Data-Driven Modeling of the Cellular Pharmacokinetics of Degradable Chitosan-Based Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Nanoparticle Preparation, and Image Acquisition
2.2. Nanoparticle Characterization
2.3. Nanoparticle Supply Kinetics
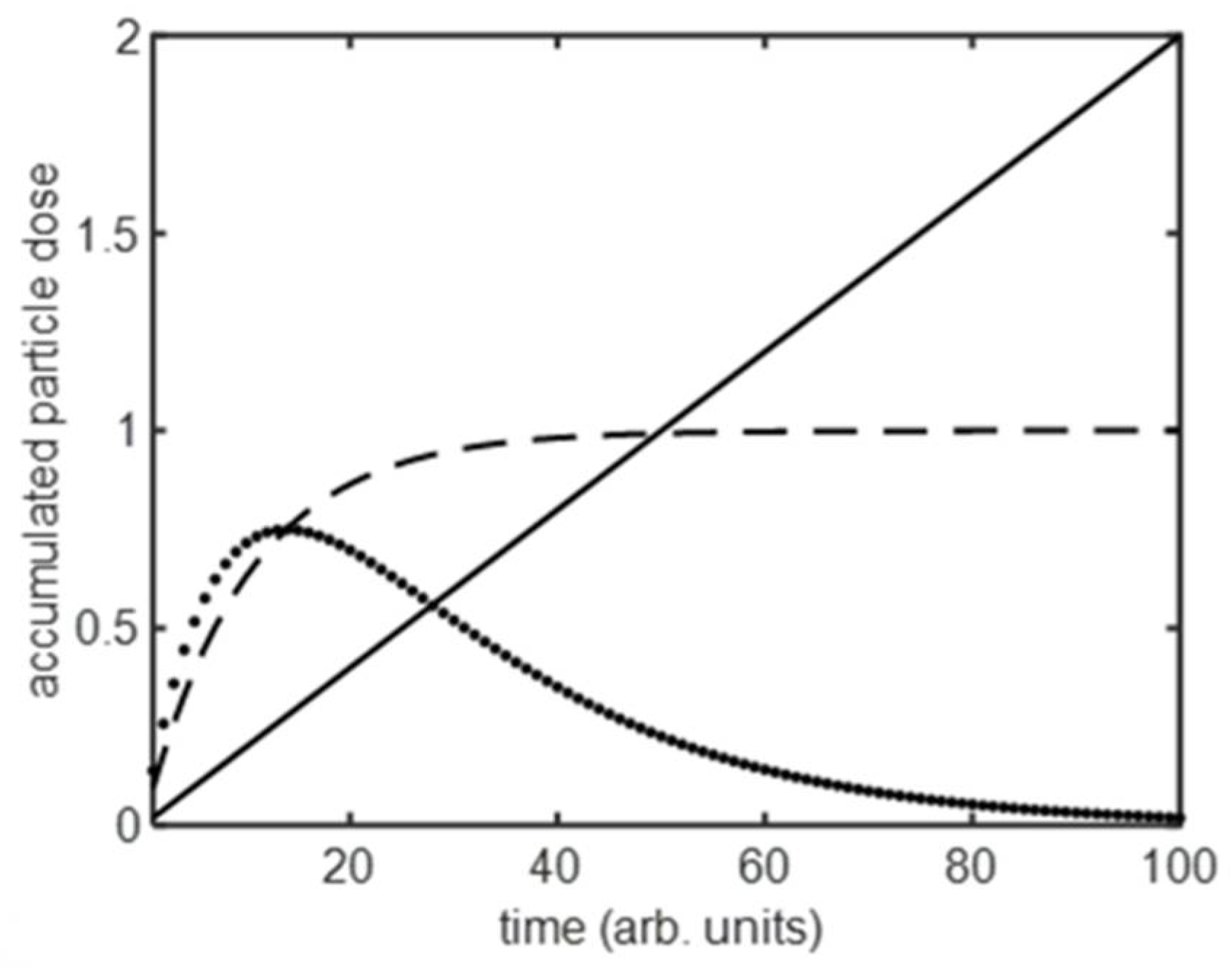
2.4. Analytical Models
