Deep Learning Based Instance Segmentation of Titanium Dioxide Particles in the Form of Agglomerates in Scanning Electron Microscopy
Abstract
1. Introduction
2. Deep Learning Based Instance Segmentation
3. Adaptation of the Mask RCNN for the Detection of TiO Particles Measured by SEM
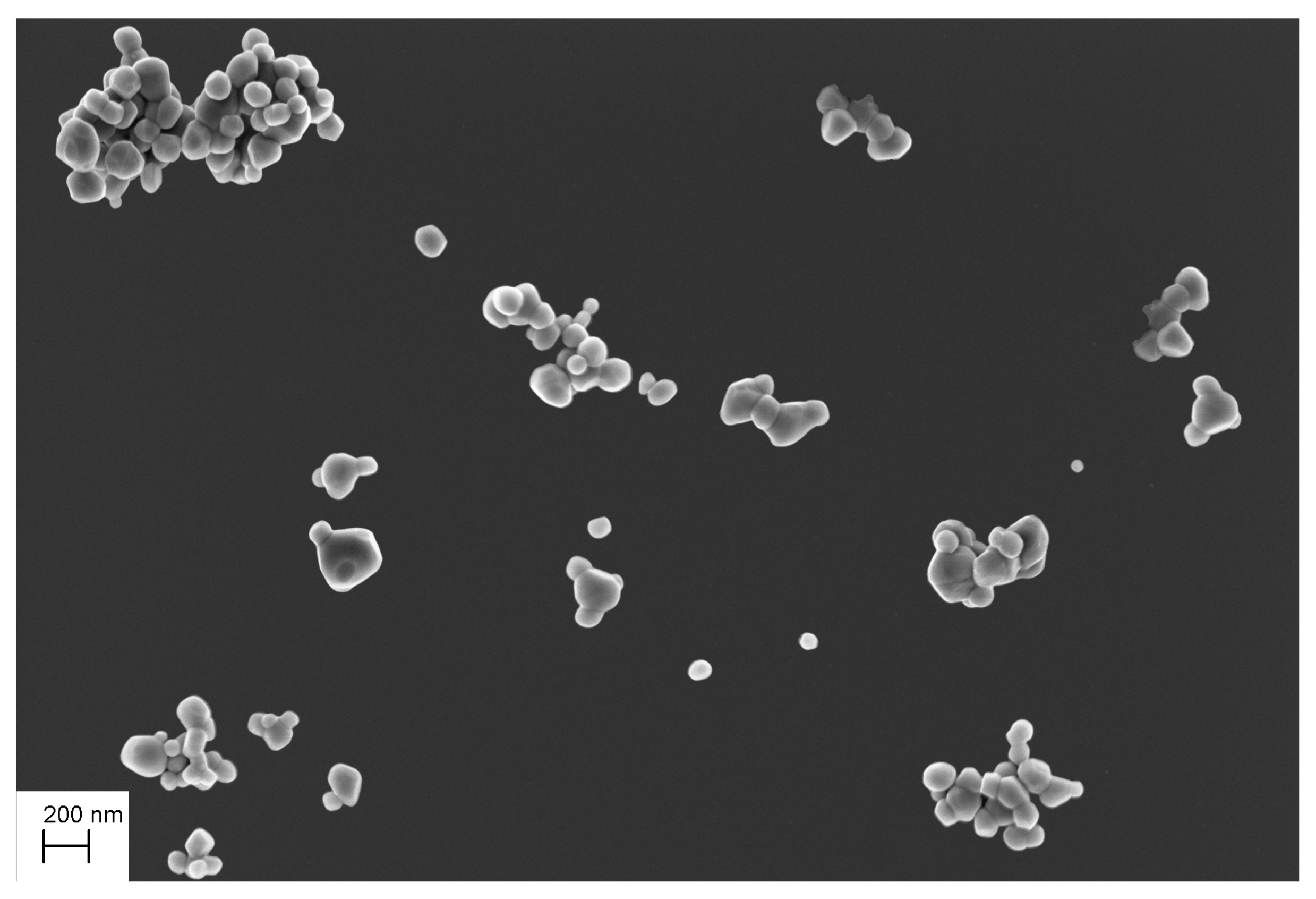
3.1. Creating the Function-Specific Database
3.2. Data Augmentation
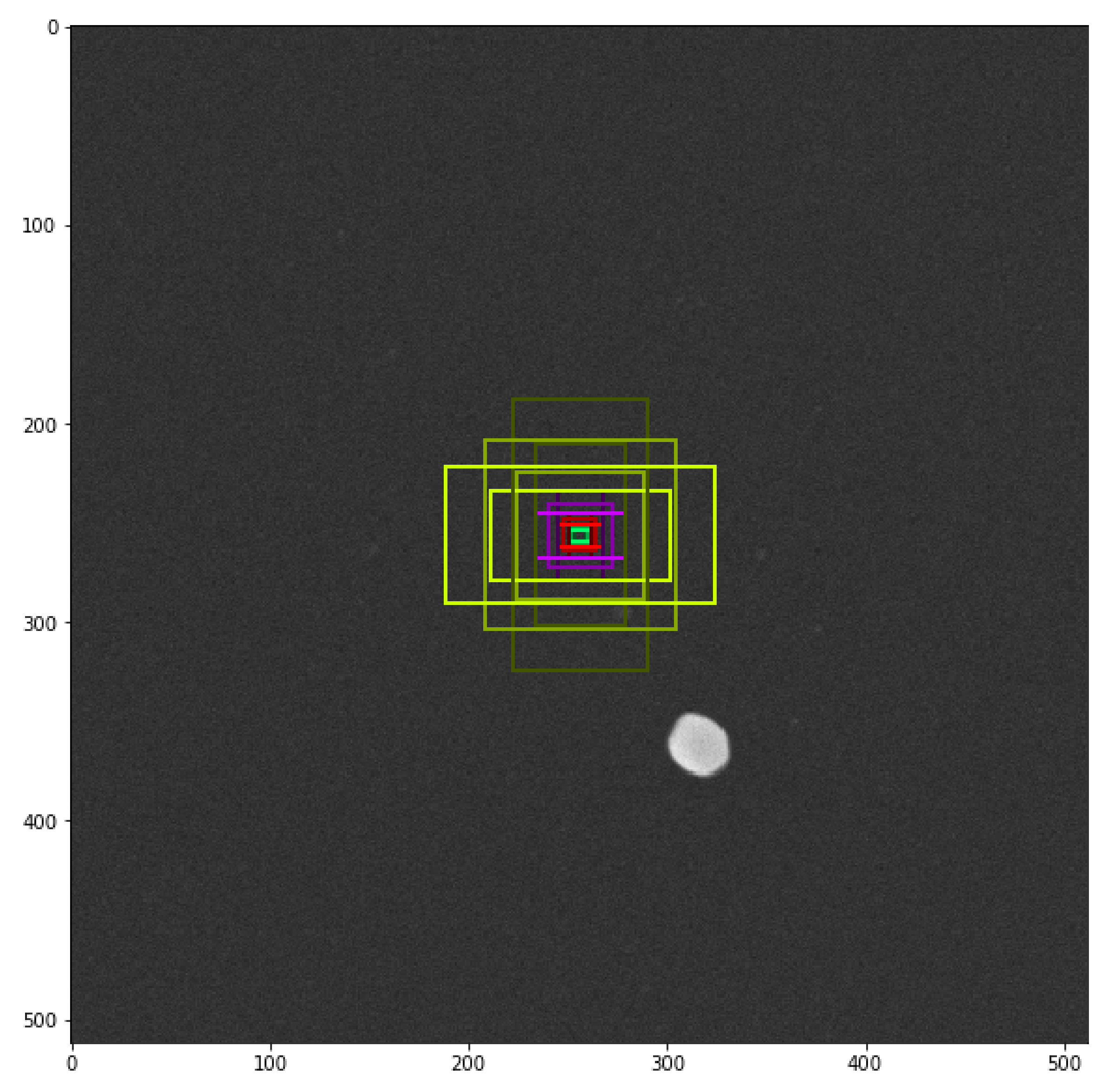
3.3. Hyper-Parameters and Network Architecture Adjustments
- The sizes of anchors were modified according to the minimum and maximum size of TiO particles: [8, 16, 32, 64, 96] (in pixels);
- The number of trained anchors was modified according to the maximum number of particle in one image: 1024;
- The stride between two consecutive anchors was set to 1 due to the agglomeration phenomenon.
