Prenatal Exposure to Heavy Metals Affects Gestational Age by Altering DNA Methylation Patterns
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. DNA Methylation Assay
2.3. Data Quality Control
2.4. Data Adjustment
2.5. Bioinformatics and Statistical Analyses
3. Results
3.1. Sample and Data Preparation
3.2. Assessment of Exposure
3.3. EWAS on Heavy Metal Exposures and Birth Outcomes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wild, C.P. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardenas, A.; Rifas-Shiman, S.L.; Agha, G.; Hivert, M.-F.; Litonjua, A.A.; DeMeo, D.L.; Lin, X.; Amarasiriwardena, C.J.; Oken, E.; Gillman, M.W. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Visser, G.H. Developmental Origins of Health and Disease (DOHaD). Early Human Dev. 2006, 82, 1–7. [Google Scholar]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Mol. Clin. Environ. Toxicol. 2012, 101, 133–164. [Google Scholar]
- Risk Assessment of Lead. Available online: http://www.nifds.go.kr/cont/down.do?gubn=en&fileNm=17%20Risk%20Assessment%20of%20Lead(web).pdf (accessed on 12 October 2021).
- Risk Assessment of Mercury and Methylmercury. Available online: http://www.nifds.go.kr/cont/down.do?gubn=en&fileNm=18%20Risk%20Assessment%20of%20Mercury%20and%20Methylmercury(web).pdf (accessed on 12 October 2021).
- Risk Assessment of Cadmium. Available online: http://www.nifds.go.kr/cont/down.do?gubn=en&fileNm=16%20Risk%20Assessment%20of%20Cadmium(web).pdf (accessed on 12 October 2021).
- Kim, M.J.; Kim, C.-H.; An, M.-J.; Shin, G.-S.; Lee, H.-M.; Kim, J.-Y.; Hwang, J.Y.; Lee, J.-H.; Kim, J.-W. Exposure to mercury induced early apoptotic signals in human placental BeWo cells through alteration of cell cycle regulation. Mol. Cell Toxicol. 2020, 16, 419–429. [Google Scholar] [CrossRef]
- Espart, A.; Artime, S.; Tort-Nasarre, G.; Yara-Varón, E. Cadmium exposure during pregnancy and lactation: Materno-fetal and newborn repercussions of Cd (II), and Cd–metallothionein complexes. Metallomics 2018, 10, 1359–1367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, J.; Kim, E.; Kim, W.J.; Won, S. Prenatal lead exposure and cord blood DNA methylation in the Korean Exposome Study. Environ. Res. 2021, 195, 110767. [Google Scholar] [CrossRef]
- WHO Preterm Birth Report. Available online: https://www.who.int/news-room/fact-sheets/detail/preterm-birth (accessed on 12 October 2021).
- KOSTAT Birth Statistics 2020. Available online: https://kostat.go.kr/assist/synap/preview/skin/miri.html?fn=c1d13184443522230214713&rs=/assist/synap/preview (accessed on 12 October 2021).
- Parets, S.E.; Bedient, C.E.; Menon, R.; Smith, A.K. Preterm birth and its long-term effects: Methylation to mechanisms. Biology 2014, 3, 498–513. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.; Faes, C.; Debevec, T.; Rytz, C.; Millet, G.; Pialoux, V. Preterm birth and oxidative stress: Effects of acute physical exercise and hypoxia physiological responses. Redox Biol. 2018, 17, 315–322. [Google Scholar] [CrossRef]
- Ahmed, S.; Khoda, S.M.-e.; Rekha, R.S.; Gardner, R.M.; Ameer, S.S.; Moore, S.; Ekström, E.-C.; Vahter, M.; Raqib, R. Arsenic-associated oxidative stress, inflammation, and immune disruption in human placenta and cord blood. Environ. Health Perspect 2011, 119, 258–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arpino, C.; Compagnone, E.; Montanaro, M.L.; Cacciatore, D.; De Luca, A.; Cerulli, A.; Di Girolamo, S.; Curatolo, P. Preterm birth and neurodevelopmental outcome: A review. Childs Nerv. Syst. 2010, 26, 1139–1149. [Google Scholar] [CrossRef]
- Jeong, K.S.; Ha, E.; Shin, J.Y.; Park, H.; Hong, Y.-C.; Ha, M.; Kim, S.; Lee, S.-J.; Lee, K.Y.; Kim, J.H. Blood heavy metal concentrations in pregnant Korean women and their children up to age 5 years: Mothers’ and Children’s Environmental Health (MOCEH) birth cohort study. Sci. Total Environ. 2017, 605, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Spindola, L.M.; Santoro, M.L.; Pan, P.M.; Ota, V.K.; Xavier, G.; Carvalho, C.M.; Talarico, F.; Sleiman, P.; March, M.; Pellegrino, R. Detecting multiple differentially methylated CpG sites and regions related to dimensional psychopathology in youths. Clin. Epigenet. 2019, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.E.; Irizarry, R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014, 15, 1–9. [Google Scholar] [CrossRef]
- Houseman, E.A.; Kile, M.L.; Christiani, D.C.; Ince, T.A.; Kelsey, K.T.; Marsit, C.J. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinf. 2016, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]
- FlowSorted.CordBloodCombined.450k. Available online: https://github.com/immunomethylomics/FlowSorted.CordBloodCombined.450k (accessed on 12 October 2021).
