Protein Supramolecular Structures: From Self-Assembly to Nanovaccine Design
Abstract
1. Introduction
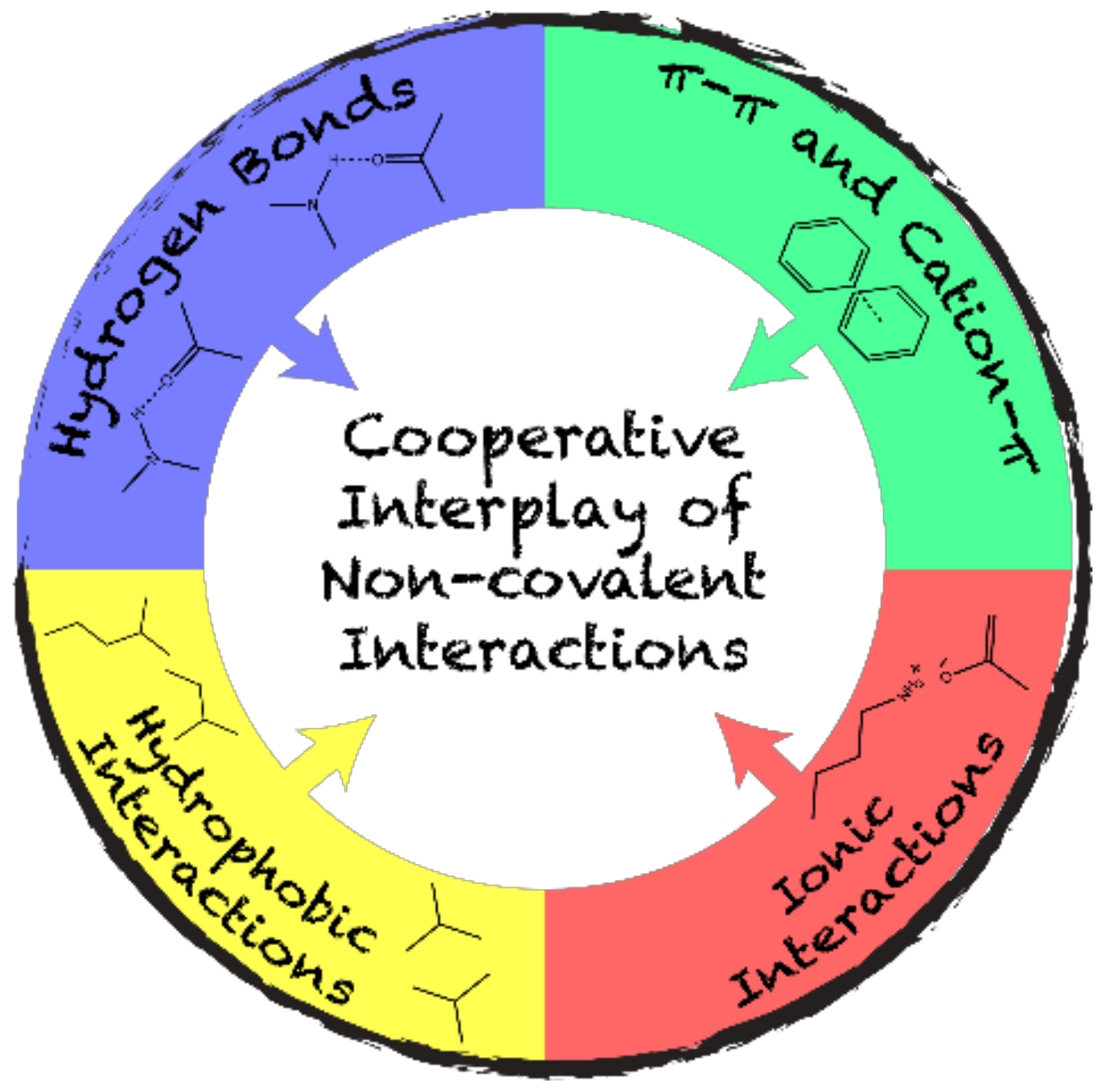
2. Interactions Guiding Protein Self-Assembly
2.1. Fundamentals of Molecular Self-Assembly
2.2. The Interplay of Non-Covalent Interactions
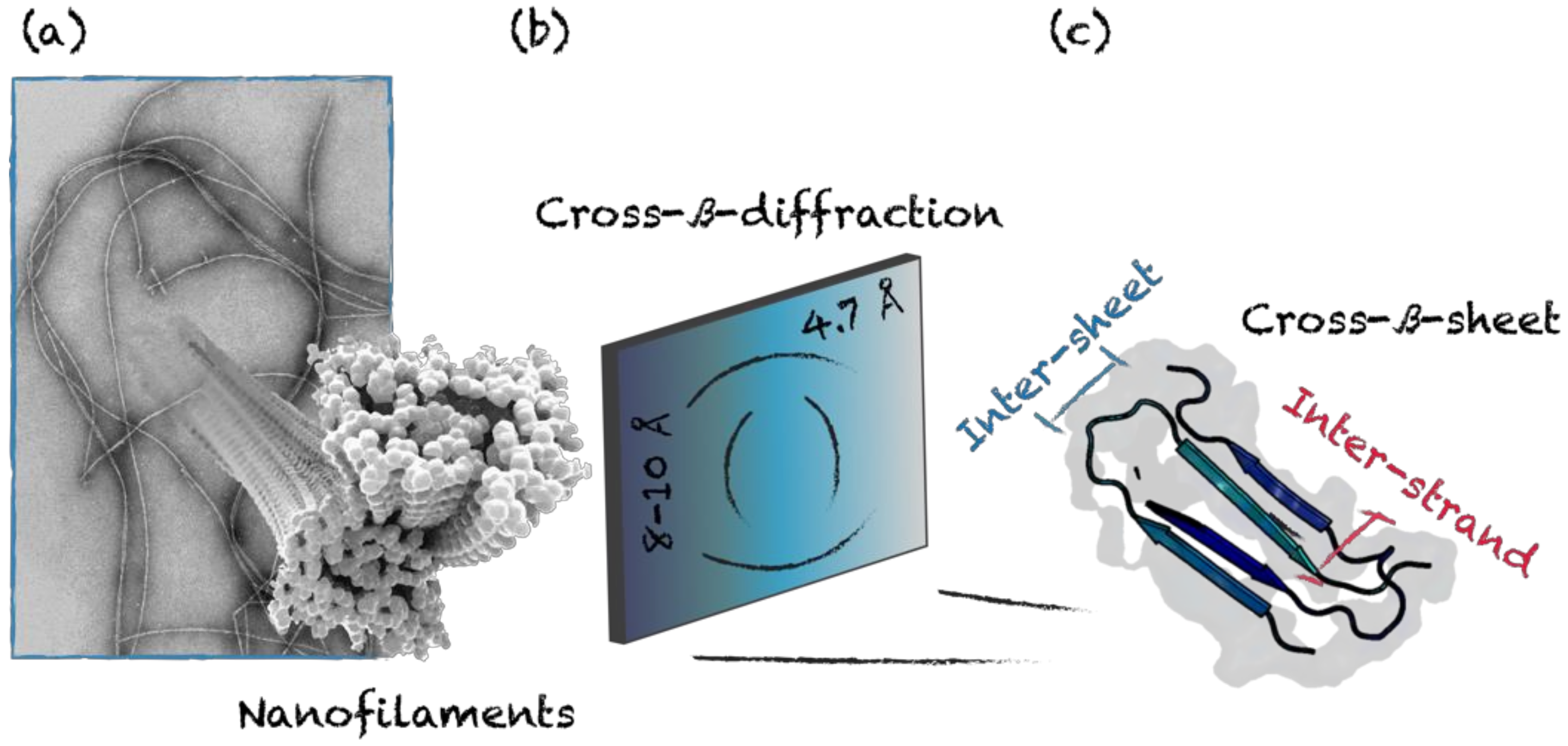
3. Self-Assembling Motifs
3.1. Coiled-Coil Motifs
3.2. Cross-β-Sheet and Other β-Sheet-Rich Quaternary Motifs
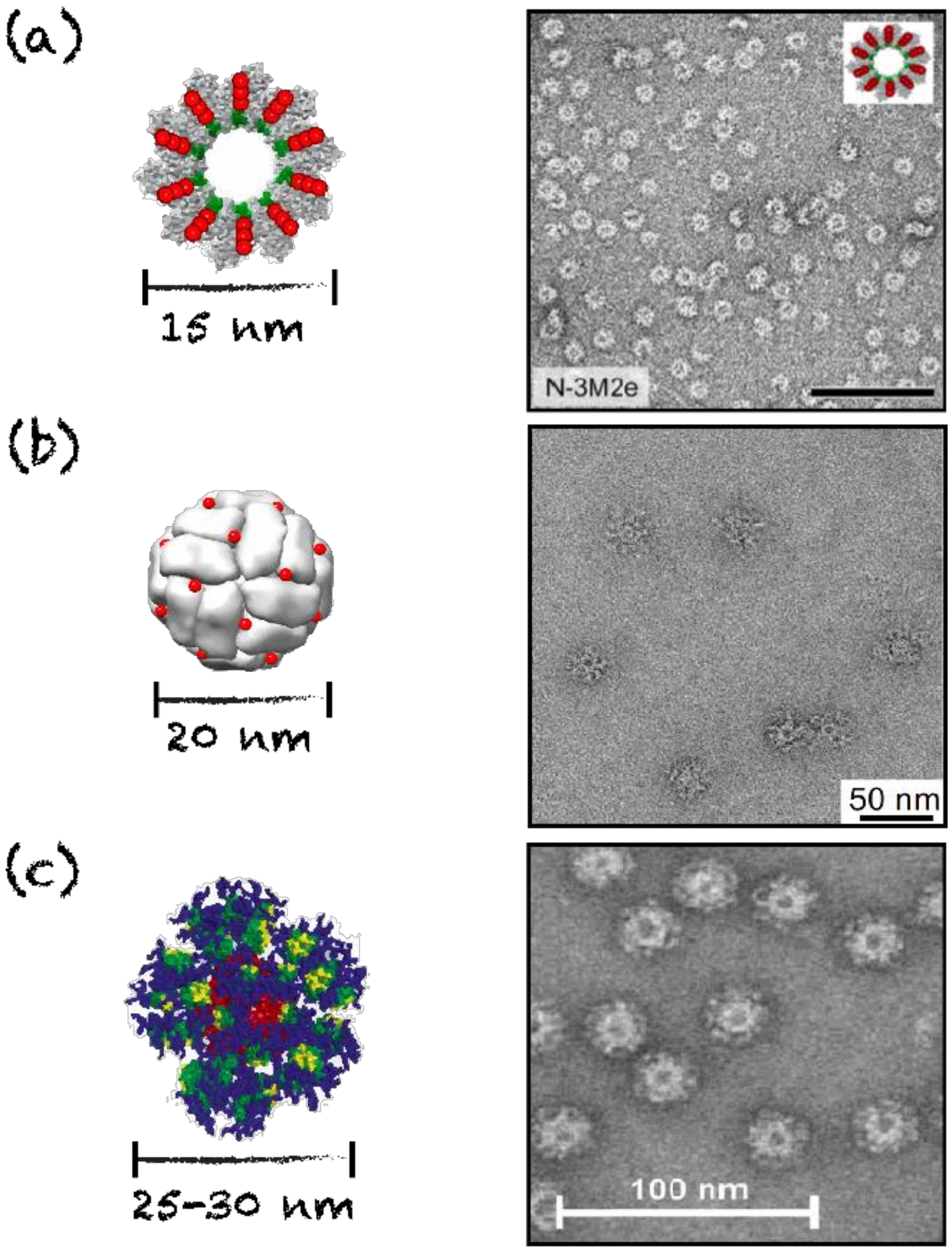
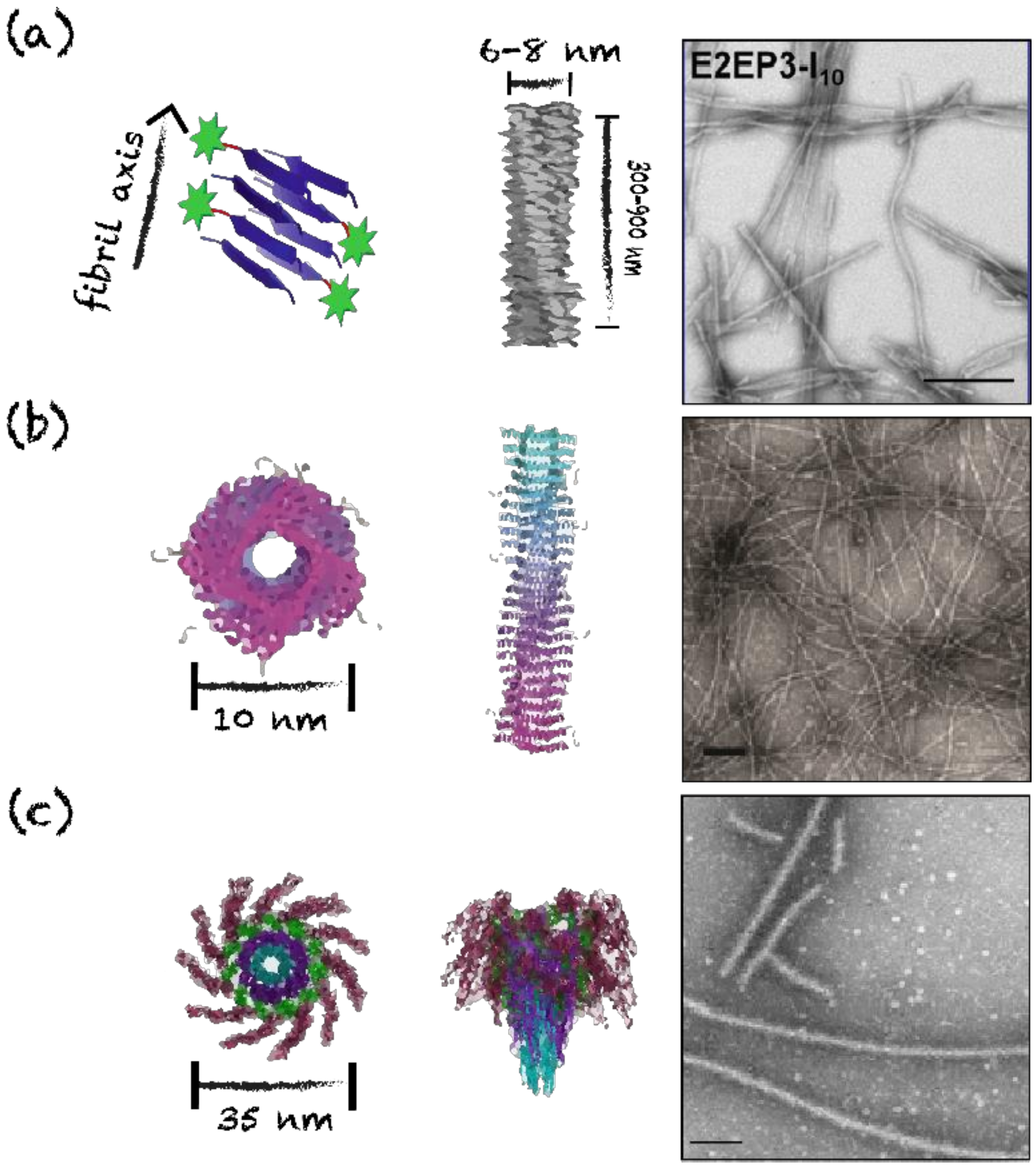
4. Supramolecular Protein Assemblies as Nanovaccine Scaffolds
4.1. Symmetry-Based Design
4.2. Nanorings, Polyhedral Cages and Nanoparticles
4.3. Nanofilaments and Nanotubes
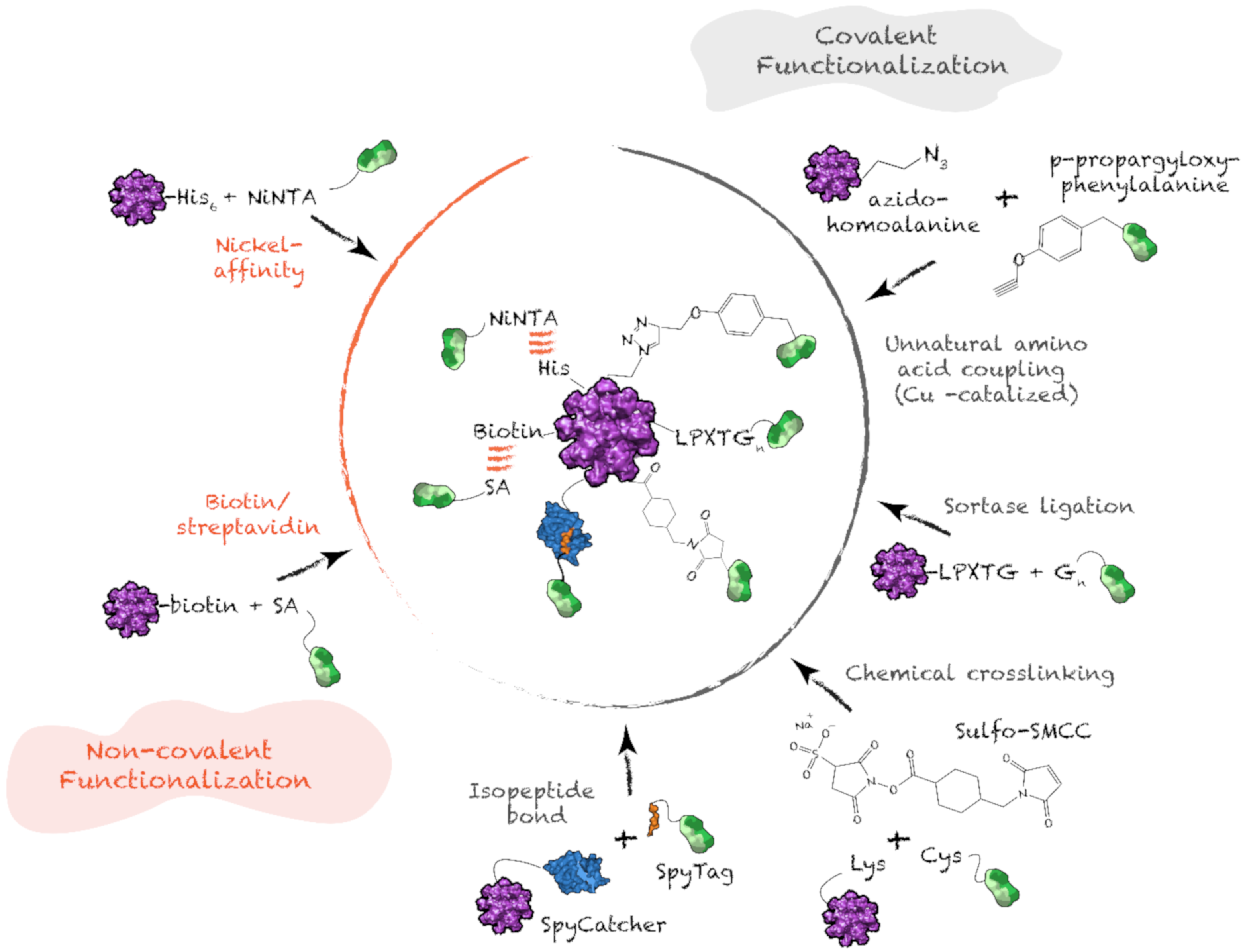
4.4. Antigen Functionalization on Protein Supramolecular Structure
5. Tuning the Immune Response with Protein Nanostructures
5.1. Induction of an Adaptive Immune Response with Protein Nanostructures
5.2. Modulating Immune Responses with Tailored Protein Nanostructures
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Plotkin, S.A. Vaccination against the major infectious diseases. Comptes Rendus Acad. Sci. III 1999, 322, 943–951. [Google Scholar] [CrossRef]
- McCullers, J.A.; Dunn, J.D. Advances in vaccine technology and their impact on managed care. Pharm. Ther. 2008, 33, 35–41. [Google Scholar]
- Shen, Y.; Hao, T.; Ou, S.; Hu, C.; Chen, L. Applications and perspectives of nanomaterials in novel vaccine development. MedChemComm 2018, 9, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Foged, C. Subunit vaccines of the future: The need for safe, customized and optimized particulate delivery systems. Ther. Deliv. 2011, 2, 1057–1077. [Google Scholar] [CrossRef] [PubMed]
- AWATE, S.; Babiuk, L.; Mutwiri, G. Mechanisms of Action of Adjuvants. Front. Immunol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- Karch, C.P.; Burkhard, P. Vaccine technologies: From whole organisms to rationally designed protein assemblies. Biochem. Pharm. 2016, 120, 1–14. [Google Scholar] [CrossRef]
- Al-Halifa, S.; Gauthier, L.; Arpin, D.; Bourgault, S.; Archambault, D. Nanoparticle-Based Vaccines Against Respiratory Viruses. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Han, J.; Zhao, D.; Li, D.; Wang, X.; Jin, Z.; Zhao, K. Polymer-Based Nanomaterials and Applications for Vaccines and Drugs. Polymers 2018, 10, 31. [Google Scholar] [CrossRef]
- Wang, L.; Gong, C.; Yuan, X.; Wei, G. Controlling the Self-Assembly of Biomolecules into Functional Nanomaterials through Internal Interactions and External Stimulations: A Review. Nanomaterials 2019, 9, 285. [Google Scholar] [CrossRef]
- Webber, M.J.; Appel, E.A.; Meijer, E.W.; Langer, R. Supramolecular biomaterials. Nat. Mater. 2016, 15, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Lehn, J.-M. Towards Complex Matter: Supramolecular Chemistry and Self-organization. Eur. Rev. 2009, 17, 263–280. [Google Scholar] [CrossRef]
- Amit, M.; Yuran, S.; Gazit, E.; Reches, M.; Ashkenasy, N. Tailor-Made Functional Peptide Self-Assembling Nanostructures. Adv. Mater. 2018, 30, 1707083. [Google Scholar] [CrossRef] [PubMed]
- Müller-Dethlefs, K.; Hobza, P. Noncovalent Interactions: A Challenge for Experiment and Theory. Chem. Rev. 2000, 100, 143–168. [Google Scholar] [CrossRef]
- Biedermann, F.; Nau, W.M.; Schneider, H.-J. The Hydrophobic Effect Revisited—Studies with Supramolecular Complexes Imply High-Energy Water as a Noncovalent Driving Force. Angew. Chem. Int. Ed. 2014, 53, 11158–11171. [Google Scholar] [CrossRef]
- Dill, K.A.; Truskett, T.M.; Vlachy, V.; Hribar-Lee, B. Modeling Water, the Hydrophobic Effect, and Ion Solvation. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 173–199. [Google Scholar] [CrossRef]
- Malleshappa Gowder, S.; Chatterjee, J.; Chaudhuri, T.; Paul, K. Prediction and Analysis of Surface Hydrophobic Residues in Tertiary Structure of Proteins. Sci. World J. 2014, 2014, 7. [Google Scholar] [CrossRef]
- Tanford, C. The hydrophobic effect and the organization of living matter. Science 1978, 200, 1012. [Google Scholar] [CrossRef]
- Chandler, D. Interfaces and the driving force of hydrophobic assembly. Nature 2005, 437, 640–647. [Google Scholar] [CrossRef]
- Southall, N.T.; Dill, K.A.; Haymet, A.D.J. A View of the Hydrophobic Effect. J. Phys. Chem. B 2002, 106, 521–533. [Google Scholar] [CrossRef]
- Santoso, S.; Hwang, W.; Hartman, H.; Zhang, S. Self-assembly of Surfactant-like Peptides with Variable Glycine Tails to Form Nanotubes and Nanovesicles. Nano Lett. 2002, 2, 687–691. [Google Scholar] [CrossRef]
- Vauthey, S.; Santoso, S.; Gong, H.; Watson, N.; Zhang, S. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. Proc. Natl. Acad. Sci. USA 2002, 99, 5355. [Google Scholar] [CrossRef] [PubMed]
