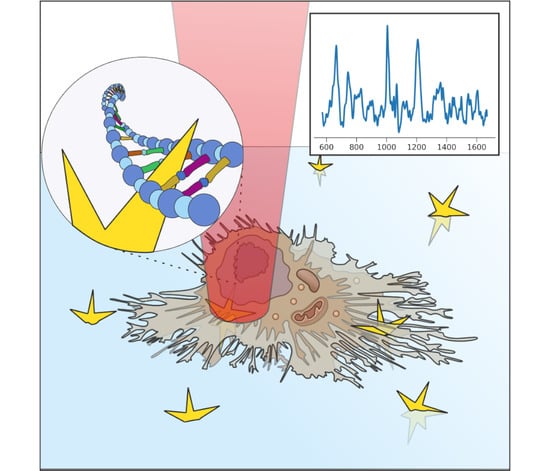
Live Cell Poration by Au Nanostars to Probe Intracellular Molecular Composition with SERS
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparing of SERS Substrate
2.3. Characterization
2.4. Cell Culture and Targeting
2.5. Living Cell Imaging
2.6. Processing of Raman Imaging Data
3. Results and Discussion
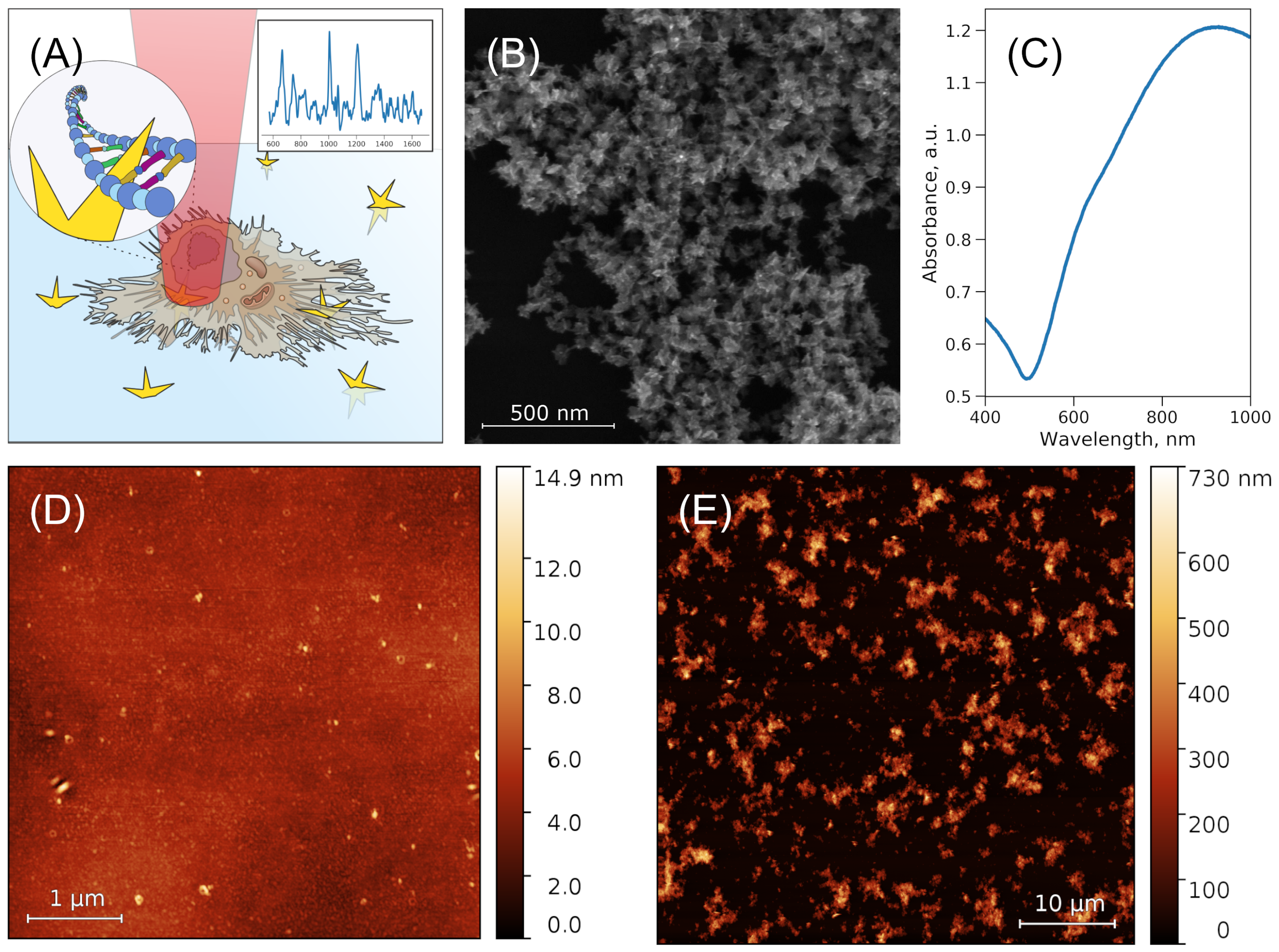
3.1. Nanostructure Characterization
3.2. Spectral Analysis
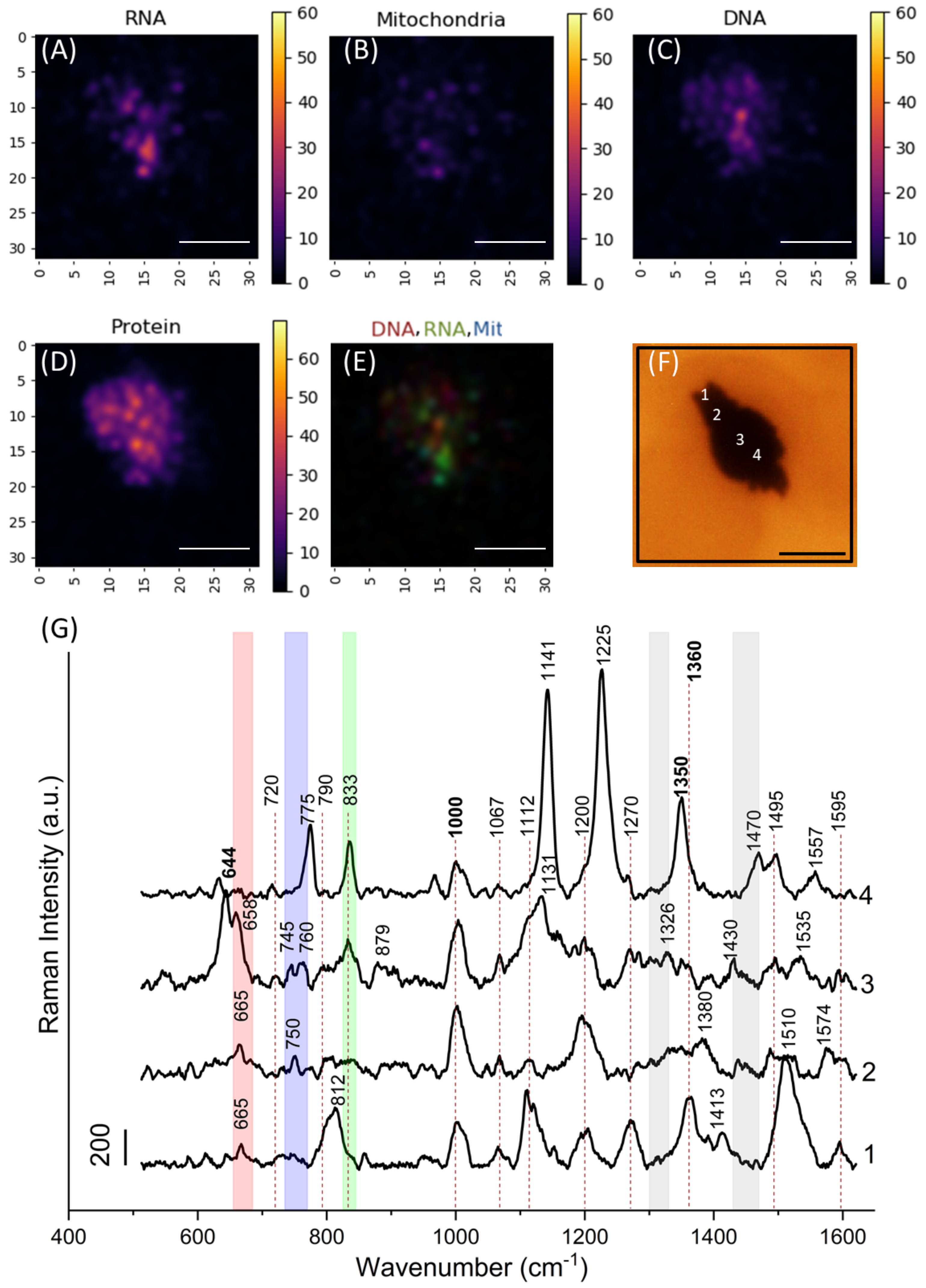
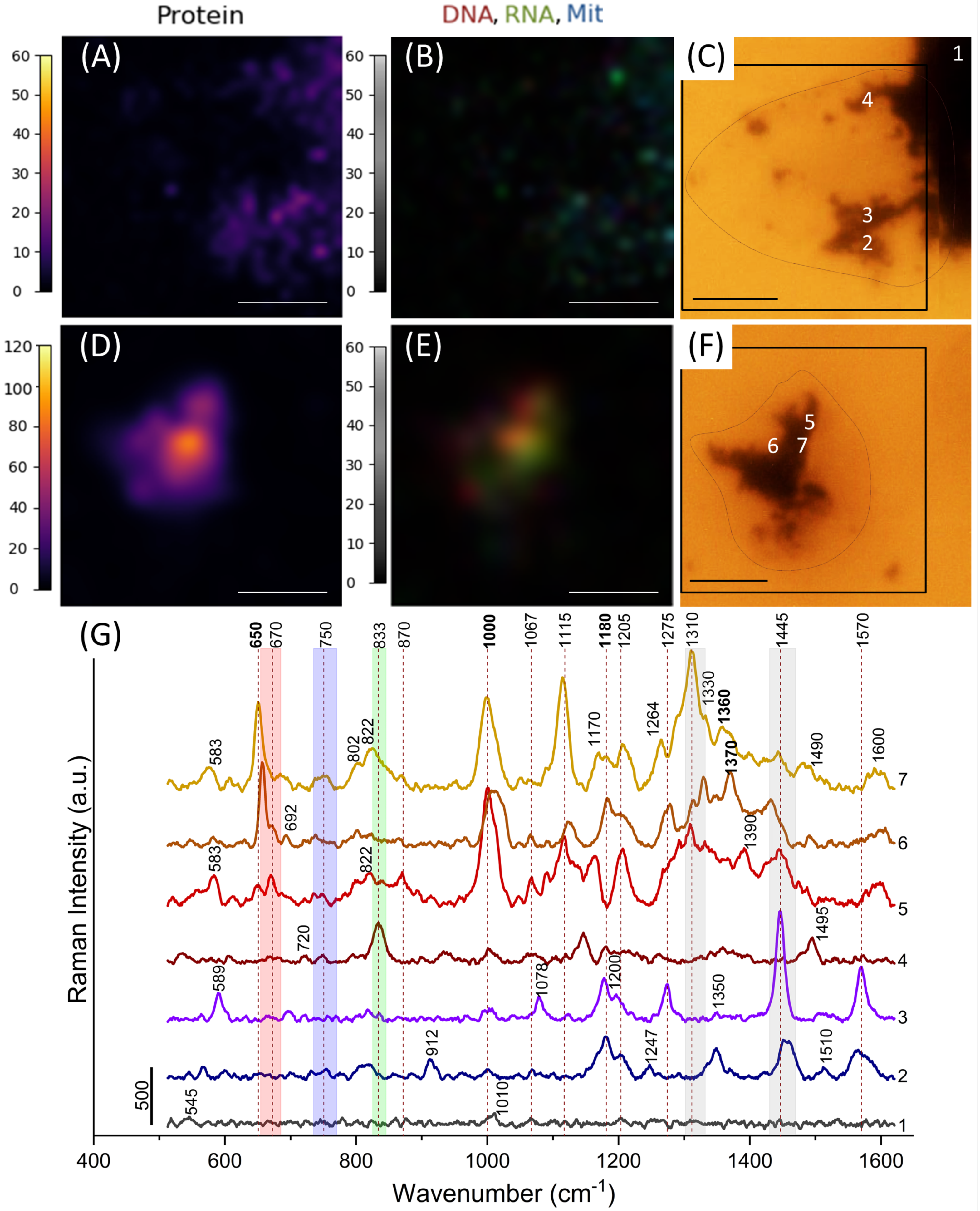
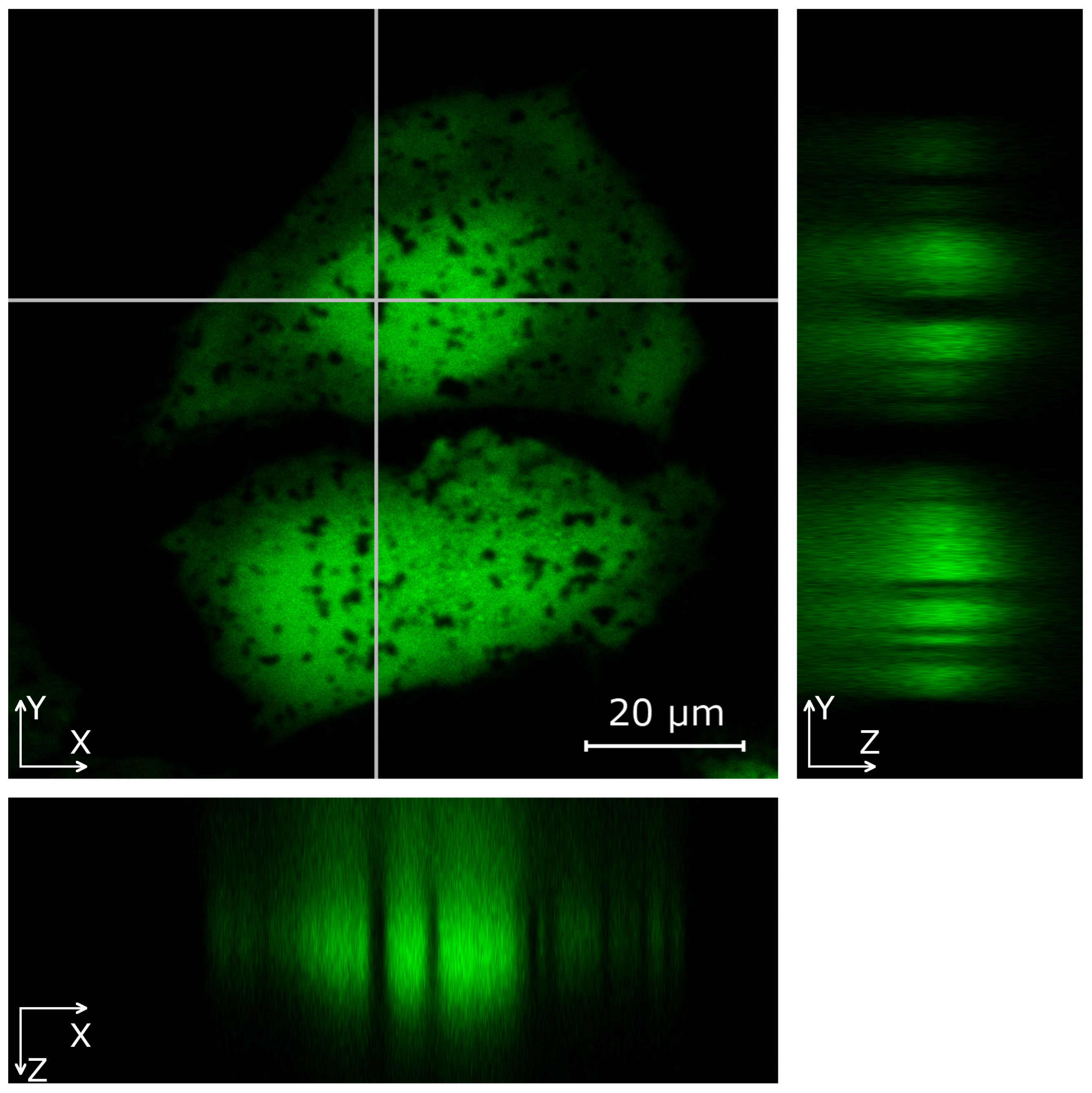
3.3. Image Analysis
3.4. Nanostars Localization Inside Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petry, R.; Schmitt, M.; Popp, J. Raman Spectroscopy-A Prospective Tool in the Life Sciences. ChemPhysChem 2003, 4, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Pahlow, S.; März, A.; Seise, B.; Hartmann, K.; Freitag, I.; Kämmer, E.; Böhme, R.; Deckert, V.; Weber, K.; Cialla, D.; et al. Bioanalytical application of surface- and tip-enhanced Raman spectroscopy. Eng. Life Sci. 2012, 12, 131–143. [Google Scholar] [CrossRef]
- Vitol, E.A.; Orynbayeva, Z.; Friedman, G.; Gogotsi, Y. Nanoprobes for intracellular and single cell surface-enhanced Raman spectroscopy (SERS). J. Raman Spectrosc. 2012, 43, 817–827. [Google Scholar] [CrossRef]
- Eremina, O.E.; Semenova, A.A.; Sergeeva, E.A.; Brazhe, N.A.; Maksimov, G.V.; Shekhovtsova, T.N.; Goodilin, E.A.; Veselova, I.A. Surface-enhanced Raman spectroscopy in modern chemical analysis: Advances and prospects. Russ. Chem. Rev. 2018, 87, 741–770. [Google Scholar] [CrossRef]