3. Results
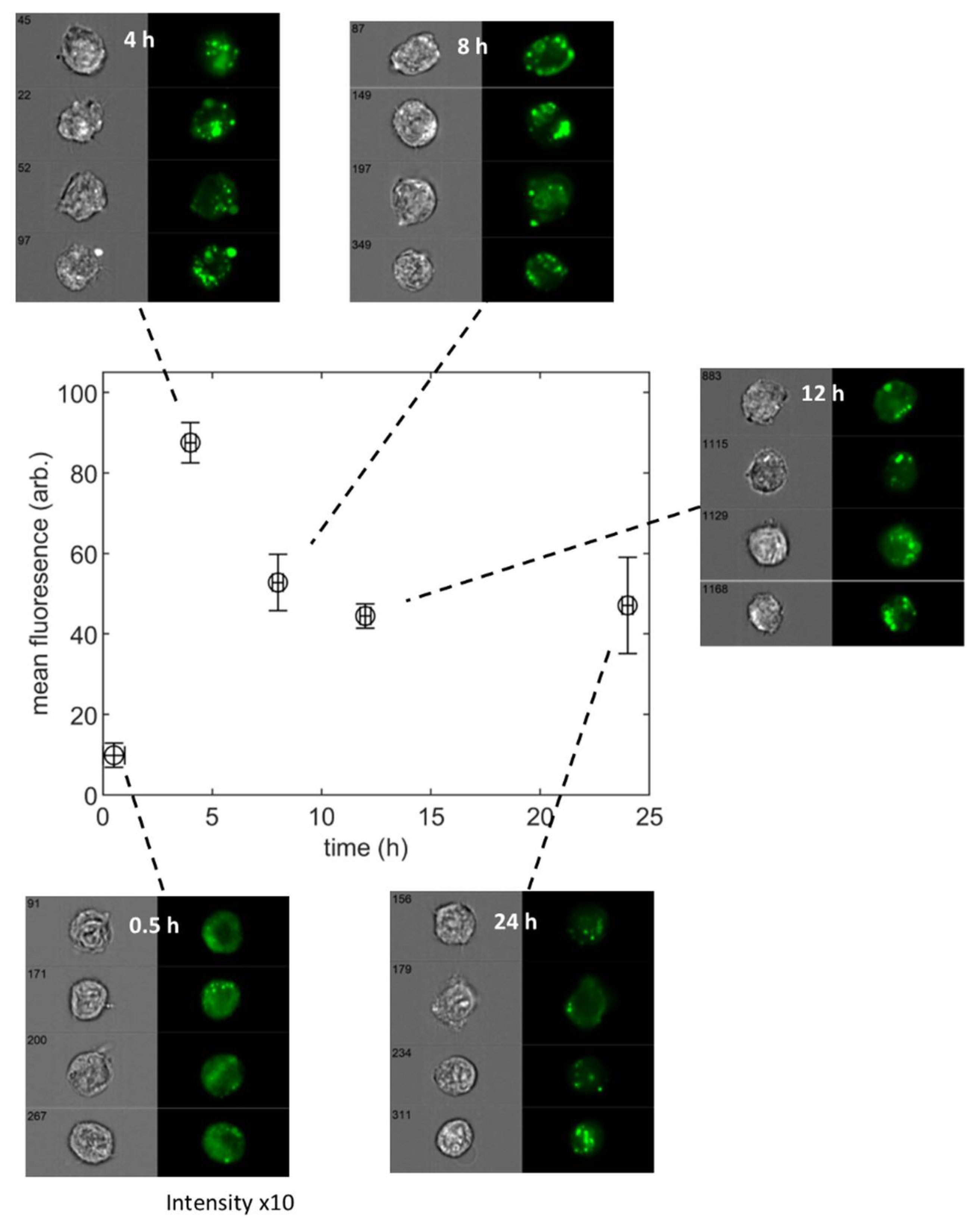
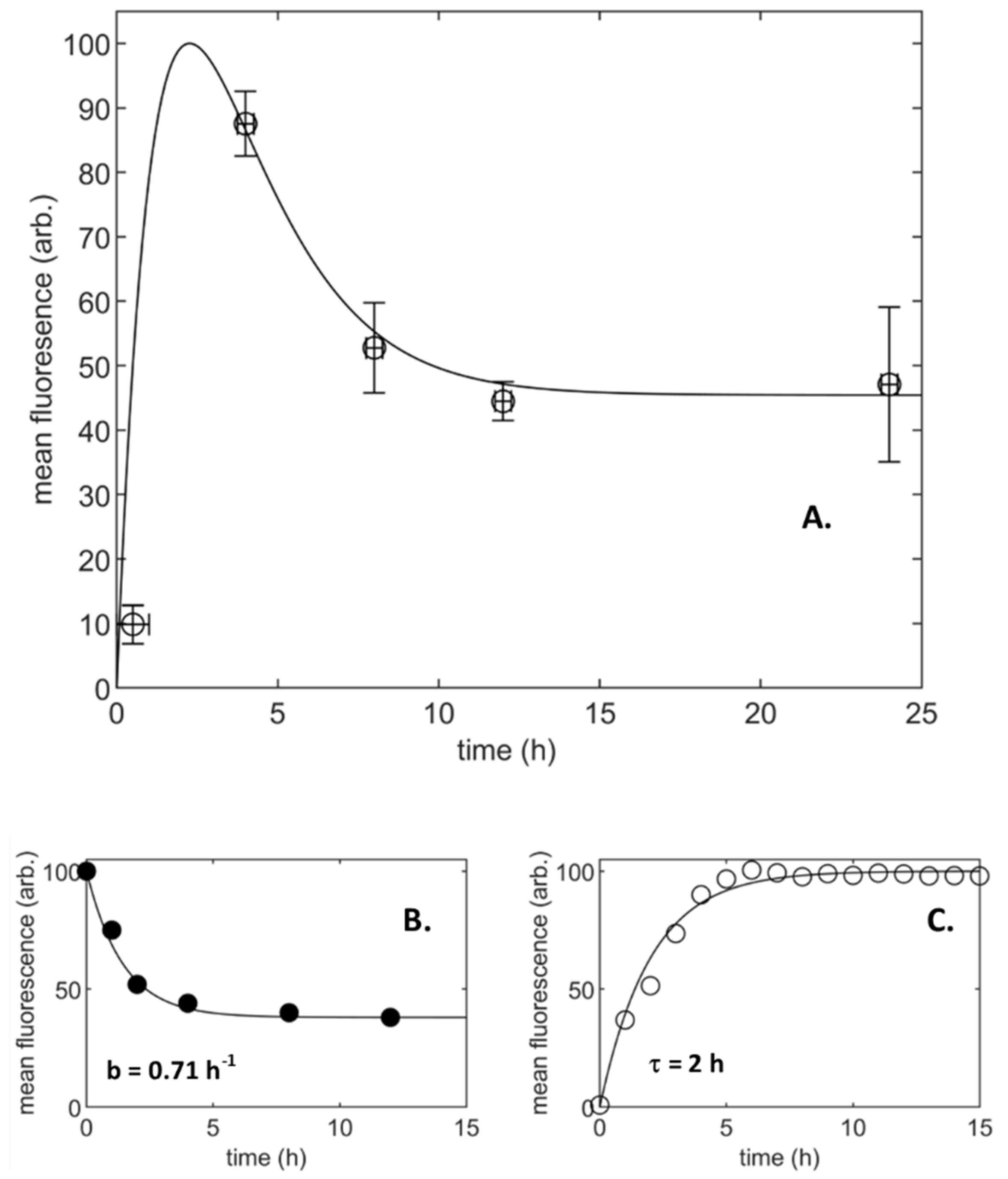
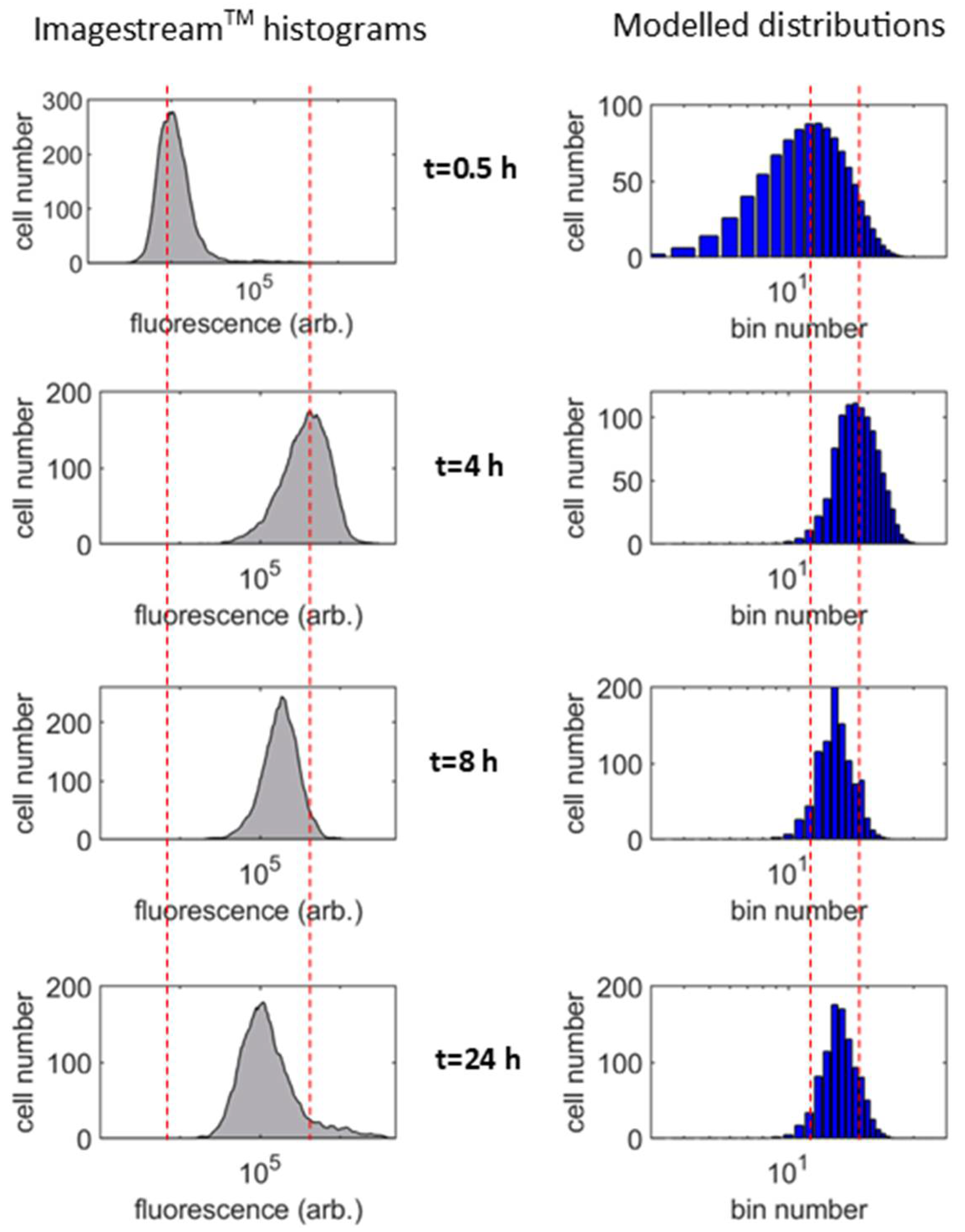
3.1. Population-Averaged Pharmacokinetics
- 1.
- Particle uptake at a constant rate (A1)
- 2.
- Particle uptake with a limiting process to supply or accumulation (A2 and A3)
- 3.