- Von Maltzahn, G.; Vauthey, S.; Santoso, S.; Zhang, S. Positively Charged Surfactant-like Peptides Self-assemble into Nanostructures. Langmuir 2003, 19, 4332–4337. [Google Scholar] [CrossRef]
- Hubbard, R.E.; Kamran Haider, M. Hydrogen Bonds in Protein: Role ans Strength. Encycl. Life Sci. 2010. [Google Scholar] [CrossRef]
- Pauling, L.; Corey, R.B. Configurations of Polypeptide Chains With Favored Orientations Around Single Bonds: Two New Pleated Sheets. Proc. Natl. Acad. Sci. USA 1951, 37, 729–740. [Google Scholar] [CrossRef]
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins: Two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205. [Google Scholar] [CrossRef]
- Cormier, A.R.; Pang, X.; Zimmerman, M.I.; Zhou, H.-X.; Paravastu, A.K. Molecular Structure of RADA16-I Designer Self-Assembling Peptide Nanofibers. ACS Nano 2013, 7, 7562–7572. [Google Scholar] [CrossRef]
- Zhang, H.; Park, J.; Jiang, Y.; Woodrow, K.A. Rational design of charged peptides that self-assemble into robust nanofibers as immune-functional scaffolds. Acta Biomater. 2017, 55, 183–193. [Google Scholar] [CrossRef]
- Restuccia, A.; Seroski, D.T.; Kelley, K.L.; O’Bryan, C.S.; Kurian, J.J.; Knox, K.R.; Farhadi, S.A.; Angelini, T.E.; Hudalla, G.A. Hierarchical self-assembly and emergent function of densely glycosylated peptide nanofibers. Commun. Chem. 2019, 2, 53. [Google Scholar] [CrossRef]
- Rudra, J.S.; Tian, Y.F.; Jung, J.P.; Collier, J.H. A self-assembling peptide acting as an immune adjuvant. Proc. Natl. Acad. Sci. USA 2010, 107, 622–627. [Google Scholar] [CrossRef]
- Baker, E.G.; Bartlett, G.J.; Crump, M.P.; Sessions, R.B.; Linden, N.; Faul, C.F.J.; Woolfson, D.N. Local and macroscopic electrostatic interactions in single α-helices. Nat. Chem. Biol. 2015, 11, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Zottig, X.; Al-Halifa, S.; Babych, M.; Quittot, N.; Archambault, D.; Bourgault, S. Guiding the Morphology of Amyloid Assemblies by Electrostatic Capping: From Polymorphic Twisted Fibrils to Uniform Nanorods. Small 2019, 15, 1901806. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.R.; Iverson, B.L. Rethinking the term “pi-stacking”. Chem. Sci. 2012, 3, 2191–2201. [Google Scholar] [CrossRef]
- Aida, T.; Meijer, E.W.; Stupp, S.I. Functional Supramolecular Polymers. Science 2012, 335, 813. [Google Scholar] [CrossRef]
- Kar, K.; Ibrar, S.; Nanda, V.; Getz, T.M.; Kunapuli, S.P.; Brodsky, B. Aromatic interactions promote self-association of collagen triple-helical peptides to higher-order structures. Biochemistry 2009, 48, 7959–7968. [Google Scholar] [CrossRef]
- Kar, K.; Wang, Y.-H.; Brodsky, B. Sequence dependence of kinetics and morphology of collagen model peptide self-assembly into higher order structures. Protein Sci. 2008, 17, 1086–1095. [Google Scholar] [CrossRef]
- Gebbie, M.A.; Wei, W.; Schrader, A.M.; Cristiani, T.R.; Dobbs, H.A.; Idso, M.; Chmelka, B.F.; Waite, J.H.; Israelachvili, J.N. Tuning underwater adhesion with cation–π interactions. Nat. Chem. 2017, 9, 473–479. [Google Scholar] [CrossRef]
- Lehn, J.-M. Supramolecular Chemistry—Scope and Perspectives Molecules, Supermolecules, and Molecular Devices (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 1988, 27, 89–112. [Google Scholar] [CrossRef]
- Whitesides, G.M.; Mathias, J.P.; Seto, C.T. Molecular self-assembly and nanochemistry: A chemical strategy for the synthesis of nanostructures. Science 1991, 254, 1312. [Google Scholar] [CrossRef]
- Gordon, D.J.; Meredith, S.C. Probing the Role of Backbone Hydrogen Bonding in β-Amyloid Fibrils with Inhibitor Peptides Containing Ester Bonds at Alternate Positions. Biochemistry 2003, 42, 475–485. [Google Scholar] [CrossRef]
- Hills, R.D., Jr.; Brooks, C.L., 3rd. Hydrophobic cooperativity as a mechanism for amyloid nucleation. J. Mol. Biol. 2007, 368, 894–901. [Google Scholar] [CrossRef]
- Westergard, L.; True, H.L. Extracellular environment modulates the formation and propagation of particular amyloid structures. Mol. Microbiol. 2014, 92, 698–715. [Google Scholar] [CrossRef] [PubMed]
- Turley, C.B.; Rupp, R.E.; Johnson, C.; Taylor, D.N.; Wolfson, J.; Tussey, L.; Kavita, U.; Stanberry, L.; Shaw, A. Safety and immunogenicity of a recombinant M2e-flagellin influenza vaccine (STF2.4xM2e) in healthy adults. Vaccine 2011, 29, 5145–5152. [Google Scholar] [CrossRef] [PubMed]
- Abram, D.; Koffler, H. In vitro formation of flagella-like filaments and other structures from flagellin. J. Mol. Biol. 1964, 9, 168–185. [Google Scholar] [CrossRef]
- Božič, S.; Doles, T.; Gradišar, H.; Jerala, R. New designed protein assemblies. Curr. Opin. Chem. Biol. 2013, 17, 940–945. [Google Scholar] [CrossRef]
- King, N.P.; Lai, Y.-T. Practical approaches to designing novel protein assemblies. Curr. Opin. Struct. Biol. 2013, 23, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Walshaw, J.; Woolfson, D.N. SOCKET: A program for identifying and analysing coiled-coil motifs within protein structures11Edited by J. Thornton. J. Mol. Biol. 2001, 307, 1427–1450. [Google Scholar] [CrossRef]
