef]
- Sutter, M.; Melnicki, M.R.; Schulz, F.; Woyke, T.; Kerfeld, C.A. A Catalog of the Diversity and Ubiquity of Metabolic Organelles in Bacteria. bioRxiv 2021. [Google Scholar] [CrossRef]
- Nichols, R.J.; Cassidy-Amstutz, C.; Chaijarasphong, T.; Savage, D.F. Encapsulins: Molecular biology of the shell. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 583–594. [Google Scholar] [CrossRef]
- Gabashvili, A.N.; Chmelyuk, N.S.; Efremova, M.V.; Malinovskaya, J.A.; Semkina, A.S.; Abakumov, M.A. Encapsulins-Bacterial Protein Nanocompartments: Structure, Properties, and Application. Biomolecules 2020, 10, 966. [Google Scholar] [CrossRef]
- Yang, M.; Simpson, D.M.; Wenner, N.; Brownridge, P.; Harman, V.M.; Hinton, J.C.D.; Beynon, R.J.; Liu, L.N. Decoding the stoichiometric composition and organisation of bacterial metabolosomes. Nat. Commun. 2020, 11, 1976. [Google Scholar] [CrossRef]
- Rae, B.D.; Long, B.M.; Whitehead, L.F.; Forster, B.; Badger, M.R.; Price, G.D. Cyanobacterial carboxysomes: Microcompartments that facilitate CO2 fixation. J. Mol. Microbiol. Biotechnol. 2013, 23, 300–307. [Google Scholar] [CrossRef]
- Long, B.M.; Rae, B.D.; Rolland, V.; Forster, B.; Price, G.D. Cyanobacterial CO2-concentrating mechanism components: Function and prospects for plant metabolic engineering. Curr. Opin. Plant Biol. 2016, 31, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dou, Z.; Heinhorst, S.; Williams, E.B.; Murin, C.D.; Shively, J.M.; Cannon, G.C. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 2008, 283, 10377–10384. [Google Scholar] [CrossRef] [Green Version]
- Cai, F.; Menon, B.B.; Cannon, G.C.; Curry, K.J.; Shively, J.M.; Heinhorst, S. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS ONE 2009, 4, e7521. [Google Scholar] [CrossRef] [Green Version]
- Demchuk, A.M.; Patel, T.R. The biomedical and bioengineering potential of protein nanocompartments. Biotechnol. Adv. 2020, 41, 107547. [Google Scholar] [CrossRef]
- Azuma, Y.; Edwardson, T.G.W.; Hilvert, D. Tailoring lumazine synthase assemblies for bionanotechnology. Chem. Soc. Rev. 2018, 47, 3543–3557. [Google Scholar] [CrossRef]
- He, D.; Marles-Wright, J. Ferritin family proteins and their use in bionanotechnology. New Biotechnol. 2015, 32, 651–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Ruiter, M.V.; Klem, R.; Luque, D.; Cornelissen, J.; Caston, J.R. Structural nanotechnology: Three-dimensional cryo-EM and its use in the development of nanoplatforms for in vitro catalysis. Nanoscale 2019, 11, 4130–4146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rome, L.H.; Kickhoefer, V.A. Development of the vault particle as a platform technology. ACS Nano 2013, 7, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Bu, W.; Joyce, M.G.; Meng, G.; Whittle, J.R.; Baxa, U.; Yamamoto, T.; Narpala, S.; Todd, J.P.; Rao, S.S.; et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 2015, 162, 1090–1100. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.J.; Lee, N.K.; Kim, I.S. Bioengineered protein-based nanocage for drug delivery. Adv. Drug Deliv. Rev. 2016, 106, 157–171. [Google Scholar] [CrossRef]