3.4. Transfer Learning
3.5. Network Training
4. Results
4.1. Test Set
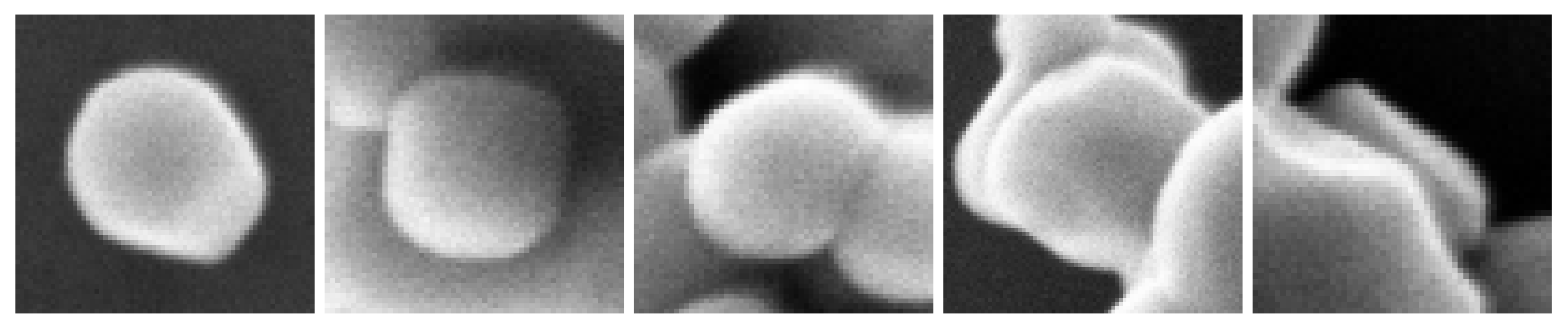
- Isolated: the particle is completely imaged and located outside of an agglomerate,
- Complete: the particle is completely imaged and located in or near an agglomerate,
- Touch complete: the particle is completely imaged but interlocked with another particle (between the complete state and the masked state),
- Masked: the particle is partly hidden by an other particle,
- Unusable: the particle is masked by an other particle with a very small visible area (less than 40 percent of its area is imaged and, therefore, does not constitute an interest for our purpose of size distribution measurement)
4.2. Result Analysis
4.2.1. Performances
- GPU NVIDIA GeForce RTX 2080 with 8 GB of memory
- CPU Intel Core i9 3.60 GHz
- RAM 32Go
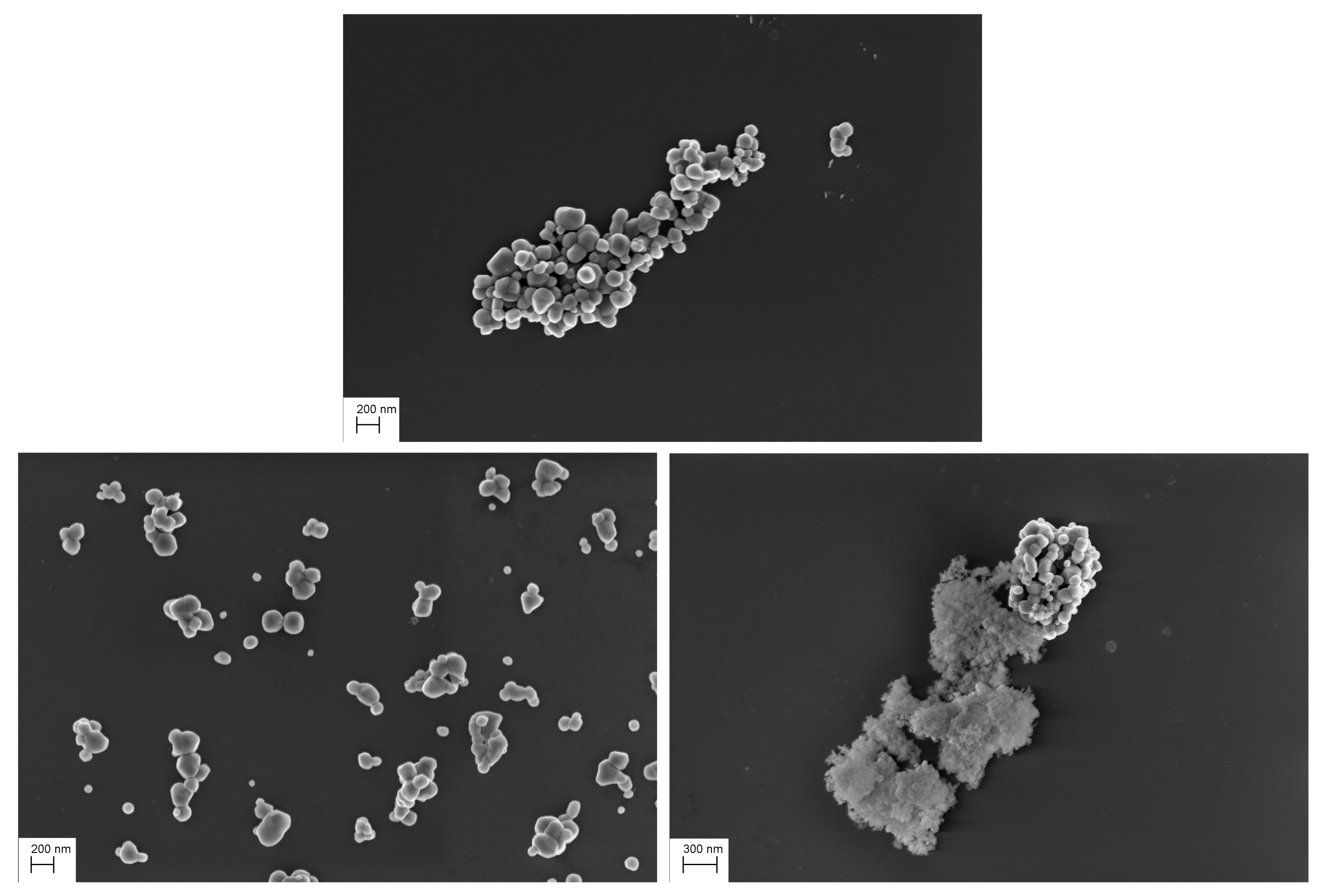
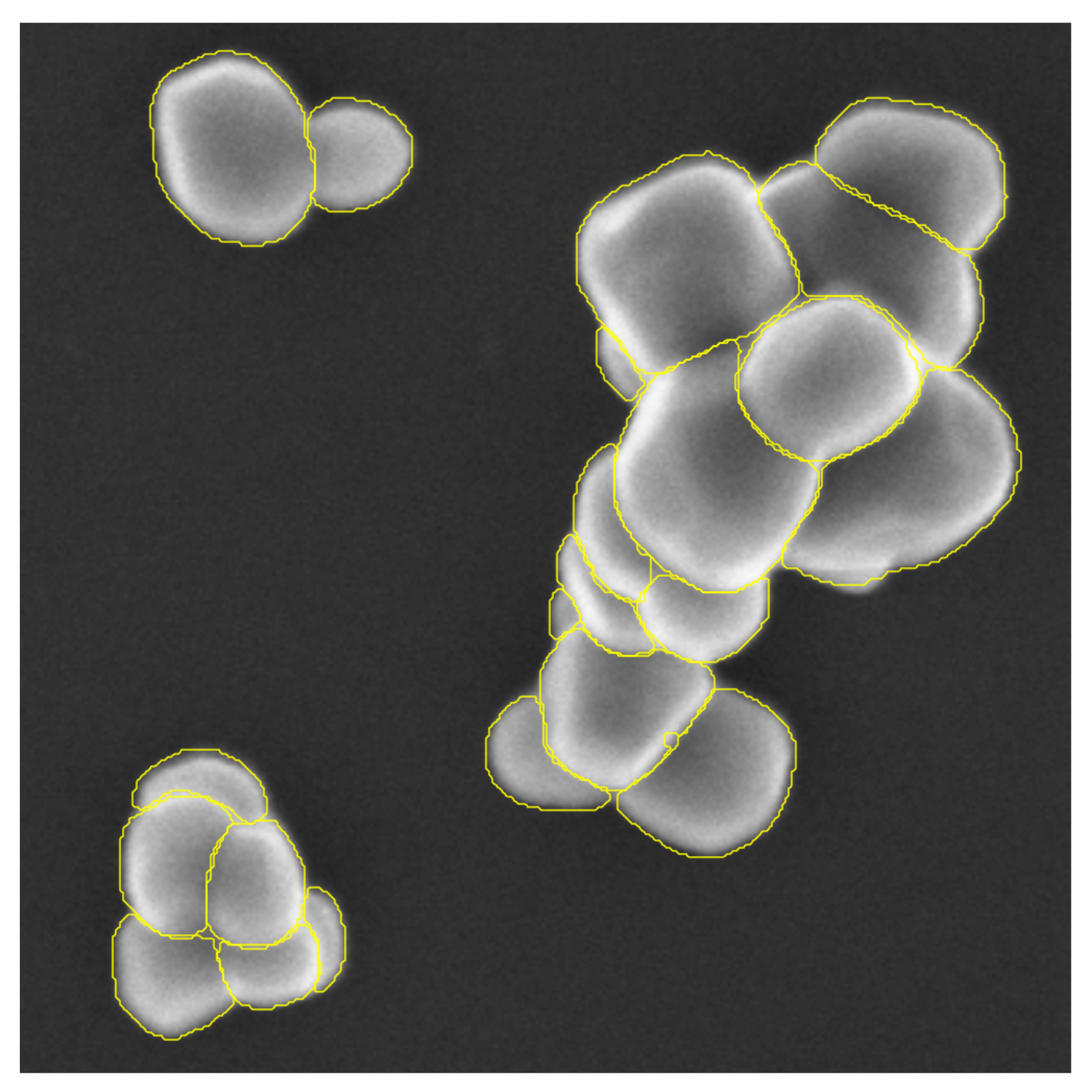
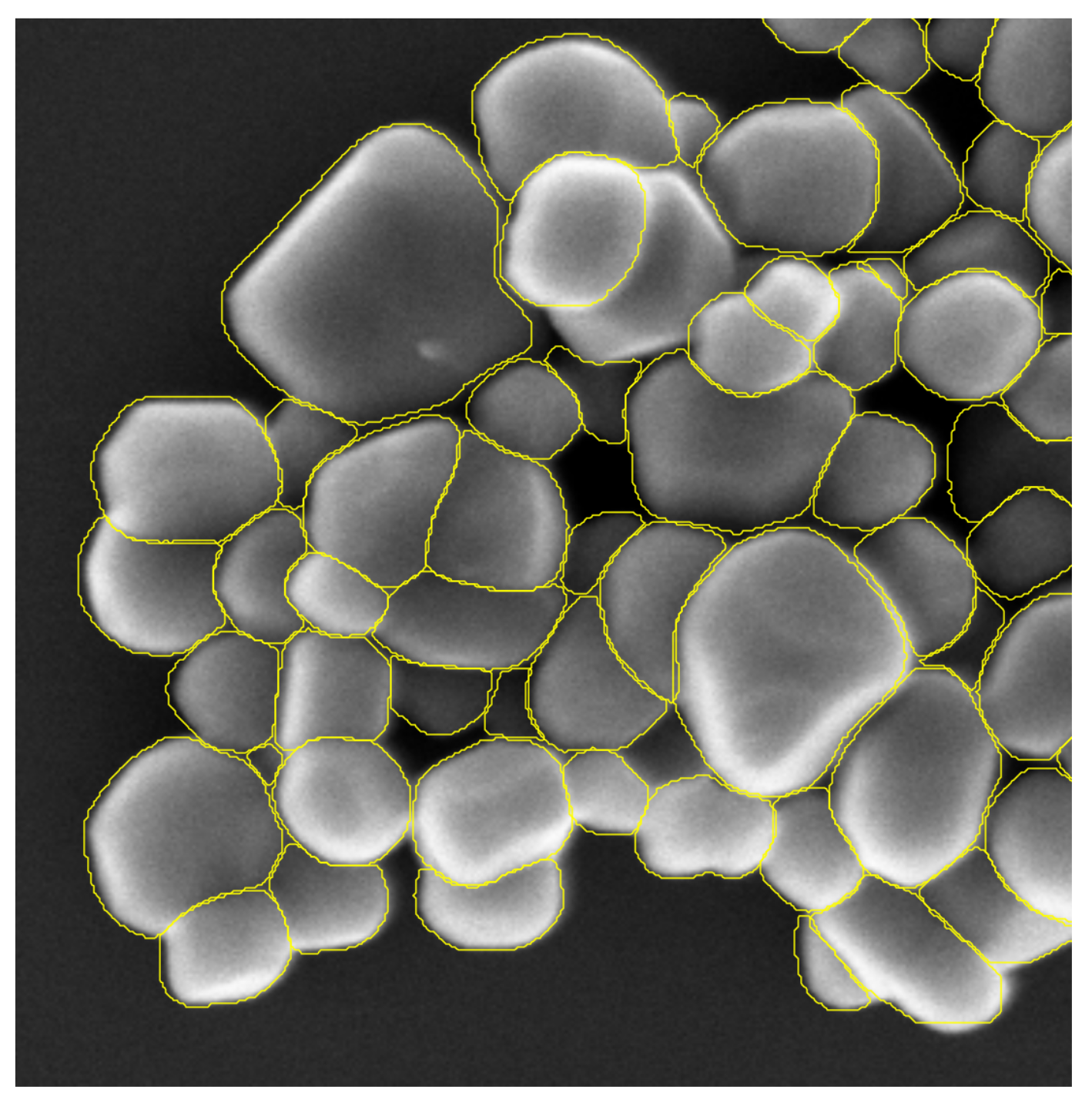
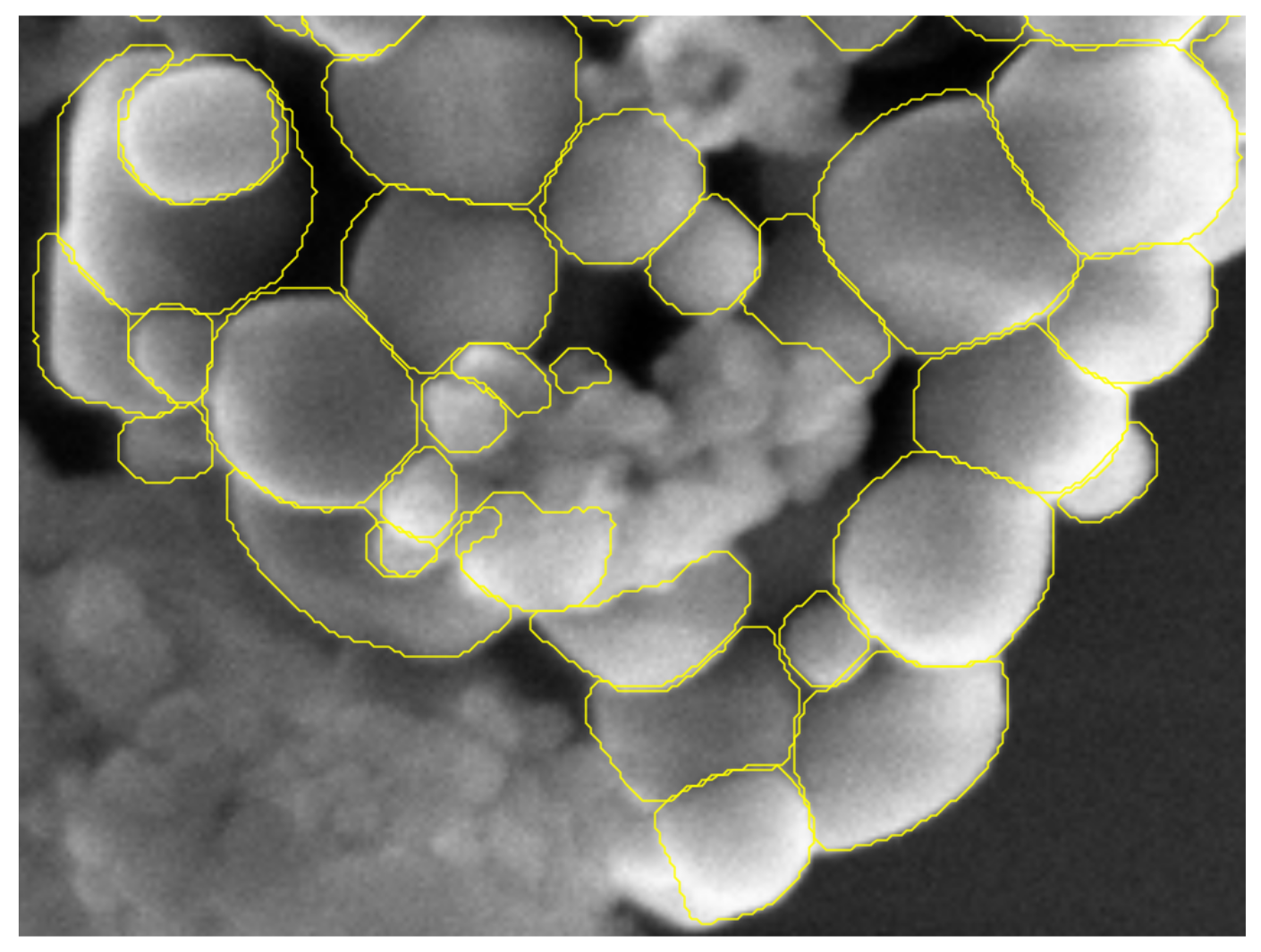
4.2.2. Visual Analysis
4.2.3. Detection Results
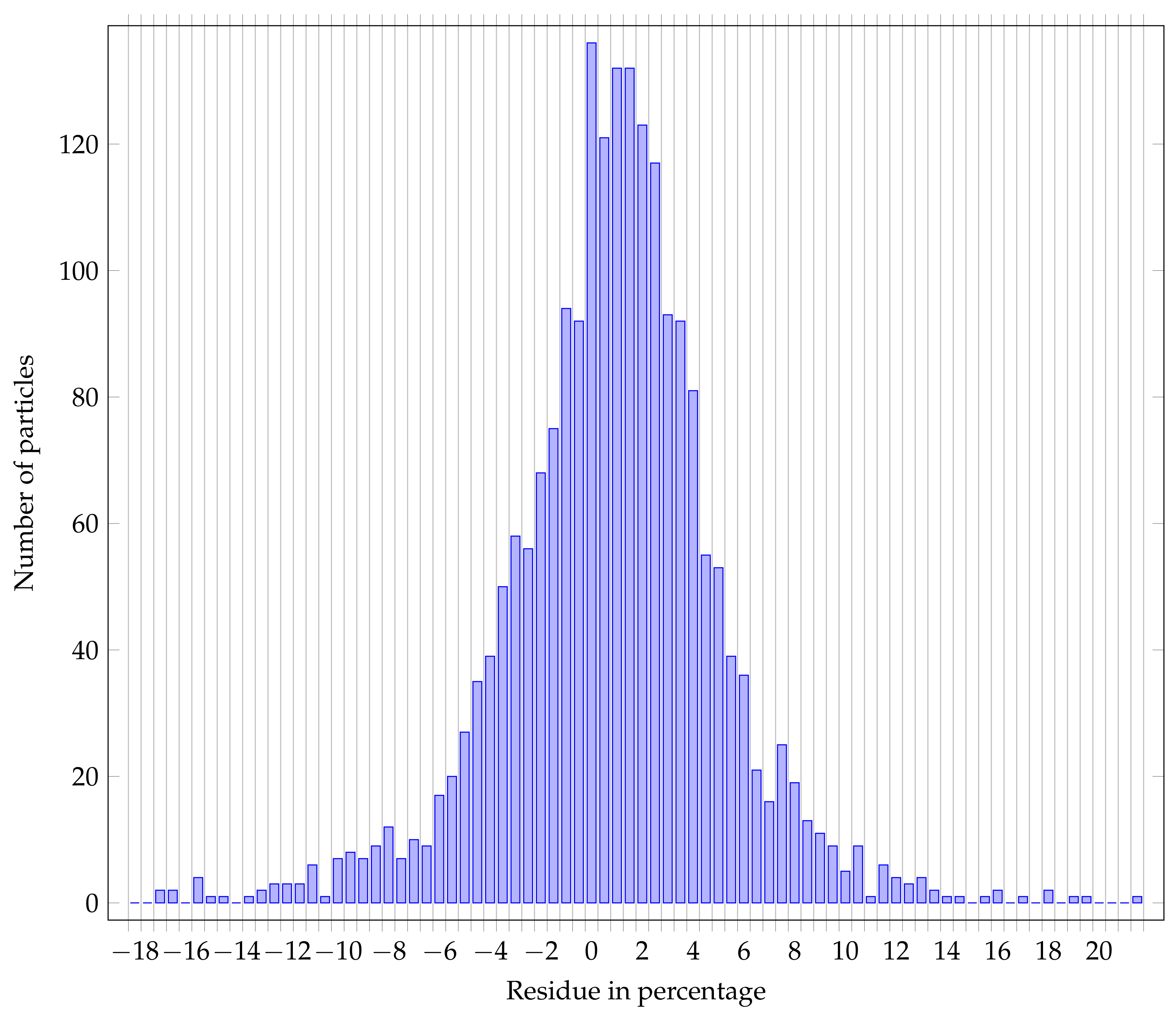
4.2.4. Segmentation Results
- Bias: the residual distributions are globally centered on 0, the bias could come from the network itself, or from a bias in the reference annotations.
- Variance: the variance of the residual distribution is higher over the area measurand than for the Feret min diameter; in fact, a small error over the radius has a squared influence over the calculated area,
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SEM | Scanning Electron Microscopy |