- Woolfson, D.N. Building fibrous biomaterials from α-helical and collagen-like coiled-coil peptides. Pept. Sci. 2010, 94, 118–127. [Google Scholar] [CrossRef]
- Fletcher, N.L.; Lockett, C.V.; Dexter, A.F. A pH-responsive coiled-coil peptide hydrogel. Soft Matter 2011, 7, 10210–10218. [Google Scholar] [CrossRef]
- Yu, Y.B. Coiled-coils: Stability, specificity, and drug delivery potential. Adv. Drug Deliv. Rev. 2002, 54, 1113–1129. [Google Scholar] [CrossRef]
- Pandya, M.J.; Spooner, G.M.; Sunde, M.; Thorpe, J.R.; Rodger, A.; Woolfson, D.N. Sticky-End Assembly of a Designed Peptide Fiber Provides Insight into Protein Fibrillogenesis. Biochemistry 2000, 39, 8728–8734. [Google Scholar] [CrossRef] [PubMed]
- Woolfson, D.N. The Design of Coiled-Coil Structures and Assemblies. In Advances in Protein Chemistry; Academic Press: Cambridge, MA, USA, 2005; Volume 70, pp. 79–112. [Google Scholar]
- Dong, H.; Paramonov, S.E.; Hartgerink, J.D. Self-Assembly of α-Helical Coiled Coil Nanofibers. J. Am. Chem. Soc. 2008, 130, 13691–13695. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Burgess, N.C.; Thomson, A.R.; Woolfson, D.N. Controlling the Assembly of Coiled–Coil Peptide Nanotubes. Angew. Chem. Int. Ed. 2016, 55, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Rudra, J.S.; Tripathi, P.K.; Hildeman, D.A.; Jung, J.P.; Collier, J.H. Immune responses to coiled coil supramolecular biomaterials. Biomaterials 2010, 31, 8475–8483. [Google Scholar] [CrossRef]
- Wu, Y.; Norberg, P.K.; Reap, E.A.; Congdon, K.L.; Fries, C.N.; Kelly, S.H.; Sampson, J.H.; Conticello, V.P.; Collier, J.H. A Supramolecular Vaccine Platform Based on α-Helical Peptide Nanofibers. ACS Biomater. Sci. Eng. 2017, 3, 3128–3132. [Google Scholar] [CrossRef]
- Pohl, G.; Beke, T.; Csizmadia, I.G.; Perczel, A. Extended Apolar β-Peptide Foldamers: The Role of Axis Chirality on β-Peptide Sheet Stability. J. Phys. Chem. B 2010, 114, 9338–9348. [Google Scholar] [CrossRef]
- Eskandari, S.; Guerin, T.; Toth, I.; Stephenson, R.J. Recent advances in self-assembled peptides: Implications for targeted drug delivery and vaccine engineering. Adv. Drug Deliv. Rev. 2017, 110–111, 169–187. [Google Scholar] [CrossRef]
- Zelzer, M.; Ulijn, R.V. Next-generation peptide nanomaterials: Molecular networks, interfaces and supramolecular functionality. Chem. Soc. Rev. 2010, 39, 3351–3357. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Debelouchina, G.T.; Bayro, M.J.; Clare, D.K.; Caporini, M.A.; Bajaj, V.S.; Jaroniec, C.P.; Wang, L.; Ladizhansky, V.; Müller, S.A.; et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc. Natl. Acad. Sci. USA 2013, 110, 5468. [Google Scholar] [CrossRef]
- Jahn, T.R.; Makin, O.S.; Morris, K.L.; Marshall, K.E.; Tian, P.; Sikorski, P.; Serpell, L.C. The Common Architecture of Cross-β Amyloid. J. Mol. Biol. 2010, 395, 717–727. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Andraka, N.; De Carufel, C.A.; Bourgault, S. Mechanistic Contributions of Biological Cofactors in Islet Amyloid Polypeptide Amyloidogenesis. J. Diabetes Res. 2015, 2015, 515307. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D.; Riek, R. Functional Amyloids. Cold Spring Harb. Perspect. Biol. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.; Mezzenga, R. Amyloid Fibrils as Building Blocks for Natural and Artificial Functional Materials. Adv. Mater 2016, 28, 6546–6561. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.; Estrada, L.D.; Diaz-Espinoza, R.; Morales-Scheihing, D.; Jara, M.C.; Castilla, J.; Soto, C. Molecular cross talk between misfolded proteins in animal models of Alzheimer’s and prion diseases. J. Neurosci. 2010, 30, 4528–4535. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.; Moreno-Gonzalez, I.; Soto, C. Cross-seeding of misfolded proteins: Implications for etiology and pathogenesis of protein misfolding diseases. PLoS Pathog. 2013, 9, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Nussinov, R. Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J. Mol. Biol. 2012, 421, 172–184. [Google Scholar] [CrossRef]
- Babych, M.; Bertheau-Mailhot, G.; Zottig, X.; Dion, J.; Gauthier, L.; Archambault, D.; Bourgault, S. Engineering and evaluation of amyloid assemblies as a nanovaccine against the Chikungunya virus. Nanoscale 2018, 10, 19547–19556. [Google Scholar] [CrossRef]
- Matsuura, K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem. Commun. 2018, 54, 8944–8959. [Google Scholar] [CrossRef]
- Matsuura, K.; Watanabe, K.; Matsuzaki, T.; Sakurai, K.; Kimizuka, N. Self-Assembled Synthetic Viral Capsids from a 24-mer Viral Peptide Fragment. Angew. Chem. Int. Ed. 2010, 49, 9662–9665. [Google Scholar] [CrossRef]