- Bruzas, I.; Lum, W.; Gorunmez, Z.; Sagle, L. Advances in surface-enhanced Raman spectroscopy (SERS) substrates for lipid and protein characterization: Sensing and beyond. Analyst 2018, 143, 3990–4008. [Google Scholar] [CrossRef]
- Willets, K.A. Surface-enhanced Raman scattering (SERS) for probing internal cellular structure and dynamics. Anal. Bioanal. Chem. 2009, 394, 85–94. [Google Scholar] [CrossRef]
- Lane, L.A.; Qian, X.; Nie, S. SERS Nanoparticles in Medicine: From Label-Free Detection to Spectroscopic Tagging. Chem. Rev. 2015, 115, 10489–10529. [Google Scholar] [CrossRef]
- Khlebtsov, N.; Bogatyrev, V.; Dykman, L.; Khlebtsov, B.; Staroverov, S.; Shirokov, A.; Matora, L.; Khanadeev, V.; Pylaev, T.; Tsyganova, N.; et al. Analytical and Theranostic Applications of Gold Nanoparticles and Multifunctional Nanocomposites. Theranostics 2013, 3, 167–180. [Google Scholar] [CrossRef]
- Schlücker, S. SERS Microscopy: Nanoparticle Probes and Biomedical Applications. ChemPhysChem 2009, 10, 1344–1354. [Google Scholar] [CrossRef]
- Kneipp, J.; Kneipp, H.; McLaughlin, M.; Brown, D.; Kneipp, K. In Vivo Molecular Probing of Cellular Compartments with Gold Nanoparticles and Nanoaggregates. Nano Lett. 2006, 6, 2225–2231. [Google Scholar] [CrossRef]