- Stekhoven, D.J. Using the missForest Package. 2011. Available online: https://stat.ethz.ch/education/semesters/ss2013/ams/paper/missForest_1.2.pdf (accessed on 13 May 2021).
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Pidsley, R.; Wong, C.C.; Volta, M.; Lunnon, K.; Mill, J.; Schalkwyk, L.C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genom. 2013, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- maxprobe. Available online: https://github.com/markgene/maxprobes (accessed on 12 October 2021).
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Peters, T.J.; Buckley, M.J.; Statham, A.L.; Pidsley, R.; Samaras, K.; Lord, R.V.; Clark, S.J.; Molloy, P.L. De novo identification of differentially methylated regions in the human genome. Epigenet. Chromatin 2015, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Tingley, D.; Yamamoto, T.; Hirose, K.; Keele, L.; Imai, K. Mediation: R package for causal mediation analysis. J. Stat. Softw. 2014, 59, 1–38. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.-J.; Lee, S.Y.; Park, D.; Ryu, S.-H.; Yoon, J.; Jung, S.; Lee, E.; Yang, S.-I.; Hong, S.-J. Early-life exposure to humidifier disinfectant determines the prognosis of lung function in children. BMC Pulm Med. 2019, 19, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; He, Y.; Lin, Q.; Huang, L.; Zhang, Q.; Xu, Y. Adverse effects of subchronic exposure to cooking oil fumes on the gonads and the GPR30-mediated signaling pathway in female rats. Mol. Cell. Toxicol. 2020, 16, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Sabra, S.; Malmqvist, E.; Saborit, A.; Gratacós, E.; Gomez Roig, M.D. Heavy metals exposure levels and their correlation with different clinical forms of fetal growth restriction. PLoS ONE 2017, 12, e0185645. [Google Scholar] [CrossRef] [PubMed]