- Matsuura, K.; Murasato, K.; Kimizuka, N. Artificial Peptide-Nanospheres Self-Assembled from Three-Way Junctions of β-Sheet-Forming Peptides. J. Am. Chem. Soc. 2005, 127, 10148–10149. [Google Scholar] [CrossRef] [PubMed]
- Hervé, P.-L.; Raliou, M.; Bourdieu, C.; Dubuquoy, C.; Petit-Camurdan, A.; Bertho, N.; Eléouët, J.-F.; Chevalier, C.; Riffault, S. A novel subnucleocapsid nanoplatform for mucosal vaccination against influenza virus that targets the ectodomain of matrix protein 2. J. Virol. 2014, 88, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Brune, K.D.; Buldun, C.M.; Li, Y.; Taylor, I.J.; Brod, F.; Biswas, S.; Howarth, M. Dual Plug-and-Display Synthetic Assembly Using Orthogonal Reactive Proteins for Twin Antigen Immunization. Bioconjugate Chem. 2017, 28, 1544–1551. [Google Scholar] [CrossRef] [PubMed]
- Kaba, S.A.; Brando, C.; Guo, Q.; Mittelholzer, C.; Raman, S.; Tropel, D.; Aebi, U.; Burkhard, P.; Lanar, D.E. A nonadjuvanted polypeptide nanoparticle vaccine confers long-lasting protection against rodent malaria. J. Immunol. 2009, 183, 7268–7277. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Bu, W.; Joyce, M.G.; Meng, G.; Whittle, J.R.R.; Baxa, U.; Yamamoto, T.; Narpala, S.; Todd, J.-P.; Rao, S.S.; et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 2015, 162, 1090–1100. [Google Scholar] [CrossRef]
- Jardine, J.; Julien, J.-P.; Menis, S.; Ota, T.; Kalyuzhniy, O.; McGuire, A.; Sok, D.; Huang, P.-S.; MacPherson, S.; Jones, M.; et al. Rational HIV Immunogen Design to Target Specific Germline B Cell Receptors. Science 2013, 340, 711. [Google Scholar] [CrossRef]
- Jardine, J.G.; Ota, T.; Sok, D.; Pauthner, M.; Kulp, D.W.; Kalyuzhniy, O.; Skog, P.D.; Thinnes, T.C.; Bhullar, D.; Briney, B.; et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 2015, 349, 156–161. [Google Scholar] [CrossRef]
- Molino, N.M.; Neek, M.; Tucker, J.A.; Nelson, E.L.; Wang, S.-W. Viral-mimicking protein nanoparticle vaccine for eliciting anti-tumor responses. Biomaterials 2016, 86, 83–91. [Google Scholar] [CrossRef]
- Rudra, J.S.; Mishra, S.; Chong, A.S.; Mitchell, R.A.; Nardin, E.H.; Nussenzweig, V.; Collier, J.H. Self-assembled peptide nanofibers raising durable antibody responses against a malaria epitope. Biomaterials 2012, 33, 6476–6484. [Google Scholar] [CrossRef]
- Bennett, K.M.; Gorham, R.D., Jr.; Gusti, V.; Trinh, L.; Morikis, D.; Lo, D.D. Hybrid flagellin as a T cell independent vaccine scaffold. BMC Biotechnol. 2015, 15, 71. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Olson, A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 105–153. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.A.; Teichmann, S.A. Structure, Dynamics, Assembly, and Evolution of Protein Complexes. Annu. Rev. Biochem. 2015, 84, 551–575. [Google Scholar] [CrossRef] [PubMed]
- Padilla, J.E.; Colovos, C.; Yeates, T.O. Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl. Acad. Sci. USA 2001, 98, 2217. [Google Scholar] [CrossRef] [PubMed]
- Yeates, T.O. Geometric Principles for Designing Highly Symmetric Self-Assembling Protein Nanomaterials. Annu. Rev. Biophys. 2017, 46, 23–42. [Google Scholar] [CrossRef]
- Yeates, T.O.; Liu, Y.; Laniado, J. The design of symmetric protein nanomaterials comes of age in theory and practice. Curr. Opin. Struct. Biol. 2016, 39, 134–143. [Google Scholar] [CrossRef]
- Indelicato, G.; Burkhard, P.; Twarock, R. Classification of self-assembling protein nanoparticle architectures for applications in vaccine design. R. Soc. Open Sci. 2017, 4, 161092. [Google Scholar] [CrossRef]
- King, N.P.; Sheffler, W.; Sawaya, M.R.; Vollmar, B.S.; Sumida, J.P.; Andre, I.; Gonen, T.; Yeates, T.O.; Baker, D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 2012, 336, 1171–1174. [Google Scholar] [CrossRef]
- Audette, G.F.; Yaseen, A.; Bragagnolo, N.; Bawa, R. Protein Nanotubes: From Bionanotech towards Medical Applications. Biomedicines 2019, 7, 46. [Google Scholar] [CrossRef]
- Miranda, F.F.; Iwasaki, K.; Akashi, S.; Sumitomo, K.; Kobayashi, M.; Yamashita, I.; Tame, J.R.H.; Heddle, J.G. A Self-Assembled Protein Nanotube with High Aspect Ratio. Small 2009, 5, 2077–2084. [Google Scholar] [CrossRef]
- Miller, R.A.; Presley, A.D.; Francis, M.B. Self-Assembling Light-Harvesting Systems from Synthetically Modified Tobacco Mosaic Virus Coat Proteins. J. Am. Chem. Soc. 2007, 129, 3104–3109. [Google Scholar] [CrossRef]
- Maki-Yonekura, S.; Yonekura, K.; Namba, K. Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat. Struct. Mol. Biol. 2010, 17, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.-M.C.; Buldun, C.M.; Pattinson, D.J.; Draper, S.J.; Howarth, M. SnoopLigase peptide-peptide conjugation enables modular vaccine assembly. Sci. Rep. 2019, 9, 4625. [Google Scholar] [CrossRef] [PubMed]