- Particle uptake with a limiting process to supply and accumulation (A4)
3.2. Single Cell Pharmacokinetics
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilczewska, A.Z.; Niemirowicz, K.; Markiewicz, K.H.; Car, H. Nanoparticles as drug delivery systems. Pharmacol. Rep. 2012, 64, 1020–1037. [Google Scholar] [CrossRef]
- Yang, J.; Hendricks, W.; Liu, G.; McCaffery, J.M.; Kinzler, K.W.; Huso, D.L.; Vogelstein, B.; Zhou, S. A nanoparticle formulation that selectively transfects metastatic tumors in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 14717–14722. [Google Scholar] [CrossRef] [PubMed]
- Blanco, J.L.J.; Benito, J.M.; Mellet, C.O.; Fernández, J.M.G. Molecular nanoparticle-based gene delivery systems. J. Drug Deliv. Sci. Technol. 2017, 42, 18–37. [Google Scholar] [CrossRef]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Wang, Z.; Feng, M. Nanocarriers with tunable surface properties to unblock bottlenecks in systemic drug and gene delivery. J. Control. Release 2015, 214, 121–133. [Google Scholar] [CrossRef]
- Zhang, X.-Q.; Xu, X.; Bertrand, N.; Pridgen, E.; Swami, A.; Farokhzad, O.C. Interactions of nanomaterials and biological systems: Implications to personalized nanomedicine. Adv. Drug Deliv. Rev. 2012, 64, 1363–1384. [Google Scholar] [CrossRef] [PubMed]
- Hinde, E.; Thammasiraphop, K.; Duong, H.; Yeow, J.; Karagoz, B.; Boyer, C.; Gooding, J.J.; Gaus, K. Pair correlation microscopy reveals the role of nanoparticle shape in intracellular transport and site of drug release. Nat. Nanotechnol. 2016, 12, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, A.; Fenaroli, F.; Monopoli, M.P.; Åberg, C.; Dawson, K.A.; Salvati, A. Effects of the Presence or Absence of a Protein Corona on Silica Nanoparticle Uptake and Impact on Cells. ACS Nano 2012, 6, 5845–5857. [Google Scholar] [CrossRef]
- Bareford, L.M.; Swaan, P.W. Endocytic mechanisms for targeted drug delivery. Adv. Drug Deliv. Rev. 2007, 59, 748–758. [Google Scholar] [CrossRef]
- Avgoustakis, K.; Beletsi, A.; Panagi, Z.; Klepetsanis, P.; Karydas, A.G.; Ithakissios, D.S. PLGA–mPEG nanoparticles of cisplatin: In vitro nanoparticle degradation, in vitro drug release and in vivo drug residence in blood properties. J. Control. Release 2002, 79, 123–135. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Hunter, A.; Andresen, T.L. Factors Controlling Nanoparticle Pharmacokinetics: An Integrated Analysis and Perspective. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 481–503. [Google Scholar] [CrossRef]
- Higuchi, T. Mechanism of sustained-action medication. Theoretical analysis of rate of release of solid drugs dispersed in solid matrices. J. Pharm. Sci. 1963, 52, 1145–1149. [Google Scholar] [CrossRef]
- Siepmann, J.; Peppas, N.A. Higuchi equation: Derivation, applications, use and misuse. Int. J. Pharm. 2011, 418, 6–12. [Google Scholar] [CrossRef]
- Pan, R.; Liu, G.; Li, Y.; Wei, Y.; Li, S.; Tao, L. Size-dependent endocytosis and a dynamic-release model of nanoparticles. Nanoscale 2018, 10, 8269–8274. [Google Scholar] [CrossRef]
- Soares, P.I.; Sousa, A.I.; Silva, J.C.; Ferreira, I.M.; Novo, C.M.; Borges, J.P. Chitosan-based nanoparticles as drug delivery systems for doxorubicin: Optimization and modelling. Carbohydr. Polym. 2016, 147, 304–312. [Google Scholar] [CrossRef]
- Aydın, R.S.T.; Pulat, M. 5-Fluorouracil Encapsulated Chitosan Nanoparticles for pH-Stimulated Drug Delivery: Evaluation of Controlled Release Kinetics. J. Nanomater. 2012, 2012, 1–10. [Google Scholar] [CrossRef]
- Golovin, Y.; Golovin, D.; Klyachko, N.; Majouga, A.; Kabanov, A. Modeling drug release from functionalized magnetic nanoparticles actuated by non-heating low frequency magnetic field. J. Nanoparticle Res. 2017, 19, 64. [Google Scholar] [CrossRef]