| R-CNN | Regional Convolutional Neural Network |
| ROI | Region of Interest |
| RPN | Regional Proposal Network |
| FPN | Feature Pyramid Network |
| TiO2 | Titanium Dioxide |
| TEM | Transmission Electron Microscope |
| STM | Scanning Tunneling Microscope |
| FCN | Fully Connected Network |
| ResNet | Residual Network |
| SGD | Stochastic Gradient Descent |
References
- Delvallée, A.; Feltin, N.; Ducourtieux, S.; Trabelsi, M.; Hochepied, J. Direct comparison of AFM and SEM measurements on the same set of nanoparticles. Meas. Sci. Technol. 2015, 26, 085601. [Google Scholar] [CrossRef]
- Coquelin, L.; Fischer, N.; Feltin, N.; Devoille, L.; Felhi, G. Towards the use of deep generative models for the characterization in size of aggregated TiO2 nanoparticles measured by Scanning Electron Microscopy (SEM). Mater. Res. Express 2019, 6, 085001. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, W.; Zhang, F.; Xiao, Z.; Wu, J.; Geng, L. Nanoparticle size measurement method based on improved watershed segmentation. In Proceedings of the 2018 International Conference on Electronics and Electrical Engineering Technology, Tianjin, China, 19–21 September 2018; pp. 232–237. [Google Scholar]
- Tek, F.B.; Dempster, A.G.; Kale, I. Blood cell segmentation using minimum area watershed and circle radon transformations. In Mathematical Morphology: 40 Years on 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 441–454. [Google Scholar]