- Landeira, D.; Bagci, H.; Malinowski, A.R.; Brown, K.E.; Soza-Ried, J.; Feytout, A.; Webster, Z.; Ndjetehe, E.; Cantone, I.; Asenjo, H.G. Jarid2 coordinates nanog expression and PCP/Wnt signaling required for efficient ESC differentiation and early embryo development. Cell Rep. 2015, 12, 573–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hambiliki, F.; Ström, S.; Zhang, P.; Stavreus-Evers, A. Co-localization of NANOG and OCT4 in human pre-implantation embryos and in human embryonic stem cells. J. Assist. Reprod. Genet. 2012, 29, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhang, L.; Zhang, L.; Jia, L.; Wang, P.; Gao, Y. Association of Wnt2 and sFRP4 expression in the third trimester placenta in women with severe preeclampsia. Reprod. Sci. 2013, 20, 981–989. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, H.; Zhang, L.; Jia, L.; Wang, P.; Endocrinology. Differential expression of beta-catenin and dickkopf-1 in the third trimester placentas from normal and preeclamptic pregnancies: A comparative study. Reprod. Biol. Endocrinol. 2013, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Davies, E.L.; Bell, J.S.; Bhattacharya, S. Preeclampsia and preterm delivery: A population-based case–control study. Hypertens. Pregnancy 2016, 35, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Hannon, E.; Schendel, D.; Ladd-Acosta, C.; Grove, J.; Hansen, C.S.; Hougaard, D.M.; Bresnahan, M.; Mors, O.; Hollegaard, M.V.; Bækvad-Hansen, M. Variable DNA methylation in neonates mediates the association between prenatal smoking and birth weight. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180120. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.W.; Ahn, K.; Yang, H.J.; Lee, S.; Park, J.D.; Kim, W.K.; Kim, J.-T.; Kim, H.H.; Rha, Y.H.; Park, Y.M. Humidifier disinfectant–associated children’s interstitial lung disease. Am. J. Respir. Crit. Care Med. 2014, 189, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Jenkins, T.G.; Jung, A.M.; Jeong, K.S.; Zhai, J.; Jacobs, E.T.; Griffin, S.C.; Dearmon-Moore, D.; Littau, S.R.; Peate, W.F. DNA methylation among firefighters. PLoS ONE 2019, 14, e0214282. [Google Scholar]