- Schwarz, B.; Uchida, M.; Douglas, T. Biomedical and Catalytic Opportunities of Virus-Like Particles in Nanotechnology. Adv. Virus Res. 2017, 97, 1–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhen, Z.; Tang, W.; Guo, C.; Chen, H.; Lin, X.; Liu, G.; Fei, B.; Chen, X.; Xu, B.; Xie, J. Ferritin nanocages to encapsulate and deliver photosensitizers for efficient photodynamic therapy against cancer. ACS Nano 2013, 7, 6988–6996. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Guenther, R.H.; Sit, T.L.; Lommel, S.A.; Opperman, C.H.; Willoughby, J.A. Development of abamectin loaded plant virus nanoparticles for efficacious plant parasitic nematode control. ACS Appl. Mater. Interfaces 2015, 7, 9546–9553. [Google Scholar] [CrossRef]
- Akita, F.; Chong, K.T.; Tanaka, H.; Yamashita, E.; Miyazaki, N.; Nakaishi, Y.; Suzuki, M.; Namba, K.; Ono, Y.; Tsukihara, T.; et al. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J. Mol. Biol. 2007, 368, 1469–1483. [Google Scholar] [CrossRef]
- McHugh, C.A.; Fontana, J.; Nemecek, D.; Cheng, N.; Aksyuk, A.A.; Heymann, J.B.; Winkler, D.C.; Lam, A.S.; Wall, J.S.; Steven, A.C.; et al. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J. 2014, 33, 1896–1911. [Google Scholar] [CrossRef] [Green Version]
- Nichols, R.J.; LaFrance, B.; Phillips, N.R.; Radford, D.R.; Oltrogge, L.M.; Valentin-Alvarado, L.E.; Bischoff, A.J.; Nogales, E.; Savage, D.F. Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism. eLife 2021, 10. [Google Scholar] [CrossRef]
- Andreas, M.P.; Giessen, T.W. Large-scale computational discovery and analysis of virus-derived microbial nanocompartments. bioRxiv 2021. [Google Scholar] [CrossRef]
- Cassidy-Amstutz, C.; Oltrogge, L.; Going, C.C.; Lee, A.; Teng, P.; Quintanilla, D.; East-Seletsky, A.; Williams, E.R.; Savage, D.F. Identification of a Minimal Peptide Tag for in Vivo and in Vitro Loading of Encapsulin. Biochemistry 2016, 55, 3461–3468. [Google Scholar] [CrossRef]
- Altenburg, W.J.; Rollins, N.; Silver, P.A.; Giessen, T.W. Exploring targeting peptide-shell interactions in encapsulin nanocompartments. Sci. Rep. 2021, 11, 4951. [Google Scholar] [CrossRef]
- Giessen, T.W.; Silver, P.A. Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat. Microbiol. 2017, 2, 17029. [Google Scholar] [CrossRef]
- He, D.; Piergentili, C.; Ross, J.; Tarrant, E.; Tuck, L.R.; Mackay, C.L.; McIver, Z.; Waldron, K.J.; Clarke, D.J.; Marles-Wright, J. Conservation of the structural and functional architecture of encapsulated ferritins in bacteria and archaea. Biochem. J. 2019, 476, 975–989. [Google Scholar] [CrossRef] [Green Version]
- Valdes-Stauber, N.; Scherer, S. Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens. Appl. Environ. Microbiol. 1994, 60, 3809–3814. [Google Scholar] [CrossRef] [Green Version]
- Valdes-Stauber, N.; Scherer, S. Nucleotide sequence and taxonomical distribution of the bacteriocin gene lin cloned from Brevibacterium linens M18. Appl. Environ. Microbiol. 1996, 62, 1283–1286. [Google Scholar] [CrossRef] [Green Version]
- Winter, N.; Triccas, J.A.; Rivoire, B.; Pessolani, M.C.; Eiglmeier, K.; Lim, E.M.; Hunter, S.W.; Brennan, P.J.; Britton, W.J. Characterization of the gene encoding the immunodominant 35 kDa protein of Mycobacterium leprae. Mol. Microbiol. 1995, 16, 865–876. [Google Scholar] [CrossRef]
- Triccas, J.A.; Roche, P.W.; Winter, N.; Feng, C.G.; Butlin, C.R.; Britton, W.J. A 35-kilodalton protein is a major target of the human immune response to Mycobacterium leprae. Infect. Immun. 1996, 64, 5171–5177. [Google Scholar] [CrossRef] [Green Version]
- Rosenkrands, I.; Rasmussen, P.B.; Carnio, M.; Jacobsen, S.; Theisen, M.; Andersen, P. Identification and characterization of a 29-kilodalton protein from Mycobacterium tuberculosis culture filtrate recognized by mouse memory effector cells. Infect. Immun. 1998, 66, 2728–2735. [Google Scholar] [CrossRef] [Green Version]