- Baiyasi, R.; Gallagher, M.J.; McCarthy, L.A.; Searles, E.K.; Zhang, Q.; Link, S.; Landes, C.F. Quantitative Analysis of Nanorod Aggregation and Morphology from Scanning Electron Micrographs Using SEMseg. J. Phys. Chem. A 2020, 124, 5262–5270. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Asadizanjani, N.; Forte, D.; Woodard, D.L. Histogram-based Auto Segmentation: A Novel Approach to Segmenting Integrated Circuit Structures from SEM Images. arXiv 2020, arXiv:2004.13874. [Google Scholar]
- Felzenszwalb, P.F.; Huttenlocher, D.P. Efficient graph-based image segmentation. Int. J. Comput. Vis. 2004, 59, 167–181. [Google Scholar] [CrossRef]
- Nath, S.K.; Palaniappan, K.; Bunyak, F. Cell segmentation using coupled level sets and graph-vertex coloring. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Copenhagen, Denmark, 1–6 October 2006; pp. 101–108. [Google Scholar]
- Rasband, W.S. ImageJ; U. S. National Institutes of Health: Bethesda, MD, USA, 1997–2018. [Google Scholar]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Park, C.; Huang, J.Z.; Ji, J.X.; Ding, Y. Segmentation, inference and classification of partially overlapping nanoparticles. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 35. [Google Scholar] [CrossRef]
- Muneesawang, P.; Sirisathitkul, C. Size measurement of nanoparticle assembly using multilevel segmented TEM images. J. Nanomater. 2015, 2015, 790508. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Weng, Y.; Zhou, T.; Li, Y.; Qiu, X. Nas-unet: Neural architecture search for medical image segmentation. IEEE Access 2019, 7, 44247–44257. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Jing, H.; Wang, L.; Zhao, S. Ore image segmentation method using U-Net and Res_Unet convolutional networks. RSC Adv. 2020, 10, 9396–9406. [Google Scholar] [CrossRef]
- Jiao, L.; Huo, L.; Hu, C.; Tang, P. Refined UNet: UNet-Based Refinement Network for Cloud and Shadow Precise Segmentation. Remote. Sens. 2020, 12, 2001. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Zhang, R.; Cheng, C.; Zhao, X.; Li, X. Multiscale Mask R-CNN–Based Lung Tumor Detection Using PET Imaging. Mol. Imaging 2019, 18, 1536012119863531. [Google Scholar] [CrossRef]
- Johnson, J.W. Adapting mask-rcnn for automatic nucleus segmentation. arXiv 2018, arXiv:1805.00500. [Google Scholar]
- Liu, M.; Dong, J.; Dong, X.; Yu, H.; Qi, L. Segmentation of lung nodule in CT images based on mask R-CNN. In Proceedings of the 2018 9th International Conference on Awareness Science and Technology (iCAST), Fukuoka, Japan, 19–21 September 2018; pp. 1–6. [Google Scholar]
- Zhao, K.; Kang, J.; Jung, J.; Sohn, G. Building Extraction From Satellite Images Using Mask R-CNN With Building Boundary Regularization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 2018, Salt Lake City, UT, USA, 18–22 June 2018; pp. 247–251. [Google Scholar]
- Zhang, W.; Witharana, C.; Liljedahl, A.K.; Kanevskiy, M. Deep convolutional neural networks for automated characterization of arctic ice-wedge polygons in very high spatial resolution aerial imagery. Remote. Sens. 2018, 10, 1487. [Google Scholar] [CrossRef]