- Yashchenok, A.M.; Masic, A.; Gorin, D.A.; Shim, B.S.; Kotov, N.A.; Fratzl, P.; Möhwald, H.; Skirtach, A.G. Nanoengineered Colloidal Probes for Raman-based Detection of Biomolecules inside Living Cells. Small 2013, 9, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Cupil-Garcia, V.; Strobbia, P.; Crawford, B.M.; Wang, H.; Ngo, H.; Liu, Y.; Vo-Dinh, T. Plasmonic nanoplatforms: From surface-enhanced Raman scattering sensing to biomedical applications. J. Raman Spectrosc. 2021, 52, 541–553. [Google Scholar] [CrossRef]
- Panikkanvalappil, S.R.; Hira, S.M.; Mahmoud, M.A.; El-Sayed, M.A. Unraveling the Biomolecular Snapshots of Mitosis in Healthy and Cancer Cells Using Plasmonically-Enhanced Raman Spectroscopy. J. Am. Chem. Soc. 2014, 136, 15961–15968. [Google Scholar] [CrossRef]
- Sathuluri, R.R.; Yoshikawa, H.; Shimizu, E.; Saito, M.; Tamiya, E. Gold Nanoparticle-Based Surface-Enhanced Raman Scattering for Noninvasive Molecular Probing of Embryonic Stem Cell Differentiation. PLoS ONE 2011, 6, e22802. [Google Scholar] [CrossRef][Green Version]
- Jun, B.H.; Noh, M.S.; Kim, J.; Kim, G.; Kang, H.; Kim, M.S.; Seo, Y.T.; Baek, J.; Kim, J.H.; Park, J.; et al. Multifunctional Silver-Embedded Magnetic Nanoparticles as SERS Nanoprobes and Their Applications. Small 2010, 6, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Stetciura, I.Y.; Yashchenok, A.M.; Masic, A.; Lyubin, E.V.; Inozemtseva, O.A.; Drozdova, M.G.; Markvichova, E.A.; Khlebtsov, B.N.; Fedyanin, A.A.; Sukhorukov, G.B.; et al. Composite SERS-based satellites navigated by optical tweezers for single cell analysis. Analyst 2015, 140, 4981–4986. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bai, X.; Su, L.; Du, Z.; Shen, A.; Materny, A.; Hu, J. Combined Labelled and Label-free SERS Probes for Triplex Three-dimensional Cellular Imaging. Sci. Rep. 2016, 6, 19173. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Abdali, S.; Brazhe, A.R.; Luneva, O.G.; Bryzgalova, N.Y.; Parshina, E.Y.; Sosnovtseva, O.V.; Maksimov, G.V. New Insight into Erythrocyte through In Vivo Surface-Enhanced Raman Spectroscopy. Biophys. J. 2009, 97, 3206–3214. [Google Scholar] [CrossRef] [PubMed]
- Sarycheva, A.S.; Brazhe, N.A.; Baizhumanov, A.A.; Nikelshparg, E.I.; Semenova, A.A.; Garshev, A.V.; Baranchikov, A.E.; Ivanov, V.K.; Maksimov, G.V.; Sosnovtseva, O.; et al. New nanocomposites for SERS studies of living cells and mitochondria. J. Mater. Chem. B 2016, 4, 539–546. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Evlyukhin, A.B.; Goodilin, E.A.; Semenova, A.A.; Novikov, S.M.; Bozhevolnyi, S.I.; Chichkov, B.N.; Sarycheva, A.S.; Baizhumanov, A.A.; Nikelshparg, E.I.; et al. Probing cytochrome c in living mitochondria with surface-enhanced Raman spectroscopy. Sci. Rep. 2015, 5, 13793. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Nikelshparg, E.I.; Baizhumanov, A.A.; Grivennikova, V.G.; Semenova, A.A.; Novikov, S.M.; Volkov, V.S.; Arsenin, A.V.; Yakubovsky, D.I.; Evlyukhin, A.B.; et al. SERS uncovers the link between conformation of cytochrome c heme and mitochondrial membrane potential. bioRxiv 2021. [Google Scholar] [CrossRef]
- Liang, O.; Wang, P.; Xia, M.; Augello, C.; Yang, F.; Niu, G.; Liu, H.; Xie, Y.H. Label-free distinction between p53+/+ and p53−/− colon cancer cells using a graphene based SERS platform. Biosens. Bioelectron. 2018, 118, 108–114. [Google Scholar] [CrossRef]