- Tawar, R.G.; Duquerroy, S.; Vonrhein, C.; Varela, P.F.; Damier-Piolle, L.; Castagné, N.; MacLellan, K.; Bedouelle, H.; Bricogne, G.; Bhella, D.; et al. Crystal Structure of a Nucleocapsid-Like Nucleoprotein-RNA Complex of Respiratory Syncytial Virus. Science 2009, 326, 1279. [Google Scholar] [CrossRef] [PubMed]
- El Omari, K.; Dhaliwal, B.; Ren, J.; Abrescia, N.G.A.; Lockyer, M.; Powell, K.L.; Hawkins, A.R.; Stammers, D.K. Structures of respiratory syncytial virus nucleocapsid protein from two crystal forms: Details of potential packing interactions in the native helical form. Acta Cryst. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1179–1183. [Google Scholar] [CrossRef]
- Bakker, S.E.; Duquerroy, S.; Galloux, M.; Loney, C.; Conner, E.; Eléouët, J.-F.; Rey, F.A.; Bhella, D. The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J. Gen. Virol. 2013, 94, 1734–1738. [Google Scholar] [CrossRef]
- Klug, A. The tobacco mosaic virus particle: Structure and assembly. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 531–535. [Google Scholar] [CrossRef]
- Beniac, D.R.; Melito, P.L.; Devarennes, S.L.; Hiebert, S.L.; Rabb, M.J.; Lamboo, L.L.; Jones, S.M.; Booth, T.F. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS ONE 2012, 7, e29608. [Google Scholar] [CrossRef]
- Ogun, S.A.; Dumon-Seignovert, L.; Marchand, J.-B.; Holder, A.A.; Hill, F. The oligomerization domain of C4-binding protein (C4bp) acts as an adjuvant, and the fusion protein comprised of the 19-kilodalton merozoite surface protein 1 fused with the murine C4bp domain protects mice against malaria. Infect. Immun. 2008, 76, 3817–3823. [Google Scholar] [CrossRef]
- Kask, L.; Hillarp, A.; Ramesh, B.; Dahlbäck, B.; Blom, A.M. Structural Requirements for the Intracellular Subunit Polymerization of the Complement Inhibitor C4b-Binding Protein. Biochemistry 2002, 41, 9349–9357. [Google Scholar] [CrossRef]
- Blom, A.M.; Villoutreix, B.O.; Dahlbäck, B. Complement inhibitor C4b-binding protein—friend or foe in the innate immune system? Mol. Immunol. 2004, 40, 1333–1346. [Google Scholar] [CrossRef]
- Raman, S.; Machaidze, G.; Lustig, A.; Aebi, U.; Burkhard, P. Structure-based design of peptides that self-assemble into regular polyhedral nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2006, 2, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Sanner, M.F.; Stolz, M.; Burkhard, P.; Kong, X.-P.; Min, G.; Sun, T.-T.; Driamov, S.; Aebi, U.; Stoffler, D. Visualizing nature at work from the nano to the macro scale. NanoBiotechnology 2005, 1, 7–21. [Google Scholar] [CrossRef]
- López-Sagaseta, J.; Malito, E.; Rappuoli, R.; Bottomley, M.J. Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 2016, 14, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Ra, J.-S.; Shin, H.-H.; Kang, S.; Do, Y. Lumazine synthase protein cage nanoparticles as antigen delivery nanoplatforms for dendritic cell-based vaccine development. Clin. Exp. Vaccine Res. 2014, 3, 227–234. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Wei, C.-J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.R.; Rao, S.S.; Kong, W.-P.; Wang, L.; Nabel, G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef]
- Zhang, Y.; Orner, B.P. Self-assembly in the ferritin nano-cage protein superfamily. Int. J. Mol. Sci. 2011, 12, 5406–5421. [Google Scholar] [CrossRef]
- Molino, N.M.; Anderson, A.K.L.; Nelson, E.L.; Wang, S.-W. Biomimetic protein nanoparticles facilitate enhanced dendritic cell activation and cross-presentation. ACS Nano 2013, 7, 9743–9752. [Google Scholar] [CrossRef]
- Nemerow, G.R.; Stewart, P.L.; Reddy, V.S. Structure of human adenovirus. Curr. Opin. Virol. 2012, 2, 115–121. [Google Scholar] [CrossRef]
- Brown, J.C.; Newcomb, W.W. Herpesvirus capsid assembly: Insights from structural analysis. Curr. Opin. Virol. 2011, 1, 142–149. [Google Scholar] [CrossRef]
- Tükel, Ç.; Nishimori, J.H.; Wilson, R.P.; Winter, M.G.; Keestra, A.M.; Van Putten, J.P.M.; Bäumler, A.J. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell. Microbiol. 2010, 12, 1495–1505. [Google Scholar] [CrossRef]
- Kolappan, S.; Coureuil, M.; Yu, X.; Nassif, X.; Egelman, E.H.; Craig, L. Structure of the Neisseria meningitidis Type IV pilus. Nat. Commun. 2016, 7, 13015. [Google Scholar] [CrossRef] [PubMed]
- Chesson, C.B.; Huelsmann, E.J.; Lacek, A.T.; Kohlhapp, F.J.; Webb, M.F.; Nabatiyan, A.; Zloza, A.; Rudra, J.S. Antigenic peptide nanofibers elicit adjuvant-free CD8+ T cell responses. Vaccine 2014, 32, 1174–1180. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-H.; Shi, L.; Ma, J.-W.; Sun, Z.-Y.; Cai, H.; Chen, Y.-X.; Zhao, Y.-F.; Li, Y.-M. a Totally Synthetic, Self-Assembling, Adjuvant-Free MUC1 Glycopeptide Vaccine for Cancer Therapy. J. Am. Chem. Soc. 2012, 134, 8730–8733. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-T.; Lin, Y.; Spencer, R.K.; Thomas, M.R.; Nguyen, A.I.; Amdursky, N.; Pashuck, E.T.; Skaalure, S.C.; Song, C.Y.; Parmar, P.A.; et al. Sequence-Dependent Self-Assembly and Structural Diversity of Islet Amyloid Polypeptide-Derived β-Sheet Fibrils. ACS Nano 2017, 11, 8579–8589. [Google Scholar] [CrossRef] [PubMed]