- Burke, C.J.; Aleo, P.D.; Chen, Y.C.; Liu, X.; Peterson, J.R.; Sembroski, G.H.; Lin, J.Y.Y. Deblending and classifying astronomical sources with Mask R-CNN deep learning. Mon. Not. R. Astron. Soc. 2019, 490, 3952–3965. [Google Scholar] [CrossRef]
- Okunev, A.G.; Mashukov, M.Y.; Nartova, A.V.; Matveev, A.V. Nanoparticle Recognition on Scanning Probe Microscopy Images Using Computer Vision and Deep Learning. Nanomaterials 2020, 10, 1285. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Shrestha, A.; Mahmood, A. Review of deep learning algorithms and architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Bengio, Y. Practical recommendations for gradient-based training of deep architectures. In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2012; pp. 437–478. [Google Scholar]
- Soydaner, D. A Comparison of Optimization Algorithms for Deep Learning. Int. J. Pattern Recognit. Artif. Intell. 2020, 34, 2052013. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 740–755. [Google Scholar]
- Krogh, A.; Hertz, J.A. A simple weight decay can improve generalization. In Advances in Neural Information Processing Systems; NeurIPS: San Diego, CA, USA, 1992; pp. 950–957. [Google Scholar]
- Sorensen, T.A. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skar. 1948, 5, 1–34. [Google Scholar]
- Everingham, M.; Van Gool, L.; Williams, C.K.; Winn, J.; Zisserman, A. The Pascal Visual Object Classes (VOC) Challenge. Int. J. Comput. Vis. 2010, 88, 303–338. [Google Scholar] [CrossRef]