- Barbosa, S.; Agrawal, A.; Rodríguez-Lorenzo, L.; Pastoriza-Santos, I.; Alvarez-Puebla, R.A.; Kornowski, A.; Weller, H.; Liz-Marzán, L.M. Tuning Size and Sensing Properties in Colloidal Gold Nanostars. Langmuir 2010, 26, 14943–14950. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Lorenzo, L.; Álvarez-Puebla, R.A.; Pastoriza-Santos, I.; Mazzucco, S.; Stéphan, O.; Kociak, M.; Liz-Marzán, L.M.; García de Abajo, F.J. Zeptomol Detection Through Controlled Ultrasensitive Surface-Enhanced Raman Scattering. J. Am. Chem. Soc. 2009, 131, 4616–4618. [Google Scholar] [CrossRef]
- Rodríguez-Lorenzo, L.; Krpetic, Z.; Barbosa, S.; Alvarez-Puebla, R.A.; Liz-Marzán, L.M.; Prior, I.A.; Brust, M. Intracellular mapping with SERS-encoded gold nanostars. Integr. Biol. 2011, 3, 922. [Google Scholar] [CrossRef]
- Khoury, C.G.; Vo-Dinh, T. Gold Nanostars For Surface-Enhanced Raman Scattering: Synthesis, Characterization and Optimization. J. Phys. Chem. C Nanomater. Interfaces 2008, 2008, 18849–18859. [Google Scholar] [CrossRef]
- Cao, X.; Shi, C.; Lu, W.; Zhao, H.; Wang, M.; Tong, W.; Dong, J.; Han, X.; Qian, W. Synthesis of Au Nanostars and Their Application as Surface Enhanced Raman Scattering-Activity Tags Inside Living Cells. J. Nanosci. Nanotechnol. 2015, 15, 4829–4836. [Google Scholar] [CrossRef]
- Shiohara, A.; Novikov, S.M.; Solís, D.M.; Taboada, J.M.; Obelleiro, F.; Liz-Marzán, L.M. Plasmon Modes and Hot Spots in Gold Nanostar–Satellite Clusters. J. Phys. Chem. C 2015, 119, 10836–10843. [Google Scholar] [CrossRef]