- Azmi, F.; Ahmad Fuaad, A.A.H.; Giddam, A.K.; Batzloff, M.R.; Good, M.F.; Skwarczynski, M.; Toth, I. Self-adjuvanting vaccine against group A streptococcus: Application of fibrillized peptide and immunostimulatory lipid as adjuvant. Bioorganic Med. Chem. 2014, 22, 6401–6408. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Burrage, A.M.; Postel, S.; Clark, R.E.; Orlova, A.; Sundberg, E.J.; Kearns, D.B.; Egelman, E.H. A structural model of flagellar filament switching across multiple bacterial species. Nat. Commun. 2017, 8, 960. [Google Scholar] [CrossRef] [PubMed]
- Huleatt, J.W.; Nakaar, V.; Desai, P.; Huang, Y.; Hewitt, D.; Jacobs, A.; Tang, J.; McDonald, W.; Song, L.; Evans, R.K.; et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 2008, 26, 201–214. [Google Scholar] [CrossRef]
- Lockner, J.W.; Eubanks, L.M.; Choi, J.L.; Lively, J.M.; Schlosburg, J.E.; Collins, K.C.; Globisch, D.; Rosenfeld-Gunn, R.J.; Wilson, I.A.; Janda, K.D. Flagellin as carrier and adjuvant in cocaine vaccine development. Mol. Pharm. 2015, 12, 653–662. [Google Scholar] [CrossRef]
- Stepanova, L.A.; Mardanova, E.S.; Shuklina, M.A.; Blokhina, E.A.; Kotlyarov, R.Y.; Potapchuk, M.V.; Kovaleva, A.A.; Vidyaeva, I.G.; Korotkov, A.V.; Eletskaya, E.I.; et al. Flagellin-fused protein targeting M2e and HA2 induces potent humoral and T-cell responses and protects mice against various influenza viruses a subtypes. J. Biomed. Sci. 2018, 25, 33. [Google Scholar] [CrossRef]
- Al-Halifa, S.; Babych, M.; Zottig, X.; Archambault, D.; Bourgault, S. Amyloid self-assembling peptides: Potential applications in nanovaccine engineering and biosensing. Pept. Sci. 2019, 111, e24095. [Google Scholar] [CrossRef]
- Walker, A.; Skamel, C.; Nassal, M. SplitCore: An exceptionally versatile viral nanoparticle for native whole protein display regardless of 3D structure. Sci. Rep. 2011, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Brune, K.D.; Howarth, M. New Routes and Opportunities for Modular Construction of Particulate Vaccines: Stick, Click, and Glue. Front. Immunol. 2018, 9, 1432. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, P.; Moise, L.; Luza, C.; Chantaralawan, K.; Lezeau, L.; Yuan, J.; Field, M.; Richer, D.; Boyle, C.; Martin, W.D.; et al. VaxCelerate II: Rapid development of a self-assembling vaccine for Lassa fever. Hum. Vaccin. Immunother. 2014, 10, 3022–3038. [Google Scholar] [CrossRef] [PubMed]
- Koho, T.; Ihalainen, T.O.; Stark, M.; Uusi-Kerttula, H.; Wieneke, R.; Rahikainen, R.; Blazevic, V.; Marjomäki, V.; Tampé, R.; Kulomaa, M.S.; et al. His-tagged norovirus-like particles: A versatile platform for cellular delivery and surface display. Eur. J. Pharm. Biopharm. 2015, 96, 22–31. [Google Scholar] [CrossRef]
- Le, D.H.T.; Commandeur, U.; Steinmetz, N.F. Presentation and Delivery of Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand via Elongated Plant Viral Nanoparticle Enhances Antitumor Efficacy. ACS Nano 2019, 13, 2501–2510. [Google Scholar] [CrossRef]
- Hainfeld, J.F.; Liu, W.; Halsey, C.M.R.; Freimuth, P.; Powell, R.D. Ni–NTA–Gold Clusters Target His-Tagged Proteins. J. Struct. Biol. 1999, 127, 185–198. [Google Scholar] [CrossRef]
- Thrane, S.; Janitzek, C.M.; Agerbæk, M.Ø.; Ditlev, S.B.; Resende, M.; Nielsen, M.A.; Theander, T.G.; Salanti, A.; Sander, A.F. A Novel Virus-Like Particle Based Vaccine Platform Displaying the Placental Malaria Antigen VAR2CSA. PLoS ONE 2015, 10, e0143071. [Google Scholar] [CrossRef]
- Chackerian, B.; Lowy, D.R.; Schiller, J.T. Conjugation of a self-antigen to papillomavirus-like particles allows for efficient induction of protective autoantibodies. J. Clin. Invest. 2001, 108, 415–423. [Google Scholar] [CrossRef]
- Miller, K.D.; Roque, R.; Clegg, C.H. Novel Anti-Nicotine Vaccine Using a Trimeric Coiled-Coil Hapten Carrier. PLoS ONE 2014, 9, e114366. [Google Scholar] [CrossRef]
- Prakasam, T.; Matosiuk, D. Click Chemistry for Drug Development and Diverse Chemical-Biology Applications. Chem. Rev. 2013, 113. [Google Scholar] [CrossRef]
- Patel, K.G.; Swartz, J.R. Surface Functionalization of Virus-Like Particles by Direct Conjugation Using Azide−Alkyne Click Chemistry. Bioconjugate Chem. 2011, 22, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, D.C.; Lee, J.J.; Link, A.J.; Graumann, J.; Tirrell, D.A.; Schuman, E.M. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2007, 2, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Xuan, B.; Ye, X.; Huang, Z.; Qian, Z. A Modular Vaccine Development Platform Based on Sortase-Mediated Site-Specific Tagging of Antigens onto Virus-Like Particles. Sci. Rep. 2016, 6, 25741. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, Q.; Molino, N.M.; Wang, S.-W.; Boder, E.T.; Chen, W. Sortase A-mediated multi-functionalization of protein nanoparticles. Chem. Commun. 2015, 51, 12107–12110. [Google Scholar] [CrossRef]