- Feret, L. La Grosseur des grains des matières pulvérulent, Association Internationale Pour l’essai des Matériaux. 1931. [Google Scholar]
- Crouzier, L.; Delvallée, A.; Ducourtieux, S.; Devoille, L.; Noircler, G.; Ulysse, C.; Tache, O.; Barruet, E.; Tromas, C.; Feltin, N. Development of a new hybrid approach combining AFM and SEM for the nanoparticle dimensional metrology. Beilstein J. Nanotechnol. 2019, 10, 1523–1536. [Google Scholar] [CrossRef]
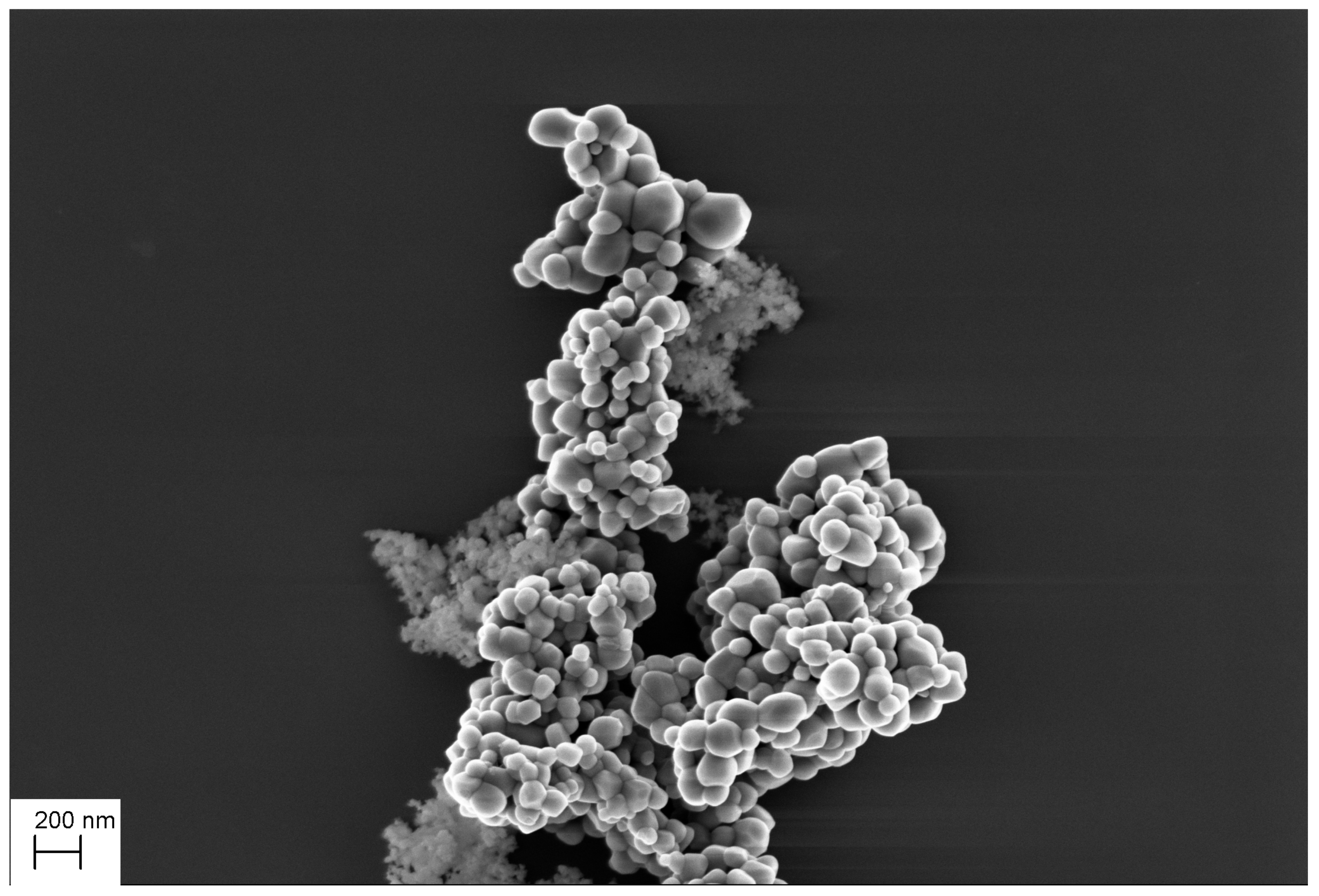
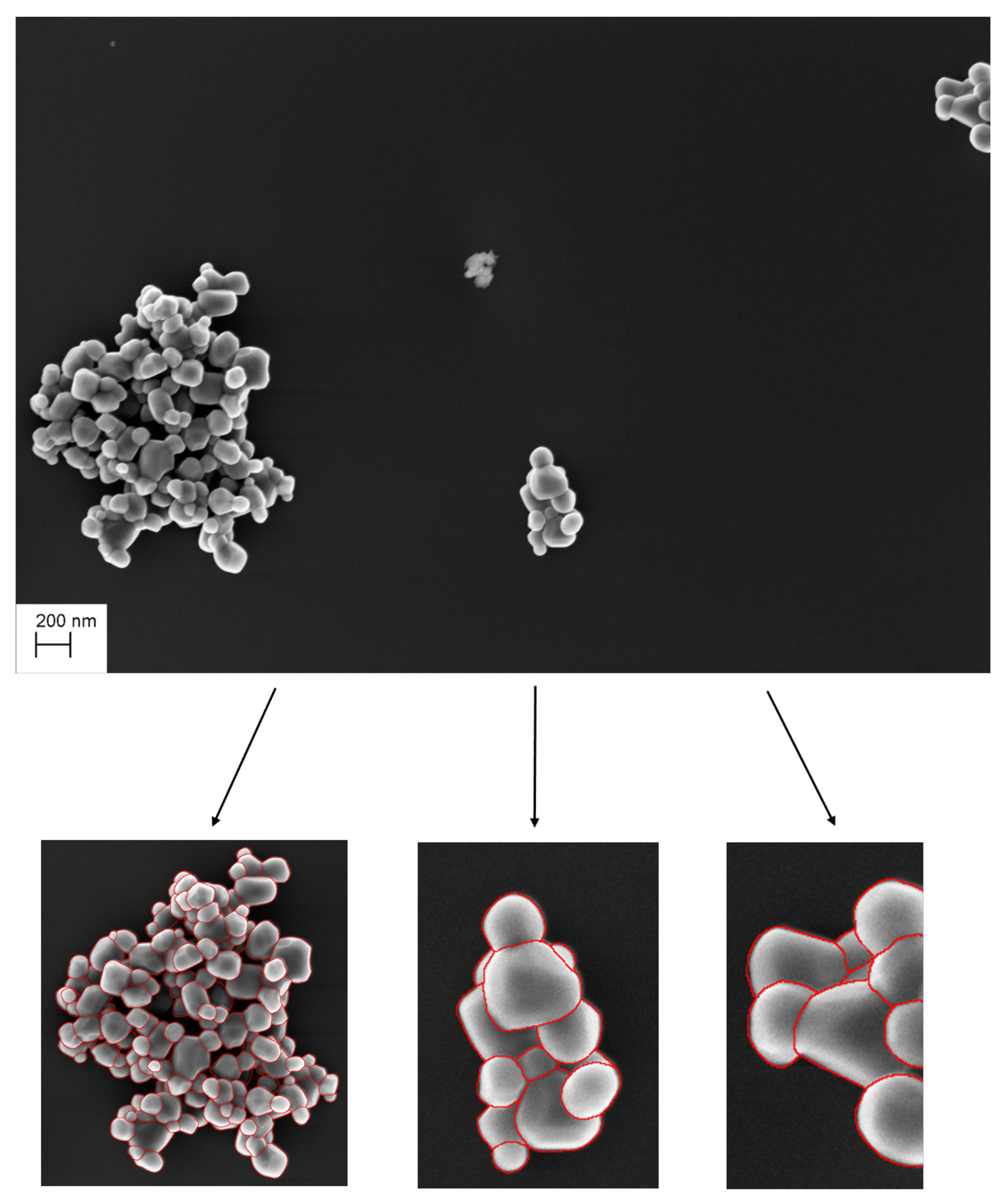

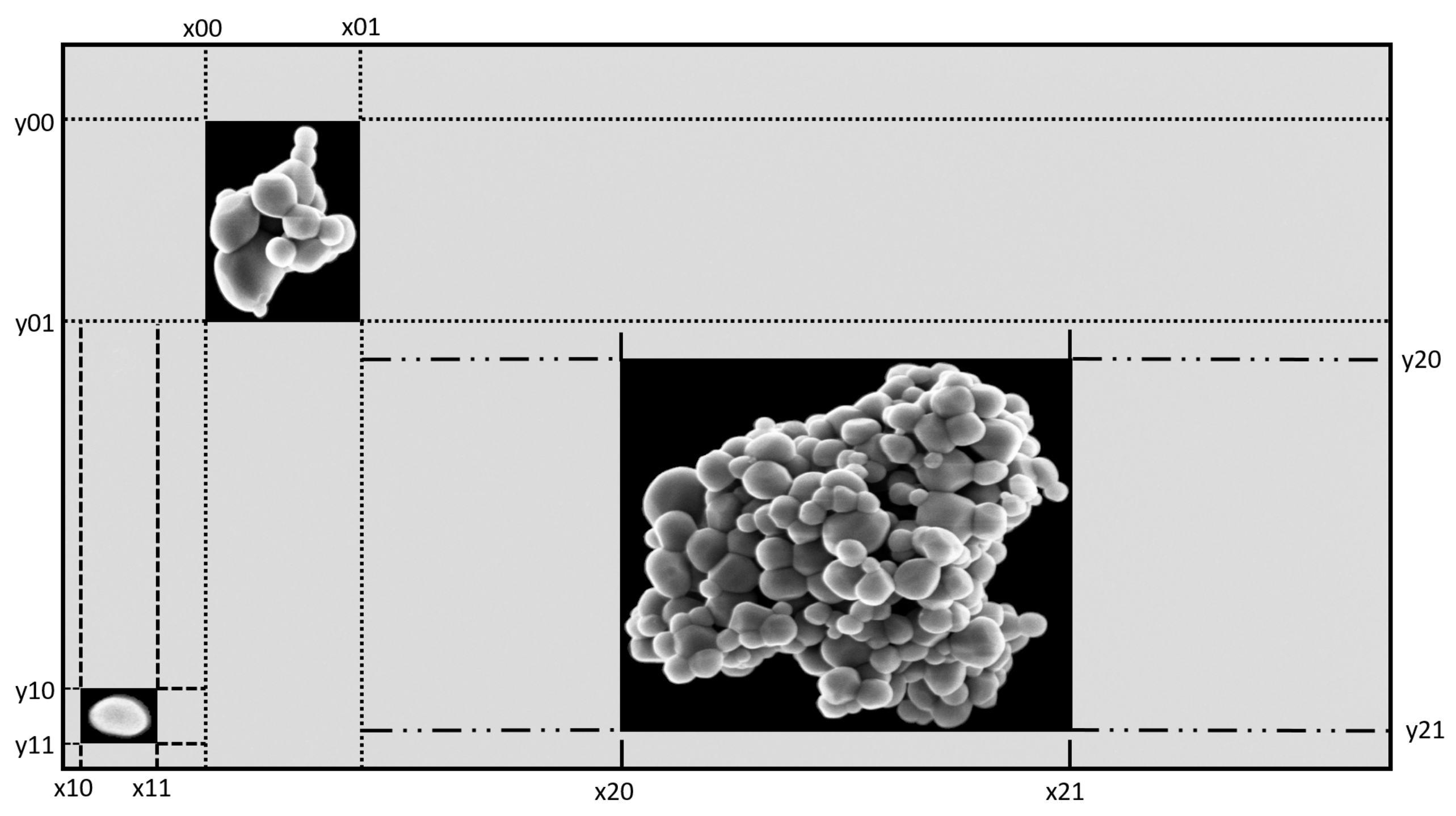

| Parameter | Value | Description |
|---|---|---|
| Resizing mode | Pad 64 | No resizing of the image at inference time |
| Minimum detection confidence | 0.5 | Threshold over class probability above which we keep a detection |
| Detection NMS Threshold | 0.3 | Threshold for deciding whether boxes overlap too much w.r.t IOU. |
| Detection post NMS ROIs | 1000 | Maximum number of ROIs to keep after NMS |
| Detection maximum instances | 512 | Maximum number of final detection |
| Number of image per GPU | 1 | The number of image your GPU memory can fit |
| Detected Particles | All | Complete | Touch Complete | Masked | Unusable |
|---|---|---|---|---|---|
| Reference | 3741 | 341 | 515 | 1302 | 1583 |
| Measured | 3135 | 339 | 495 | 1253 | 1048 |
| Percentage | 83.80 | 99.41 | 96.11 | 96.23 | 66.2 |
| mAP | AP | AP | AP | |
|---|---|---|---|---|
| Score | 60.6 | 84.6 | 71.0 | 21.5 |
| Dice Metric | Mean | Median | Std | Min | Max |
|---|---|---|---|---|---|
| All | 0.936 | 0.948 | 0.040 | 0.750 | 0.989 |