- Khlebtsov, B.N.; Panfilova, E.; Khanadeev, V.; Khlebtsov, N. Improved size-tunable synthesis and SERS properties of Au nanostars. J. Nanopart. Res. 2014, 16, 2623. [Google Scholar] [CrossRef]
- Indrasekara, A.S.D.S.; Meyers, S.; Shubeita, S.; Feldman, L.C.; Gustafsson, T.; Fabris, L. Gold nanostar substrates for SERS-based chemical sensing in the femtomolar regime. Nanoscale 2014, 6, 8891–8899. [Google Scholar] [CrossRef]
- Register, J.K.; Fales, A.M.; Wang, H.N.; Norton, S.J.; Cho, E.H.; Boico, A.; Pradhan, S.; Kim, J.; Schroeder, T.; Wisniewski, N.A.; et al. In vivo detection of SERS-encoded plasmonic nanostars in human skin grafts and live animal models. Anal. Bioanal. Chem. 2015, 407, 8215–8224. [Google Scholar] [CrossRef]
- Malynych, S.; Luzinov, I.; Chumanov, G. Poly(Vinyl Pyridine) as a Universal Surface Modifier for Immobilization of Nanoparticles. J. Phys. Chem. B 2002, 106, 1280–1285. [Google Scholar] [CrossRef]
- Guerrero-Martínez, A.; Barbosa, S.; Pastoriza-Santos, I.; Liz-Marzán, L.M. Nanostars shine bright for you. Curr. Opin. Colloid Interface Sci. 2011, 16, 118–127. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, H.; Kersey, F.; Register, J.; Parrott, M.; Vo-Dinh, T. Plasmonic Gold Nanostars for Multi-Modality Sensing and Diagnostics. Sensors 2015, 15, 3706–3720. [Google Scholar] [CrossRef]
- Indrasekara, A.S.D.S.; Thomas, R.; Fabris, L. Plasmonic properties of regiospecific core–satellite assemblies of gold nanostars and nanospheres. Phys. Chem. Chem. Phys. 2015, 17, 21133–21142. [Google Scholar] [CrossRef]