- Hicks, P.M.; Rinker, K.D.; Baker, J.R.; Kelly, R.M. Homomultimeric protease in the hyperthermophilic bacterium Thermotoga maritima has structural and amino acid sequence homology to bacteriocins in mesophilic bacteria. FEBS Lett. 1998, 440, 393–398. [Google Scholar] [CrossRef] [Green Version]
- Kawamoto, S.; Watanabe, M.; Saito, N.; Hesketh, A.; Vachalova, K.; Matsubara, K.; Ochi, K. Molecular and functional analyses of the gene (eshA) encoding the 52-kilodalton protein of Streptomyces coelicolor A3(2) required for antibiotic production. J. Bacteriol. 2001, 183, 6009–6016. [Google Scholar] [CrossRef] [Green Version]
- Tracey, J.C.; Coronado, M.; Giessen, T.W.; Lau, M.C.Y.; Silver, P.A.; Ward, B.B. The Discovery of Twenty-Eight New Encapsulin Sequences, Including Three in Anammox Bacteria. Sci. Rep. 2019, 9, 20122. [Google Scholar] [CrossRef]
- Malik, H.S.; Henikoff, S.; Eickbush, T.H. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000, 10, 1307–1318. [Google Scholar] [CrossRef] [Green Version]
- Feschotte, C.; Gilbert, C. Endogenous viruses: Insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012, 13, 283–296. [Google Scholar] [CrossRef] [Green Version]
- Ghequire, M.G.K.; De Mot, R. The Tailocin Tale: Peeling off Phage Tails. Trends Microbiol. 2015, 23, 587–590. [Google Scholar] [CrossRef] [Green Version]
- Scholl, D. Phage tail–like bacteriocins. Annu. Rev. Virol. 2017, 4, 453–467. [Google Scholar] [CrossRef]
- Giessen, T.W.; Orlando, B.J.; Verdegaal, A.A.; Chambers, M.G.; Gardener, J.; Bell, D.C.; Birrane, G.; Liao, M.; Silver, P.A. Large protein organelles form a new iron sequestration system with high storage capacity. eLife 2019, 8. [Google Scholar] [CrossRef]
- Loncar, N.; Rozeboom, H.J.; Franken, L.E.; Stuart, M.C.A.; Fraaije, M.W. Structure of a robust bacterial protein cage and its application as a versatile biocatalytic platform through enzyme encapsulation. Biochem. Biophys. Res. Commun. 2020, 529, 548–553. [Google Scholar] [CrossRef]
- Tang, Y.; Mu, A.; Zhang, Y.; Zhou, S.; Wang, W.; Lai, Y.; Zhou, X.; Liu, F.; Yang, X.; Gong, H.; et al. Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; McIver, Z.; Lambert, T.; Piergentili, C.; Gallagher, K.J.; Bird, J.E.; Cruickshank, F.L.; Zarazúa-Arvizu, E.; Horsfall, L.E.; Waldron, K.J.; et al. Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment revealed by Cryo-EM and hydrogen/deuterium exchange mass spectrometry. bioRxiv 2021. [Google Scholar] [CrossRef]
- Putri, R.M.; Allende-Ballestero, C.; Luque, D.; Klem, R.; Rousou, K.A.; Liu, A.; Traulsen, C.H.; Rurup, W.F.; Koay, M.S.T.; Caston, J.R.; et al. Structural Characterization of Native and Modified Encapsulins as Nanoplatforms for in Vitro Catalysis and Cellular Uptake. ACS Nano 2017, 11, 12796–12804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirykowicz, A.M.; Woodward, J.D. Shotgun EM of mycobacterial protein complexes during stationary phase stress. Curr. Res. Struct. Biol. 2020, 2, 204–212. [Google Scholar] [CrossRef]
- Caspar, D.L.D.; Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962, 27, 1–24. [Google Scholar] [CrossRef]
- Luque, D.; Caston, J.R. Cryo-electron microscopy for the study of virus assembly. Nat. Chem. Biol. 2020, 16, 231–239. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Liljas, L.; Duda, R.L.; Tsuruta, H.; Hendrix, R.W.; Johnson, J.E. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 2000, 289, 2129–2133. [Google Scholar] [CrossRef] [Green Version]
- Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origin of viruses: Primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 2019, 17, 449–458. [Google Scholar] [CrossRef]
- Suhanovsky, M.M.; Teschke, C.M. Nature’s favorite building block: Deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 2015, 479–480, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Duda, R.L.; Teschke, C.M. The amazing HK97 fold: Versatile results of modest differences. Curr. Opin. Virol. 2019, 36, 9–16. [Google Scholar] [CrossRef]
- Wiryaman, T.; Toor, N. Cryo-EM structure of a thermostable bacterial nanocompartment. IUCrJ 2021, 8. [Google Scholar] [CrossRef]
- Rurup, W.F.; Snijder, J.; Koay, M.S.; Heck, A.J.; Cornelissen, J.J. Self-sorting of foreign proteins in a bacterial nanocompartment. J. Am. Chem. Soc. 2014, 136, 3828–3832. [Google Scholar] [CrossRef]
- Tamura, A.; Fukutani, Y.; Takami, T.; Fujii, M.; Nakaguchi, Y.; Murakami, Y.; Noguchi, K.; Yohda, M.; Odaka, M. Packaging guest proteins into the encapsulin nanocompartment from Rhodococcus erythropolis N771. Biotechnol. Bioeng. 2015, 112, 13–20. [Google Scholar] [CrossRef]
- Snijder, J.; Kononova, O.; Barbu, I.M.; Uetrecht, C.; Rurup, W.F.; Burnley, R.J.; Koay, M.S.; Cornelissen, J.J.; Roos, W.H.; Barsegov, V.; et al. Assembly and Mechanical Properties of the Cargo-Free and Cargo-Loaded Bacterial Nanocompartment Encapsulin. Biomacromolecules 2016, 17, 2522–2529. [Google Scholar] [CrossRef]
- Singh, R.; Eltis, L.D. The multihued palette of dye-decolorizing peroxidases. Arch. Biochem. Biophys. 2015, 574, 56–65. [Google Scholar] [CrossRef]
- Mishra, S.; Imlay, J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 2012, 525, 145–160. [Google Scholar] [CrossRef] [Green Version]
- Catucci, G.; Valetti, F.; Sadeghi, S.J.; Gilardi, G. Biochemical features of dye-decolorizing peroxidases: Current impact on lignin degradation. Biotechnol. Appl. Biochem. 2020, 67, 751–759. [Google Scholar] [CrossRef]
- Andrews, S.C. The Ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta 2010, 1800, 691–705. [Google Scholar] [CrossRef]
- He, D.; Hughes, S.; Vanden-Hehir, S.; Georgiev, A.; Altenbach, K.; Tarrant, E.; Mackay, C.L.; Waldron, K.J.; Clarke, D.J.; Marles-Wright, J. Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments. eLife 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Honarmand Ebrahimi, K.; Bill, E.; Hagedoorn, P.L.; Hagen, W.R. The catalytic center of ferritin regulates iron storage via Fe(II)-Fe(III) displacement. Nat. Chem. Biol. 2012, 8, 941–948. [Google Scholar] [CrossRef]
- Xing, C.Y.; Fan, Y.C.; Chen, X.; Guo, J.S.; Shen, Y.; Yan, P.; Fang, F.; Chen, Y.P. A self-assembled nanocompartment in anammox bacteria for resisting intracelluar hydroxylamine stress. Sci. Total Environ. 2020, 717, 137030. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R. Natural Products and the Gene Cluster Revolution. Trends Microbiol. 2016, 24, 968–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palaniappan, K.; Chen, I.A.; Chu, K.; Ratner, A.; Seshadri, R.; Kyrpides, N.C.; Ivanova, N.N.; Mouncey, N.J. IMG-ABC v.5.0: An update to the IMG/Atlas of Biosynthetic Gene Clusters Knowledgebase. Nucleic Acids Res. 2020, 48, D422–D430. [Google Scholar] [CrossRef] [PubMed]