| Complete | 0.953 | 0.959 | 0.024 | 0.819 | 0.987 |
| Touch Complete | 0.953 | 0.958 | 0.022 | 0.858 | 0.986 |
| Masked | 0.949 | 0.957 | 0.026 | 0.765 | 0.989 |
| Unusable | 0.903 | 0.913 | 0.046 | 0.756 | 0.975 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monchot, P.; Coquelin, L.; Guerroudj, K.; Feltin, N.; Delvallée, A.; Crouzier, L.; Fischer, N. Deep Learning Based Instance Segmentation of Titanium Dioxide Particles in the Form of Agglomerates in Scanning Electron Microscopy. Nanomaterials 2021, 11, 968. https://doi.org/10.3390/nano11040968
Monchot P, Coquelin L, Guerroudj K, Feltin N, Delvallée A, Crouzier L, Fischer N. Deep Learning Based Instance Segmentation of Titanium Dioxide Particles in the Form of Agglomerates in Scanning Electron Microscopy. Nanomaterials. 2021; 11(4):968. https://doi.org/10.3390/nano11040968
Chicago/Turabian StyleMonchot, Paul, Loïc Coquelin, Khaled Guerroudj, Nicolas Feltin, Alexandra Delvallée, Loïc Crouzier, and Nicolas Fischer. 2021. "Deep Learning Based Instance Segmentation of Titanium Dioxide Particles in the Form of Agglomerates in Scanning Electron Microscopy" Nanomaterials 11, no. 4: 968. https://doi.org/10.3390/nano11040968
APA StyleMonchot, P., Coquelin, L., Guerroudj, K., Feltin, N., Delvallée, A., Crouzier, L., & Fischer, N. (2021). Deep Learning Based Instance Segmentation of Titanium Dioxide Particles in the Form of Agglomerates in Scanning Electron Microscopy. Nanomaterials, 11(4), 968. https://doi.org/10.3390/nano11040968