- Son, G.-H.; Lee, B.-J.; Cho, C.-W. Mechanisms of drug release from advanced drug formulations such as polymeric-based drug-delivery systems and lipid nanoparticles. J. Pharm. Investig. 2017, 47, 287–296. [Google Scholar] [CrossRef]
- Li, M.; Al-Jamal, K.T.; Kostarelos, K.; Reineke, J. Physiologically Based Pharmacokinetic Modeling of Nanoparticles. ACS Nano 2010, 4, 6303–6317. [Google Scholar] [CrossRef] [PubMed]
- Carlander, U.; Li, D.; Jolliet, O.; Emond, C.; Johanson, G. Toward a general physiologically-based pharmacokinetic model for intravenously injected nanoparticles. Int. J. Nanomed. 2016, ume 11, 625–640. [Google Scholar] [CrossRef]
- Lin, Z.; Monteiro-Riviere, N.A.; Riviere, J.E. A physiologically based pharmacokinetic model for polyethylene glycol-coated gold nanoparticles of different sizes in adult mice. Nanotoxicology 2015, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yaehne, K.; Tekrony, A.; Clancy, A.; Gregoriou, Y.; Walker, J.; Dean, K.; Nguyen, T.; Doiron, A.; Rinker, K.; Jiang, X.Y.; et al. Nanoparticle Accumulation in Angiogenic Tissues: Towards Predictable Pharmacokinetics. Small 2013, 9, 3118–3127. [Google Scholar] [CrossRef] [PubMed]
- Salvati, A.; Åberg, C.; Santos, T.; Varela, J.A.; Pinto, P.; Lynch, I.; Dawson, K.A. Experimental and theoretical comparison of intracellular import of polymeric nanoparticles and small molecules: Toward models of uptake kinetics. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Soininen, S.K.; Vellonen, K.-S.; Heikkinen, A.T.; Auriola, S.; Ranta, V.-P.; Urtti, A.; Ruponen, M. Intracellular PK/PD Relationships of Free and Liposomal Doxorubicin: Quantitative Analyses and PK/PD Modeling. Mol. Pharm. 2016, 13, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Donahue, N.D.; Acar, H.; Wilhelm, S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv. Drug Deliv. Rev. 2019, 143, 68–96. [Google Scholar] [CrossRef]
- Lopes, C.D.; Gomes, C.P.; Neto, E.; Sampaio, P.; Aguiar, P.; Pêgo, A.P. Microfluidic-based platform to mimic the in vivo peripheral administration of neurotropic nanoparticles. Nanomedicine 2016, 11, 3205–3221. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.; Gonçalves, N.P.; Gomes, C.; Saraiva, M.J.; Pêgo, A.P. BDNF gene delivery mediated by neuron-targeted nanoparticles is neuroprotective in peripheral nerve injury. Biomaterials 2017, 121, 83–96. [Google Scholar] [CrossRef]
- Gomes, C.; Varela-Moreira, A.; Leiro, V.; Lopes, C.; Moreno, P.; Lazaro, M.G.; Pêgo, A.P. A high-throughput bioimaging study to assess the impact of chitosan-based nanoparticle degradation on DNA delivery performance. Acta Biomater. 2016, 46, 129–140. [Google Scholar] [CrossRef]
- Ware, M.J.; Godin, B.; Singh, N.; Majithia, R.; Shamsudeen, S.; Serda, R.E.; Meissner, K.E.; Rees, P.; Summers, H.D. Analysis of the Influence of Cell Heterogeneity on Nanoparticle Dose Response. ACS Nano 2014, 8, 6693–6700. [Google Scholar] [CrossRef]
- Lunov, O.; Zablotskii, V.; Syrovets, T.; Röcker, C.; Tron, K.; Nienhaus, G.U.; Simmet, T. Modeling receptor-mediated endocytosis of polymer-functionalized iron oxide nanoparticles by human macrophages. Biomaterials 2011, 32, 547–555. [Google Scholar] [CrossRef]
- Papadopoulou, V.; Kosmidis, K.; Vlachou, M.; Macheras, P. On the use of the Weibull function for the discernment of drug release mechanisms. Int. J. Pharm. 2006, 309, 44–50. [Google Scholar] [CrossRef]
- Hans, M.L.; Lowman, A.M. Biodegradable nanoparticles for drug delivery and targeting. Curr. Opin. Solid State Mater. Sci. 2002, 6, 319–327. [Google Scholar] [CrossRef]
- Oh, N.; Park, J.H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int. J. Nanomed. 2014, 9, 51–63. [Google Scholar]