- Rahmanpour, R.; Bugg, T.D. Assembly in vitro of Rhodococcus jostii RHA1 encapsulin and peroxidase DypB to form a nanocompartment. FEBS J. 2013, 280, 2097–2104. [Google Scholar] [CrossRef]
- Kunzle, M.; Mangler, J.; Lach, M.; Beck, T. Peptide-directed encapsulation of inorganic nanoparticles into protein containers. Nanoscale 2018, 10, 22917–22926. [Google Scholar] [CrossRef]
- Contreras, H.; Joens, M.S.; McMath, L.M.; Le, V.P.; Tullius, M.V.; Kimmey, J.M.; Bionghi, N.; Horwitz, M.A.; Fitzpatrick, J.A.; Goulding, C.W. Characterization of a Mycobacterium tuberculosis nanocompartment and its potential cargo proteins. J. Biol. Chem. 2014, 289, 18279–18289. [Google Scholar] [CrossRef] [Green Version]
- Lien, K.A.; Nichols, R.J.; Cassidy-Amstutz, C.; Dinshaw, K.; Knight, M.; Singh, R.; Eltis, L.D.; Savage, D.F.; Stanley, S.A. A nanocompartment containing the peroxidase DypB contributes to defense against oxidative stress in M. tuberculosis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Snijder, J.; van de Waterbeemd, M.; Damoc, E.; Denisov, E.; Grinfeld, D.; Bennett, A.; Agbandje-McKenna, M.; Makarov, A.; Heck, A.J. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by Orbitrap mass spectrometry. J. Am. Chem. Soc. 2014, 136, 7295–7299. [Google Scholar] [CrossRef]
- Putri, R.M.; Fredy, J.W.; Cornelissen, J.J.; Koay, M.S.; Katsonis, N. Labelling Bacterial Nanocages with Photo-switchable Fluorophores. ChemPhysChem 2016, 17, 1815–1818. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.X.; Rivas, G.; Minton, A.P. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008, 37, 375–397. [Google Scholar] [CrossRef] [Green Version]
- Williams, E.M.; Jung, S.M.; Coffman, J.L.; Lutz, S. Pore Engineering for Enhanced Mass Transport in Encapsulin Nanocompartments. ACS Synth. Biol. 2018, 7, 2514–2517. [Google Scholar] [CrossRef]
- Adamson, L.; Tasneem, N.; Andreas, M.P.; Close, W.; Szyszka, T.N.; Jenner, E.; Young, R.; Cheah, L.C.; Norman, A.; Sainsbury, F.; et al. Pore structure controls stability and molecular flux in engineered protein cages. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jenkins, M.C.; Lutz, S. Encapsulin Nanocontainers as Versatile Scaffolds for the Development of Artificial Metabolons. ACS Synth. Biol. 2021, 10, 857–869. [Google Scholar] [CrossRef]
- Kim, D.; Choi, J.; Lee, S.; Hyun, H.; Lee, K.; Cho, K. Mutants defective in the production of encapsulin show a tan-phase-locked phenotype in Myxococcus xanthus. J. Microbiol. 2019, 57, 795–802. [Google Scholar] [CrossRef]
- Kim, D.; Chung, J.; Hyun, H.; Lee, C.; Lee, K.; Cho, K. Operon required for fruiting body development in Myxococcus xanthus. J. Microbiol. Biotechnol. 2009, 19, 1288–1294. [Google Scholar] [CrossRef]
- Sigmund, F.; Pettinger, S.; Kube, M.; Schneider, F.; Schifferer, M.; Schneider, S.; Efremova, M.V.; Pujol-Marti, J.; Aichler, M.; Walch, A.; et al. Iron-Sequestering Nanocompartments as Multiplexed Electron Microscopy Gene Reporters. ACS Nano 2019, 13, 8114–8123. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Chu, C.; Zhou, Z.; Chen, B.; Pang, X.; Lin, G.; Lin, H.; Guo, Y.; Ren, E.; et al. Genetically engineered magnetic nanocages for cancer magneto-catalytic theranostics. Nat. Commun. 2020, 11, 5421. [Google Scholar] [CrossRef]
- Sigmund, F.; Massner, C.; Erdmann, P.; Stelzl, A.; Rolbieski, H.; Desai, M.; Bricault, S.; Worner, T.P.; Snijder, J.; Geerlof, A.; et al. Bacterial encapsulins as orthogonal compartments for mammalian cell engineering. Nat. Commun. 2018, 9, 1990. [Google Scholar] [CrossRef] [Green Version]