- Sun, Q.; Chen, Q.; Blackstock, D.; Chen, W. Post-Translational Modification of Bionanoparticles as a Modular Platform for Biosensor Assembly. ACS Nano 2015, 9, 8554–8561. [Google Scholar] [CrossRef]
- Reddington, S.C.; Howarth, M. Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher. Curr. Opin. Chem. Biol. 2015, 29, 94–99. [Google Scholar] [CrossRef]
- Kang, H.J.; Coulibaly, F.; Clow, F.; Proft, T.; Baker, E.N. Stabilizing Isopeptide Bonds Revealed in Gram-Positive Bacterial Pilus Structure. Science 2007, 318, 1625–1628. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef]
- Li, L.; Fierer, J.O.; Rapoport, T.A.; Howarth, M. Structural analysis and optimization of the covalent association between SpyCatcher and a peptide Tag. J. Mol. Biol. 2014, 426, 309–317. [Google Scholar] [CrossRef]
- Brune, K.D.; Leneghan, D.B.; Brian, I.J.; Ishizuka, A.S.; Bachmann, M.F.; Draper, S.J.; Biswas, S.; Howarth, M. Plug-and-Display: Decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234. [Google Scholar] [CrossRef]
- El Bissati, K.; Zhou, Y.; Dasgupta, D.; Cobb, D.; Dubey, J.P.; Burkhard, P.; Lanar, D.E.; McLeod, R. Effectiveness of a novel immunogenic nanoparticle platform for Toxoplasma peptide vaccine in HLA transgenic mice. Vaccine 2014, 32, 3243–3248. [Google Scholar] [CrossRef] [PubMed]
- Kaba, S.A.; McCoy, M.E.; Doll, T.A.P.F.; Brando, C.; Guo, Q.; Dasgupta, D.; Yang, Y.; Mittelholzer, C.; Spaccapelo, R.; Crisanti, A.; et al. Protective Antibody and CD8+ T-Cell Responses to the Plasmodium falciparum Circumsporozoite Protein Induced by a Nanoparticle Vaccine. PLoS ONE 2012, 7, e48304. [Google Scholar] [CrossRef] [PubMed]
- Pompano, R.R.; Chen, J.; Verbus, E.A.; Han, H.; Fridman, A.; McNeely, T.; Collier, J.H.; Chong, A.S. Titrating T-cell epitopes within self-assembled vaccines optimizes CD4+ helper T cell and antibody outputs. Adv. Healthc. Mater. 2014, 3, 1898–1908. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, L.A.; Maciaszek, J.W.; Rock, K.L. Both dendritic cells and macrophages can stimulate naive CD8 T cells in vivo to proliferate, develop effector function, and differentiate into memory cells. J. Immunol. 2005, 175, 2071–2081. [Google Scholar] [CrossRef] [PubMed]
- Blander, J.M.; Medzhitov, R. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature 2006, 440, 808–812. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef]
- Pearce, E.J.; Everts, B. Dendritic cell metabolism. Nat. Rev. Immunol. 2015, 15, 18–29. [Google Scholar] [CrossRef]
- Reddy, S.T.; van der Vlies, A.J.; Simeoni, E.; Angeli, V.; Randolph, G.J.; O’Neil, C.P.; Lee, L.K.; Swartz, M.A.; Hubbell, J.A. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat. Biotechnol. 2007, 25, 1159–1164. [Google Scholar] [CrossRef]
- Joffre, O.P.; Segura, E.; Savina, A.; Amigorena, S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012, 12, 557–569. [Google Scholar] [CrossRef]
- Sallusto, F.; Lanzavecchia, A. Heterogeneity of CD4+ memory T cells: Functional modules for tailored immunity. Eur. J. Immunol. 2009, 39, 2076–2082. [Google Scholar] [CrossRef]
- Bonilla, F.A.; Oettgen, H.C. Adaptive immunity. J. Allergy Clin. Immunol. 2010, 125, S33–S40. [Google Scholar] [CrossRef] [PubMed]
- Mottram, P.L.; Leong, D.; Crimeen-Irwin, B.; Gloster, S.; Xiang, S.D.; Meanger, J.; Ghildyal, R.; Vardaxis, N.; Plebanski, M. Type 1 and 2 immunity following vaccination is influenced by nanoparticle size: Formulation of a model vaccine for respiratory syncytial virus. Mol. Pharm. 2007, 4, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Toy, R.; Roy, K. Engineering nanoparticles to overcome barriers to immunotherapy. Bioeng. Transl. Med. 2016, 1, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Zinkernagel, R.M. On natural and artificial vaccinations. Annu. Rev. Immunol. 2003, 21, 515–546. [Google Scholar] [CrossRef]
- Gamvrellis, A.; Leong, D.; Hanley, J.C.; Xiang, S.D.; Mottram, P.; Plebanski, M. Vaccines that facilitate antigen entry into dendritic cells. Immunol. Cell Biol. 2004, 82, 506–516. [Google Scholar] [CrossRef]
- Kim, H.; Uto, T.; Akagi, T.; Baba, M.; Akashi, M. Amphiphilic Poly(Amino Acid) Nanoparticles Induce Size-Dependent Dendritic Cell Maturation. Adv. Funct. Mater. 2010, 20, 3925–3931. [Google Scholar] [CrossRef]
- Shima, F.; Uto, T.; Akagi, T.; Baba, M.; Akashi, M. Size effect of amphiphilic poly(gamma-glutamic acid) nanoparticles on cellular uptake and maturation of dendritic cells in vivo. Acta Biomater. 2013, 9, 8894–8901. [Google Scholar] [CrossRef]
- Sallusto, F.; Cella, M.; Danieli, C.; Lanzavecchia, A. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: Downregulation by cytokines and bacterial products. J. Exp. Med. 1995, 182, 389–400. [Google Scholar] [CrossRef]