- Tancini, B.; Buratta, S.; Delo, F.; Sagini, K.; Chiaradia, E.; Pellegrino, R.M.; Emiliani, C.; Urbanelli, L. Lysosomal Exocytosis: The Extracellular Role of an Intracellular Organelle. Membranes 2020, 10, 406. [Google Scholar] [CrossRef] [PubMed]
- Douglas, J.F.; Johnson, H.E.; Granick, S. A Simple Kinetic Model of Polymer Adsorption and Desorption. Science 1993, 262, 2010–2012. [Google Scholar] [CrossRef] [PubMed]
- Jonker, C.T.H.; Deo, C.; Zager, P.J.; Tkachuk, A.N.; Weinstein, A.M.; Rodriguez-Boulan, E.; Lavis, L.D.; Schreiner, R. Accurate measurement of fast endocytic recycling kinetics in real time. J. Cell Sci. 2019, 133. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, A.; Salvati, A.; Santos-Martinez, M.J.; Radomski, M.W.; Dawson, K.A.; Åberg, C. Nanoparticle Adhesion to the Cell Membrane and Its Effect on Nanoparticle Uptake Efficiency. J. Am. Chem. Soc. 2013, 135, 1438–1444. [Google Scholar] [CrossRef]
- Pires, L.R.; Oliveira, H.; Barrias, C.C.; Sampaio, P.; Pereira, A.J.; Maiato, H.; Simões, S.; Pêgo, A.P. Imidazole-grafted chitosan-mediated gene delivery: In vitro study on transfection, intracellular trafficking and degradation. Nanomedicine 2011, 6, 1499–1512. [Google Scholar] [CrossRef]
- Summers, H.D.; Rees, P.; Holton, M.D.; Brown, M.R.; Chappell, S.C.; Smith, P.J.; Errington, R.J. Statistical analysis of nanoparticle dosing in a dynamic cellular system. Nat. Nanotechnol. 2011, 6, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Rees, P.; Wills, J.W.; Brown, R.; Barnes, C.M.; Summers, H.D. The origin of heterogeneous nanoparticle uptake by cells. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Gaylor, D.W. The use of Haber’s Law in standard setting and risk assessment. Toxicology 2000, 149, 17–19. [Google Scholar] [CrossRef]
- Stukalin, E.B.; Aifuwa, I.; Kim, J.S.; Wirtz, D.; Sun, S.X. Age-dependent stochastic models for understanding population fluctuations in continuously cultured cells. J. R. Soc. Interface 2013, 10, 20130325. [Google Scholar] [CrossRef]
- Alonso, A.A.; Molina, I.; Theodoropoulos, C. Modeling Bacterial Population Growth from Stochastic Single-Cell Dynamics. Appl. Environ. Microbiol. 2014, 80, 5241–5253. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Amir, A. The Effects of Stochasticity at the Single-Cell Level and Cell Size Control on the Population Growth. Cell Syst. 2017, 5, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Editorial. Join the dialogue. Nat. Nanotechnol. 2012, 7, 545. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Editorial. The dialogue continues. Nat. Nanotechnol. 2013, 8, 69. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Summers, H.D.; Gomes, C.P.; Varela-Moreira, A.; Spencer, A.P.; Gomez-Lazaro, M.; Pêgo, A.P.; Rees, P. Data-Driven Modeling of the Cellular Pharmacokinetics of Degradable Chitosan-Based Nanoparticles. Nanomaterials 2021, 11, 2606. https://doi.org/10.3390/nano11102606
Summers HD, Gomes CP, Varela-Moreira A, Spencer AP, Gomez-Lazaro M, Pêgo AP, Rees P. Data-Driven Modeling of the Cellular Pharmacokinetics of Degradable Chitosan-Based Nanoparticles. Nanomaterials. 2021; 11(10):2606. https://doi.org/10.3390/nano11102606
Chicago/Turabian StyleSummers, Huw D., Carla P. Gomes, Aida Varela-Moreira, Ana P. Spencer, Maria Gomez-Lazaro, Ana P. Pêgo, and Paul Rees. 2021. "Data-Driven Modeling of the Cellular Pharmacokinetics of Degradable Chitosan-Based Nanoparticles" Nanomaterials 11, no. 10: 2606. https://doi.org/10.3390/nano11102606
APA StyleSummers, H. D., Gomes, C. P., Varela-Moreira, A., Spencer, A. P., Gomez-Lazaro, M., Pêgo, A. P., & Rees, P. (2021). Data-Driven Modeling of the Cellular Pharmacokinetics of Degradable Chitosan-Based Nanoparticles. Nanomaterials, 11(10), 2606. https://doi.org/10.3390/nano11102606