- Zusman, D.R.; Scott, A.E.; Yang, Z.; Kirby, J.R. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 2007, 5, 862–872. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Reddy, M.C.; Ioerger, T.R.; Rothchild, A.C.; Dartois, V.; Schuster, B.M.; Trauner, A.; Wallis, D.; Galaviz, S.; Huttenhower, C.; et al. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 2013, 155, 1296–1308. [Google Scholar] [CrossRef] [Green Version]
- Weldingh, K.; Andersen, P. Immunological evaluation of novel Mycobacterium tuberculosis culture filtrate proteins. FEMS Immunol. Med. Microbiol. 1999, 23, 159–164. [Google Scholar] [CrossRef] [Green Version]
- Weiss, G.; Schaible, U.E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015, 264, 182–203. [Google Scholar] [CrossRef] [Green Version]
- Kamimura, N.; Sakamoto, S.; Mitsuda, N.; Masai, E.; Kajita, S. Advances in microbial lignin degradation and its applications. Curr. Opin. Biotechnol. 2019, 56, 179–186. [Google Scholar] [CrossRef]
- Ahmad, M.; Roberts, J.N.; Hardiman, E.M.; Singh, R.; Eltis, L.D.; Bugg, T.D. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 2011, 50, 5096–5107. [Google Scholar] [CrossRef]
- Uchida, T.; Sasaki, M.; Tanaka, Y.; Ishimori, K. A Dye-Decolorizing Peroxidase from Vibrio cholerae. Biochemistry 2015, 54, 6610–6621. [Google Scholar] [CrossRef]
- Shrestha, R.; Chen, X.; Ramyar, K.X.; Hayati, Z.; Carlson, E.A.; Bossmann, S.H.; Song, L.; Geisbrecht, B.V.; Li, P. Identification of Surface-Exposed Protein Radicals and A Substrate Oxidation Site in A-Class Dye-Decolorizing Peroxidase from Thermomonospora curvata. ACS Catal. 2016, 6, 8036–8047. [Google Scholar] [CrossRef] [Green Version]
- Lau, Y.H.; Giessen, T.W.; Altenburg, W.J.; Silver, P.A. Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat. Commun. 2018, 9, 1311. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Sun, C.; Vago, F.S.; Klose, T.; Zhu, J.; Jiang, W. Cryo-EM Structure of Heterologous Protein Complex Loaded Thermotoga Maritima Encapsulin Capsid. Biomolecules 2020, 10, 1342. [Google Scholar] [CrossRef]
- Efremova, M.V.; Bodea, S.V.; Sigmund, F.; Semkina, A.; Westmeyer, G.G.; Abakumov, M.A. Genetically Encoded Self-Assembling Iron Oxide Nanoparticles as a Possible Platform for Cancer-Cell Tracking. Pharmaceutics 2021, 13, 397. [Google Scholar] [CrossRef]
- Moon, H.; Lee, J.; Min, J.; Kang, S. Developing genetically engineered encapsulin protein cage nanoparticles as a targeted delivery nanoplatform. Biomacromolecules 2014, 15, 3794–3801. [Google Scholar] [CrossRef]
- Moon, H.; Lee, J.; Kim, H.; Heo, S.; Min, J.; Kang, S. Genetically engineering encapsulin protein cage nanoparticle as a SCC-7 cell targeting optical nanoprobe. Biomater. Res. 2014, 18, 21. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.H.; Carpenter, T.S.; D’Haeseleer, P.; Savage, D.F.; Yung, M.C. Encapsulin carrier proteins for enhanced expression of antimicrobial peptides. Biotechnol. Bioeng. 2020, 117, 603–613. [Google Scholar] [CrossRef]
- Diaz, D.; Vidal, X.; Sunna, A.; Care, A. Bioengineering a Light-Responsive Encapsulin Nanoreactor: A Potential Tool for In Vitro Photodynamic Therapy. ACS Appl. Mater. Interfaces 2021, 13, 7977–7986. [Google Scholar] [CrossRef]
- Sonotaki, S.; Noguchi, K.; Yohda, M.; Murakami, Y. A zeolite as a tool for successful refolding of PEGylated proteins and their reassembly with tertiary structures. Biotechnol. Prog. 2019, 35, e2853. [Google Scholar] [CrossRef]