- Vanzha, E.; Pylaev, T.; Prilepskii, A.; Golubev, A.; Khlebtsov, B.; Bogatyrev, V.; Khlebtsov, N. Cell culture surfaces with immobilized gold nanostars: A new approach for laser-induced plasmonic cell optoporation. In Proceedings of the Saratov Fall Meeting 2016: Optical Technologies in Biophysics and Medicine XVIII, Saratov, Russia, 26–30 September 2016; p. 103360L. [Google Scholar] [CrossRef]
- Pylaev, T.; Vanzha, E.; Avdeeva, E.; Khlebtsov, B.; Khlebtsov, N. A novel cell transfection platform based on laser optoporation mediated by Au nanostar layers. J. Biophotonics 2019, 12, e201800166. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, W.; Xiong, C.; Huang, Y.; Chen, B.J.; Racioppi, L.; Chao, N.; Vo-Dinh, T. Biodistribution and sensitive tracking of immune cells with plasmonic gold nanostars. Int. J. Nanomed. 2019, 14, 3403–3411. [Google Scholar] [CrossRef]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Open Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Korolovych, V.F.; Grishina, O.A.; Inozemtseva, O.A.; Selifonov, A.V.; Bratashov, D.N.; Suchkov, S.G.; Bulavin, L.A.; Glukhova, O.E.; Sukhorukov, G.B.; Gorin, D.A. Impact of high-frequency ultrasound on nanocomposite microcapsules: In silico and in situ visualization. Phys. Chem. Chem. Phys. 2016, 18, 2389–2397. [Google Scholar] [CrossRef]
- Brazhe, A.R. Pyraman: Working with Raman Spectra in Python. 2017. Available online: https://github.com/abrazhe/pyraman (accessed on 6 August 2021).