- Yu, S.Y.; Koh, E.J.; Kim, S.H.; Lee, S.Y.; Lee, J.S.; Son, S.W.; Hwang, S.Y. Integrated analysis of multi-omics data on epigenetic changes caused by combined exposure to environmental hazards. Environ. Toxicol. 2021, 36, 1001–1010. [Google Scholar] [CrossRef]
- Hong, J.Y.; Yu, S.Y.; Kim, S.Y.; Ahn, J.J.; Kim, Y.; Kim, G.W.; Son, S.W.; Park, J.-T.; Hwang, S.Y. Association analysis of toluene exposure time with high-throughput mRNA expressions and methylation patterns using in vivo samples. Environ. Res. 2016, 146, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Hebels, D.G.; Georgiadis, P.; Keun, H.C.; Athersuch, T.J.; Vineis, P.; Vermeulen, R.; Portengen, L.; Bergdahl, I.A.; Hallmans, G.; Palli, D. Performance in omics analyses of blood samples in long-term storage: Opportunities for the exploitation of existing biobanks in environmental health research. Environ. Health Perspect. 2013, 121, 480–487. [Google Scholar] [CrossRef] [Green Version]
- Koh, E.J.; Yu, S.Y.; Kim, S.H.; Kim, S.J.; Lee, E.-I.; Hwang, S.Y. Understanding confounding effects of blood handling strategies on RNA quality and transcriptomic alteration using RNA sequencing. BioChip J. 2021, 15, 1–8. [Google Scholar] [CrossRef]
- Koh, E.J.; Hwang, S.Y. Multi-omics approaches for understanding environmental exposure and human health. Mol. Cell. Toxicol. 2019, 15, 1–7. [Google Scholar] [CrossRef]
- Park, J.; Kwon, S.O.; Kim, S.-H.; Kim, S.J.; Koh, E.J.; Won, S.; Kim, W.J.; Hwang, S.Y. Methylation quantitative trait loci analysis in Korean exposome study. Mol. Cell. Toxicol. 2020, 16, 175–183. [Google Scholar] [CrossRef]

| Newborns (n = 367) | Mothers (of 367 Newborns) | ||
|---|---|---|---|
| Boys | 186 | Age (year) | 30.4 ± 3.6 |
| Girls | 181 | BMI (kg/m2) | 22.9 ± 3.1 |
| Gestational age (day) | 275.4 ± 8.2 | Smoker (yes/no) | 38/329 |