- Sonotaki, S.; Takami, T.; Noguchi, K.; Odaka, M.; Yohda, M.; Murakami, Y. Successful PEGylation of hollow encapsulin nanoparticles from Rhodococcus erythropolis N771 without affecting their disassembly and reassembly properties. Biomater. Sci. 2017, 5, 1082–1089. [Google Scholar] [CrossRef]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef] [Green Version]
- Klem, R.; de Ruiter, M.V.; Cornelissen, J. Protecting Encapsulin Nanoparticles with Cysteine-Knot Miniproteins. Mol. Pharm. 2018, 15, 2991–2996. [Google Scholar] [CrossRef]
- Lagoutte, P.; Mignon, C.; Stadthagen, G.; Potisopon, S.; Donnat, S.; Mast, J.; Lugari, A.; Werle, B. Simultaneous surface display and cargo loading of encapsulin nanocompartments and their use for rational vaccine design. Vaccine 2018, 36, 3622–3628. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef] [Green Version]
- Bae, Y.; Kim, G.J.; Kim, H.; Park, S.G.; Jung, H.S.; Kang, S. Engineering Tunable Dual Functional Protein Cage Nanoparticles Using Bacterial Superglue. Biomacromolecules 2018, 19, 2896–2904. [Google Scholar] [CrossRef]
- Giessen, T.W.; Silver, P.A. Converting a Natural Protein Compartment into a Nanofactory for the Size-Constrained Synthesis of Antimicrobial Silver Nanoparticles. ACS Synth. Biol. 2016, 5, 1497–1504. [Google Scholar] [CrossRef]
- Torra, J.; Lafaye, C.; Signor, L.; Aumonier, S.; Flors, C.; Shu, X.; Nonell, S.; Gotthard, G.; Royant, A. Tailing miniSOG: Structural bases of the complex photophysics of a flavin-binding singlet oxygen photosensitizing protein. Sci. Rep. 2019, 9, 2428. [Google Scholar] [CrossRef]
- Shu, X.; Lev-Ram, V.; Deerinck, T.J.; Qi, Y.; Ramko, E.B.; Davidson, M.W.; Jin, Y.; Ellisman, M.H.; Tsien, R.Y. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011, 9, e1001041. [Google Scholar] [CrossRef] [Green Version]
- Kohler, A.C.; Mills, M.J.L.; Adams, P.D.; Simmons, B.A.; Sale, K.L. Structure of aryl O-demethylase offers molecular insight into a catalytic tyrosine-dependent mechanism. Proc. Natl. Acad. Sci. USA 2017, 114, E3205–E3214. [Google Scholar] [CrossRef] [Green Version]
- Manzari, M.T.; Shamay, Y.; Kiguchi, H.; Rosen, N.; Scaltriti, M.; Heller, D.A. Targeted drug delivery strategies for precision medicines. Nat. Rev. Mater. 2021, 6, 351–370. [Google Scholar] [CrossRef]
- Lo, A.; Lin, C.T.; Wu, H.C. Hepatocellular carcinoma cell-specific peptide ligand for targeted drug delivery. Mol. Cancer Ther. 2008, 7, 579–589. [Google Scholar] [CrossRef] [Green Version]
- Gomes, A.C.; Mohsen, M.; Bachmann, M.F. Harnessing Nanoparticles for Immunomodulation and Vaccines. Vaccines 2017, 5, 6. [Google Scholar] [CrossRef]
- Choi, B.; Moon, H.; Hong, S.J.; Shin, C.; Do, Y.; Ryu, S.; Kang, S. Effective Delivery of Antigen-Encapsulin Nanoparticle Fusions to Dendritic Cells Leads to Antigen-Specific Cytotoxic T Cell Activation and Tumor Rejection. ACS Nano 2016, 10, 7339–7350. [Google Scholar] [CrossRef]
- Kerppola, T.K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 2008, 37, 465–487. [Google Scholar] [CrossRef] [Green Version]
- Kerppola, T.K. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem. Soc. Rev. 2009, 38, 2876–2886. [Google Scholar]
- Ntziachristos, V. Going deeper than microscopy: The optical imaging frontier in biology. Nat. Methods 2010, 7, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Piraner, D.I.; Farhadi, A.; Davis, H.C.; Wu, D.; Maresca, D.; Szablowski, J.O.; Shapiro, M.G. Going Deeper: Biomolecular Tools for Acoustic and Magnetic Imaging and Control of Cellular Function. Biochemistry 2017, 56, 5202–5209. [Google Scholar] [CrossRef] [PubMed]