- Brazhe, N.A.; Thomsen, K.; Lønstrup, M.; Brazhe, A.R.; Nikelshparg, E.I.; Maksimov, G.V.; Lauritzen, M.; Sosnovtseva, O. Monitoring of blood oxygenation in brain by resonance Raman spectroscopy. J. Biophotonics 2018, 11, e201700311. [Google Scholar] [CrossRef]
- Khlebtsov, N.G.; Zarkov, S.V.; Khanadeev, V.A.; Avetisyan, Y.A. A novel concept of two-component dielectric function for gold nanostars: Theoretical modelling and experimental verification. Nanoscale 2020, 12, 19963–19981. [Google Scholar] [CrossRef] [PubMed]
- Prikhozhdenko, E.S.; Bratashov, D.N.; Atkin, V.S.; Kamyshinsky, R.A.; Vasiliev, A.L.; Khlebtsov, B.N.; Gorin, D.A.; Möhwald, H.; Yashchenok, A.M. Precise control of distance between plasmonic surface-enhanced Raman scattering substrate and analyte molecules with polyelectrolyte layers. J. Raman Spectrosc. 2018, 49, 1581–1593. [Google Scholar] [CrossRef]
- Cheng, W.T.; Liu, M.T.; Liu, H.N.; Lin, S.Y. Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma. Microsc. Res. Tech. 2005, 68, 75–79. [Google Scholar] [CrossRef]
- Zhu, G.; Zhu, X.; Fan, Q.; Wan, X. Raman spectra of amino acids and their aqueous solutions. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 78, 1187–1195. [Google Scholar] [CrossRef]
- Erjavec, N.; Pinato, G.; Ramser, K. Raman spectroscopy as a tool for detecting mitochondrial fitness. J. Raman Spectrosc. 2016, 47, 933–939. [Google Scholar] [CrossRef]
- Chan, J.W.; Taylor, D.S.; Zwerdling, T.; Lane, S.M.; Ihara, K.; Huser, T. Micro-Raman Spectroscopy Detects Individual Neoplastic and Normal Hematopoietic Cells. Biophys. J. 2006, 90, 648–656. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Krafft, C.; Neudert, L.; Simat, T.; Salzer, R. Near infrared Raman spectra of human brain lipids. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2005, 61, 1529–1535. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Nikelshparg, E.I.; Prats, C.; Dela, F.; Sosnovtseva, O. Raman probing of lipids, proteins, and mitochondria in skeletal myocytes: A case study on obesity. J. Raman Spectrosc. 2017, 48, 1158–1165. [Google Scholar] [CrossRef]
- Wu, H.; Volponi, J.V.; Oliver, A.E.; Parikh, A.N.; Simmons, B.A.; Singh, S. In vivo lipidomics using single-cell Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 3809–3814. [Google Scholar] [CrossRef]
- Fazio, B.; D’Andrea, C.; Foti, A.; Messina, E.; Irrera, A.; Donato, M.G.; Villari, V.; Micali, N.; Maragò, O.M.; Gucciardi, P.G. SERS detection of Biomolecules at Physiological pH via aggregation of Gold Nanorods mediated by Optical Forces and Plasmonic Heating. Sci. Rep. 2016, 6, 26952. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H.; Baranska, M. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib. Spectrosc. 2007, 43, 13–25. [Google Scholar] [CrossRef]
- Huefner, A.; Kuan, W.L.; Barker, R.A.; Mahajan, S. Intracellular SERS Nanoprobes For Distinction Of Different Neuronal Cell Types. Nano Lett. 2013, 13, 2463–2470. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikelshparg, E.I.; Prikhozhdenko, E.S.; Verkhovskii, R.A.; Atkin, V.S.; Khanadeev, V.A.; Khlebtsov, B.N.; Bratashov, D.N. Live Cell Poration by Au Nanostars to Probe Intracellular Molecular Composition with SERS. Nanomaterials 2021, 11, 2588. https://doi.org/10.3390/nano11102588
Nikelshparg EI, Prikhozhdenko ES, Verkhovskii RA, Atkin VS, Khanadeev VA, Khlebtsov BN, Bratashov DN. Live Cell Poration by Au Nanostars to Probe Intracellular Molecular Composition with SERS. Nanomaterials. 2021; 11(10):2588. https://doi.org/10.3390/nano11102588
Chicago/Turabian StyleNikelshparg, Evelina I., Ekaterina S. Prikhozhdenko, Roman A. Verkhovskii, Vsevolod S. Atkin, Vitaly A. Khanadeev, Boris N. Khlebtsov, and Daniil N. Bratashov. 2021. "Live Cell Poration by Au Nanostars to Probe Intracellular Molecular Composition with SERS" Nanomaterials 11, no. 10: 2588. https://doi.org/10.3390/nano11102588
APA StyleNikelshparg, E. I., Prikhozhdenko, E. S., Verkhovskii, R. A., Atkin, V. S., Khanadeev, V. A., Khlebtsov, B. N., & Bratashov, D. N. (2021). Live Cell Poration by Au Nanostars to Probe Intracellular Molecular Composition with SERS. Nanomaterials, 11(10), 2588. https://doi.org/10.3390/nano11102588