| Preterm birth (case/control) | 5/362 | Parity (0/>0) | 209/158 |
| Hg (μg/L) | 5.9 ± 2.8 | Hg (μg/L) | 3.6 ± 1.9 |
| Pb (μg/L) | 10 ± 4.2 | Pb (μg/L) | 13.7 ± 5.7 |
| Cd (μg/L) | 0.7 ± 0.2 | Cd (μg/L) | 1.6 ± 0.4 |
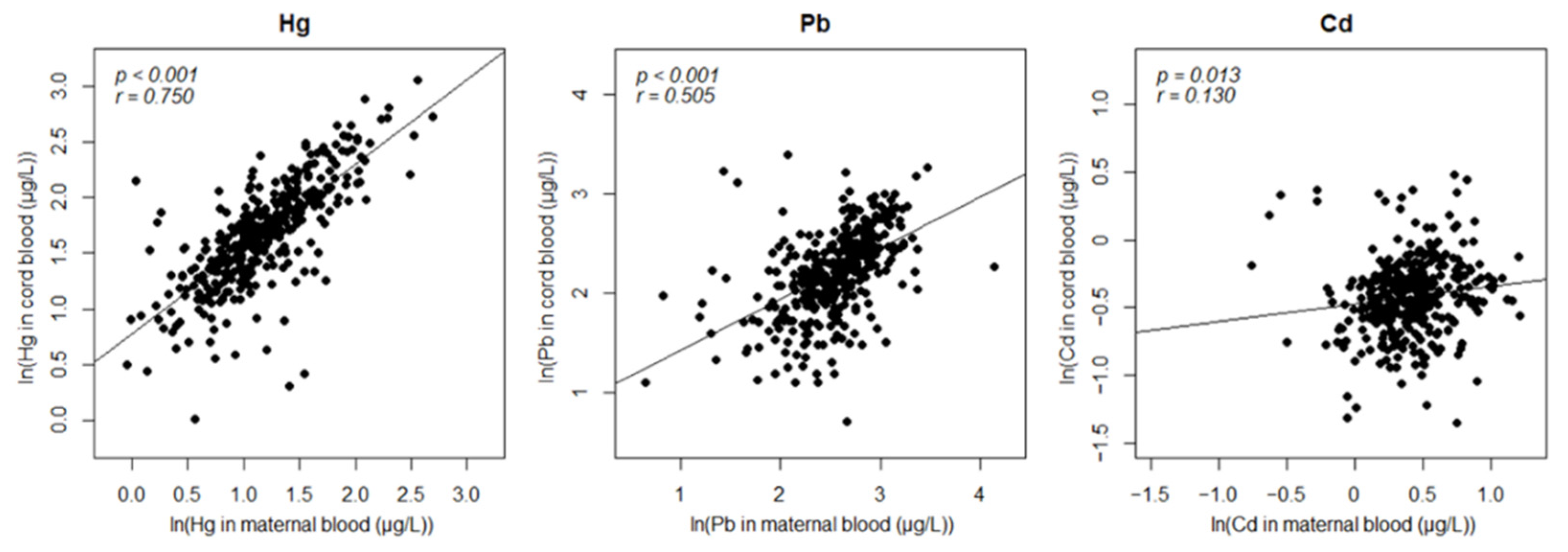
| F-Statistics (p) | Pearson’s Coefficients | ||
|---|---|---|---|
| Maternal–Prenatal exposure | |||
| Hg | 2.277 × 10−67 *** | 0.750 | |
| Pb | 3.773 × 10−25 *** | 0.505 | |
| Cd | 0.013 * | 0.130 | |
| Exposure–Gestational age | |||
| Maternal | Hg | 0.924 | 0.005 |
| Pb | 0.199 | −0.067 | |
| Cd | 0.303 | −0.054 | |
| Prenatal | Hg | 0.542 | 0.032 |
| Pb | 0.170 | −0.072 | |
| Cd | 0.983 | −0.001 | |
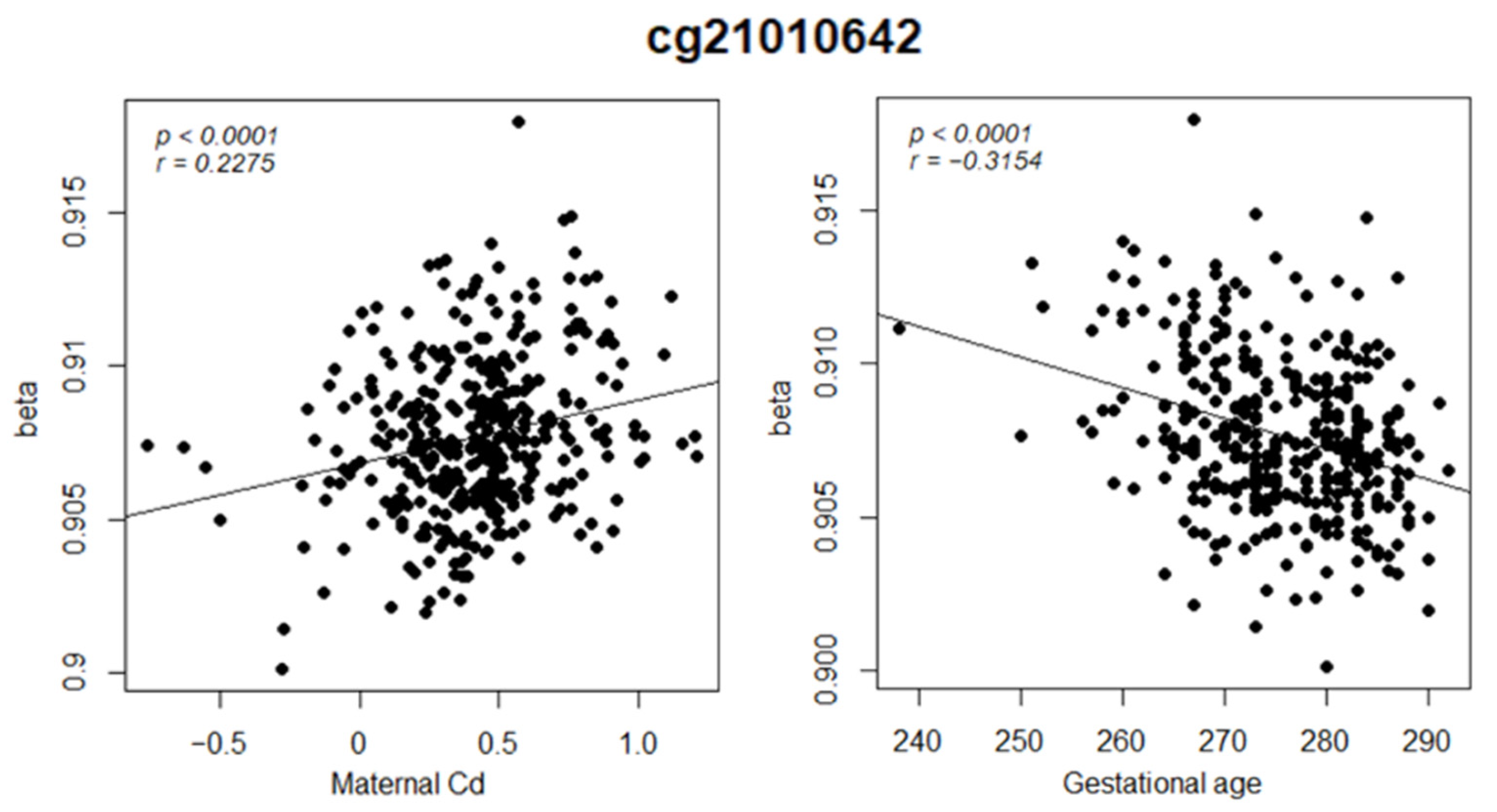
| Differential Methylation (β) | Linear Regression | |||
|---|---|---|---|---|
| p-Value | q-Value | p-Value | Correlation | |
| Maternal Cd | 8.6 × 10−6 | 0.14 | 1.07 × 10−5 | 0.2275 |
| Gestational age | 5.4 × 10−10 | 2.7 × 10−7 | 6.4 × 10−10 | −0.3154 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koh, E.J.; Yu, S.Y.; Kim, S.H.; Lee, J.S.; Hwang, S.Y. Prenatal Exposure to Heavy Metals Affects Gestational Age by Altering DNA Methylation Patterns. Nanomaterials 2021, 11, 2871. https://doi.org/10.3390/nano11112871
Koh EJ, Yu SY, Kim SH, Lee JS, Hwang SY. Prenatal Exposure to Heavy Metals Affects Gestational Age by Altering DNA Methylation Patterns. Nanomaterials. 2021; 11(11):2871. https://doi.org/10.3390/nano11112871
Chicago/Turabian StyleKoh, Eun Jung, So Yeon Yu, Seung Hwan Kim, Ji Su Lee, and Seung Yong Hwang. 2021. "Prenatal Exposure to Heavy Metals Affects Gestational Age by Altering DNA Methylation Patterns" Nanomaterials 11, no. 11: 2871. https://doi.org/10.3390/nano11112871
APA StyleKoh, E. J., Yu, S. Y., Kim, S. H., Lee, J. S., & Hwang, S. Y. (2021). Prenatal Exposure to Heavy Metals Affects Gestational Age by Altering DNA Methylation Patterns. Nanomaterials, 11(11), 2871. https://doi.org/10.3390/nano11112871