- Sendovski, M.; Kanteev, M.; Shuster Ben-Yosef, V.; Adir, N.; Fishman, A. Crystallization and preliminary X-ray crystallographic analysis of a bacterial tyrosinase from Bacillus megaterium. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1101–1103. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, T.B.; Cousins, R.J. The Multiple Faces of the Metal Transporter ZIP14 (SLC39A14). J. Nutr. 2018, 148, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Schrofel, A.; Kratosova, G.; Safarik, I.; Safarikova, M.; Raska, I.; Shor, L.M. Applications of biosynthesized metallic nanoparticles—A review. Acta Biomater. 2014, 10, 4023–4042. [Google Scholar] [CrossRef] [PubMed]
- Myrovali, E.; Maniotis, N.; Samaras, T.; Angelakeris, M. Spatial focusing of magnetic particle hyperthermia. Nanoscale Adv. 2020, 2, 408–416. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Kim, D.H.; Woo, Y.; Hur, H.G.; Lim, Y. Solution structure of peptide AG4 used to form silver nanoparticles. Biochem. Biophys. Res. Commun. 2008, 376, 595–598. [Google Scholar] [CrossRef]
- Tang, S.; Zheng, J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Healthc. Mater. 2018, 7, e1701503. [Google Scholar] [CrossRef]
- Vigderman, L.; Manna, P.; Zubarev, E.R. Quantitative replacement of cetyl trimethylammonium bromide by cationic thiol ligands on the surface of gold nanorods and their extremely large uptake by cancer cells. Angew. Chem. 2012, 51, 636–641. [Google Scholar] [CrossRef]
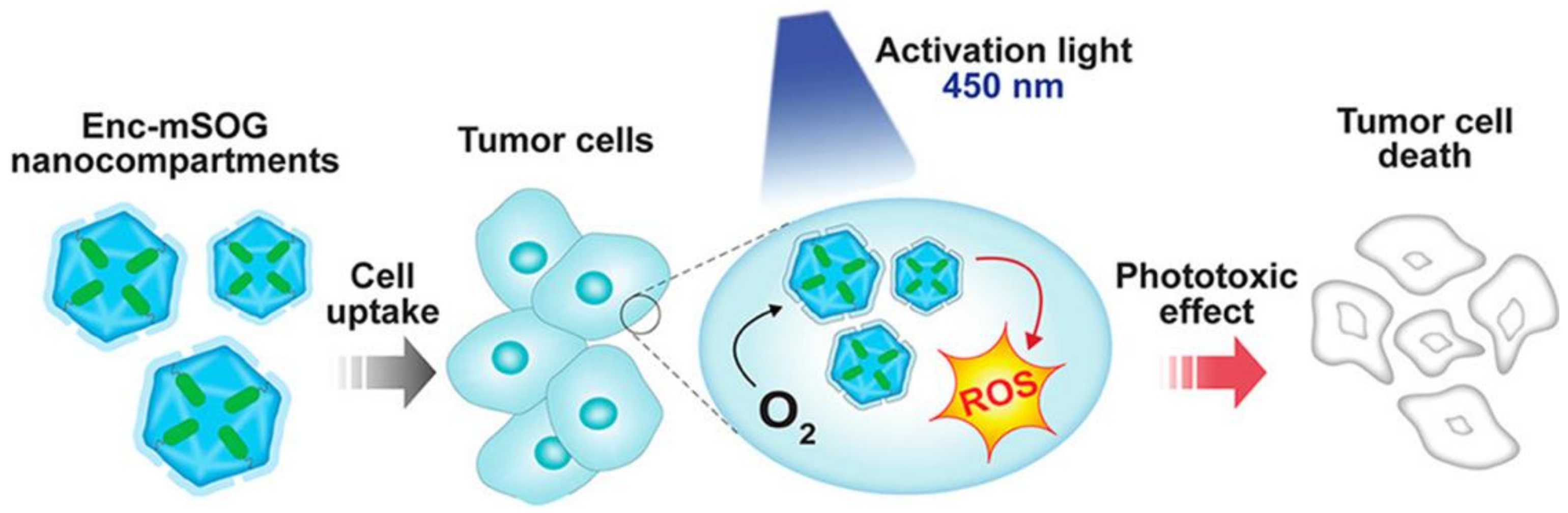
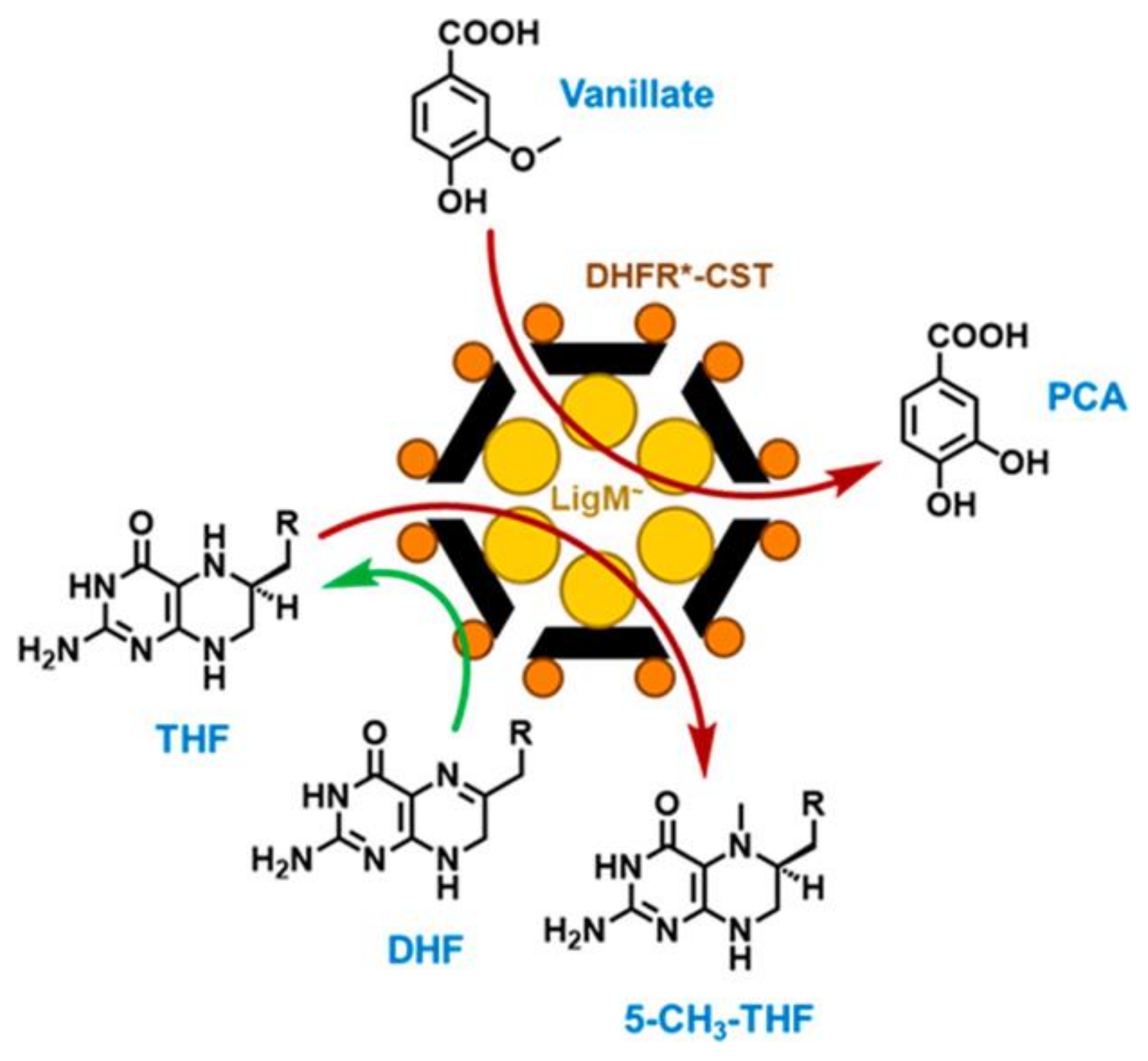
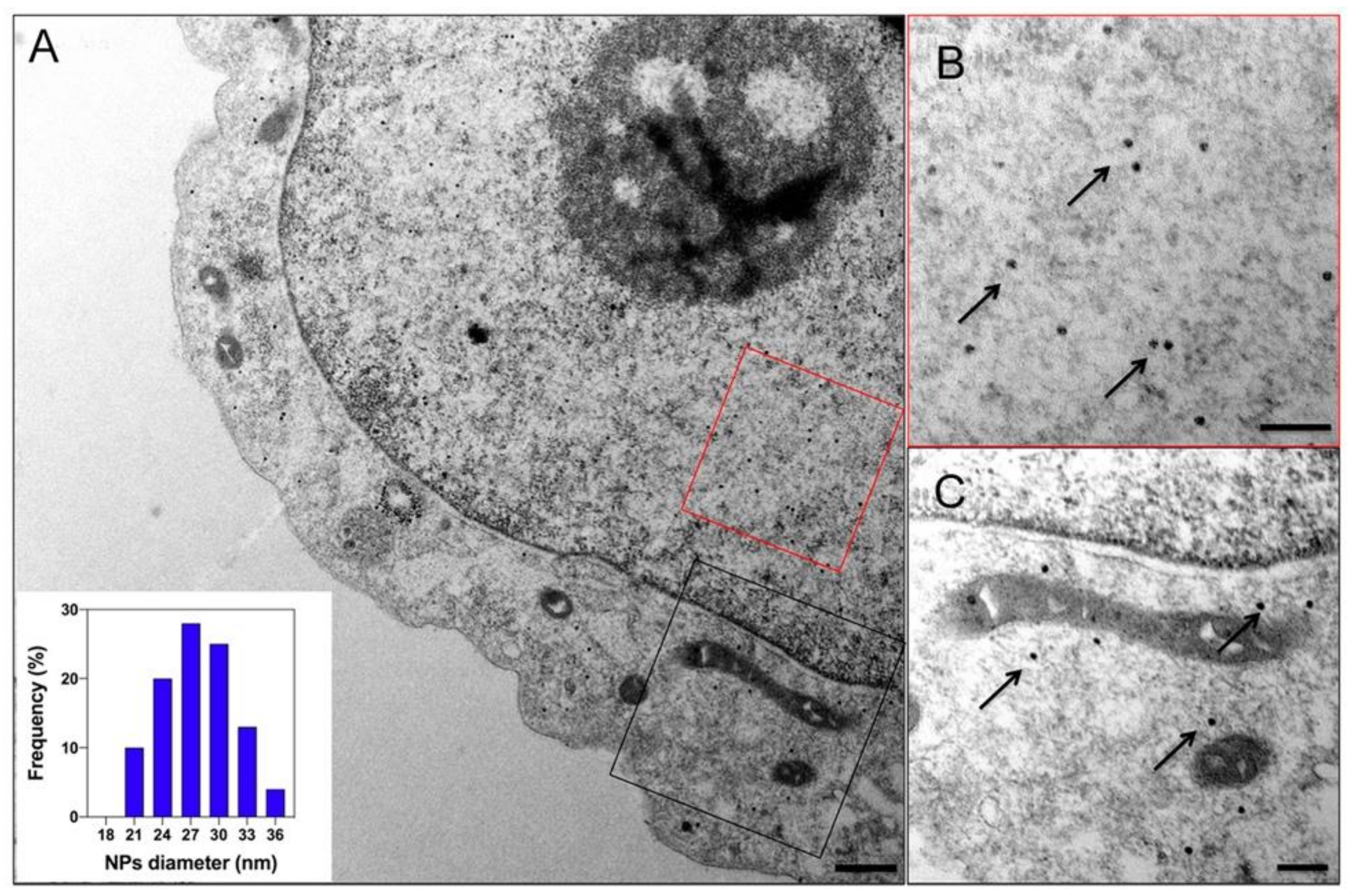
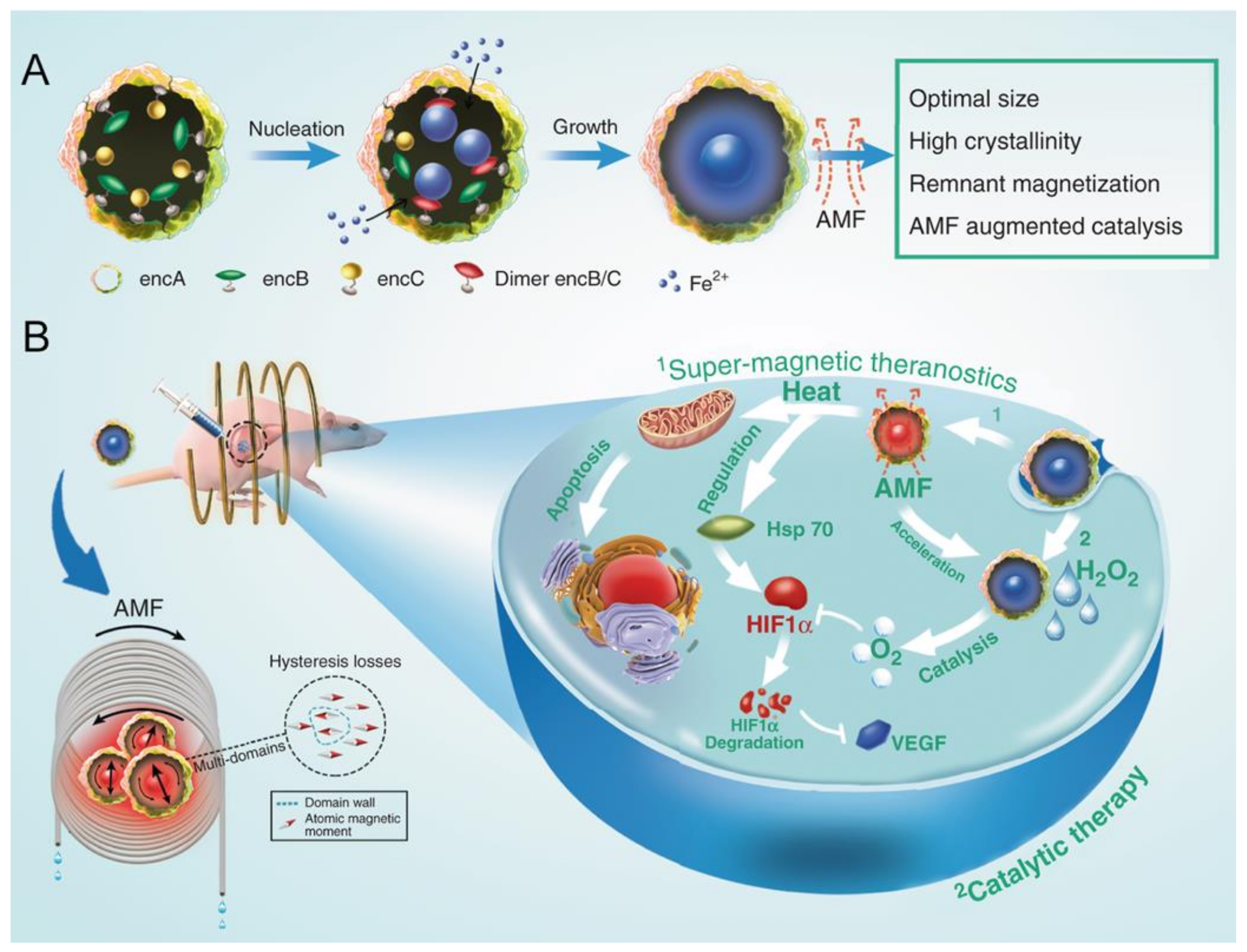
| Name | Application | References |
|---|---|---|
| Brevibacterium linens M18 | Biological imaging | [62,79] |
| Shell improvement | [107] | |
| Mixococcus xanthus | Biological imaging | [88] |
| Nanoreactor engineering | [88,97] | |
| Hyperthermia therapy | [87] | |
| Quasibacillus thermotolerans | Biological imaging | [86,99] |
| Mycolicibacterium hassiacum | Nanoreactor engineering | [50] |
| Thermotoga maritima | Targeted delivery | [100,101,110] |
| Vaccine development | [24,108,118] | |
| Nanoreactor engineering | [83,103] | |
| Microbial peptide synthesis | [102] | |
| Biometallic nanoparticle synthesis | [111] | |
| Rhodococcus erythropolis N771 | Shell improvement | [104,105] |
| Nanoreactor engineering | [63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, J.M.; Allende-Ballestero, C.; Cornelissen, J.J.L.M.; Castón, J.R. Nanotechnological Applications Based on Bacterial Encapsulins. Nanomaterials 2021, 11, 1467. https://doi.org/10.3390/nano11061467
Rodríguez JM, Allende-Ballestero C, Cornelissen JJLM, Castón JR. Nanotechnological Applications Based on Bacterial Encapsulins. Nanomaterials. 2021; 11(6):1467. https://doi.org/10.3390/nano11061467
Chicago/Turabian StyleRodríguez, Javier M., Carolina Allende-Ballestero, Jeroen J. L. M. Cornelissen, and José R. Castón. 2021. "Nanotechnological Applications Based on Bacterial Encapsulins" Nanomaterials 11, no. 6: 1467. https://doi.org/10.3390/nano11061467
APA StyleRodríguez, J. M., Allende-Ballestero, C., Cornelissen, J. J. L. M., & Castón, J. R. (2021). Nanotechnological Applications Based on Bacterial Encapsulins. Nanomaterials, 11(6), 1467. https://doi.org/10.3390/nano11061467