- Pacheco, P.; White, D.; Sulchek, T. Effects of microparticle size and Fc density on macrophage phagocytosis. PLoS ONE 2013, 8, e60989. [Google Scholar] [CrossRef]
- Mond, J.J.; Vos, Q.; Lees, A.; Snapper, C.M. T cell independent antigens. Curr. Opin. Immunol. 1995, 7, 349–354. [Google Scholar] [CrossRef]
- Werling, D.; Hope, J.C.; Chaplin, P.; Collins, R.A.; Taylor, G.; Howard, C.J. Involvement of caveolae in the uptake of respiratory syncytial virus antigen by dendritic cells. J. Leukoc. Biol. 1999, 66, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Correia, I.; Eulalio, A.; Nir, S.; Pedroso de Lima, M.C. Caveolae as an additional route for influenza virus endocytosis in MDCK cells. Cell. Mol. Biol. Lett. 2004, 9, 47–60. [Google Scholar] [PubMed]
- Fifis, T.; Gamvrellis, A.; Crimeen-Irwin, B.; Pietersz, G.A.; Li, J.; Mottram, P.L.; McKenzie, I.F.; Plebanski, M. Size-dependent immunogenicity: Therapeutic and protective properties of nano-vaccines against tumors. J. Immunol. 2004, 173, 3148–3154. [Google Scholar] [CrossRef] [PubMed]
- Slutter, B.; Jiskoot, W. Sizing the optimal dimensions of a vaccine delivery system: A particulate matter. Expert Opin. Drug Deliv. 2016, 13, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Kanchan, V.; Panda, A.K. Interactions of antigen-loaded polylactide particles with macrophages and their correlation with the immune response. Biomaterials 2007, 28, 5344–5357. [Google Scholar] [CrossRef] [PubMed]
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef]
- Vos, Q.; Lees, A.; Wu, Z.Q.; Snapper, C.M.; Mond, J.J. B-cell activation by T-cell-independent type 2 antigens as an integral part of the humoral immune response to pathogenic microorganisms. Immunol. Rev. 2000, 176, 154–170. [Google Scholar] [CrossRef]
- Gatto, D.; Martin, S.W.; Bessa, J.; Pellicioli, E.; Saudan, P.; Hinton, H.J.; Bachmann, M.F. Regulation of memory antibody levels: The role of persisting antigen versus plasma cell life span. J. Immunol. 2007, 178, 67–76. [Google Scholar] [CrossRef]
- Fang, Y.; Xu, C.; Fu, Y.X.; Holers, V.M.; Molina, H. Expression of complement receptors 1 and 2 on follicular dendritic cells is necessary for the generation of a strong antigen-specific IgG response. J. Immunol. 1998, 160, 5273–5279. [Google Scholar]
- Liu, W.; Chen, Y.-H. High epitope density in a single protein molecule significantly enhances antigenicity as well as immunogenicity: A novel strategy for modern vaccine development and a preliminary investigation about B cell discrimination of monomeric proteins. Eur. J. Immunol. 2005, 35, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Leon, M.A.; Firdessa-Fite, R.; Ruffalo, J.K.; Pickens, C.J.; Sestak, J.O.; Creusot, R.J.; Berkland, C. Soluble Antigen Arrays Displaying Mimotopes Direct the Response of Diabetogenic T Cells. ACS Chem. Biol. 2019, 14, 1436–1448. [Google Scholar] [CrossRef]
- Irvine, D.J.; Swartz, M.A.; Szeto, G.L. Engineering synthetic vaccines using cues from natural immunity. Nat. Mater. 2013, 12, 978–990. [Google Scholar] [CrossRef]
- Wang, L.; Chang, T.Z.; He, Y.; Kim, J.R.; Wang, S.; Mohan, T.; Berman, Z.; Tompkins, S.M.; Tripp, R.A.; Compans, R.W.; et al. Coated protein nanoclusters from influenza H7N9 HA are highly immunogenic and induce robust protective immunity. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 253–262. [Google Scholar] [CrossRef]
- Wang, L.; Hess, A.; Chang, T.Z.; Wang, Y.C.; Champion, J.A.; Compans, R.W.; Wang, B.Z. Nanoclusters self-assembled from conformation-stabilized influenza M2e as broadly cross-protective influenza vaccines. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, M.; Easten, A. The relationship between antigenic structure and the requirement for thymus-derived cells in the immune response. J. Exp. Med. 1971, 134, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Zinkernagel, R.M. Neutralizing antiviral B cell responses. Annu. Rev. Immunol. 1997, 15, 235–270. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Reddy, V.S.; Brooks Iii, C.L. Invariant Polymorphism in Virus Capsid Assembly. J. Am. Chem. Soc. 2009, 131, 2606–2614. [Google Scholar] [CrossRef]
- Elrad, O.M.; Hagan, M.F. Mechanisms of Size Control and Polymorphism in Viral Capsid Assembly. Nano Lett. 2008, 8, 3850–3857. [Google Scholar] [CrossRef]
- Wen, Y.; Collier, J.H. Supramolecular peptide vaccines: Tuning adaptive immunity. Curr. Opin. Immunol. 2015, 35, 73–79. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; McNeil, S.E. Immunological properties of engineered nanomaterials. Nat. Nanotechnol. 2007, 2, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Waltman, A.; Han, H.; Collier, J.H. Switching the Immunogenicity of Peptide Assemblies Using Surface Properties. ACS Nano 2016, 10, 9274–9286. [Google Scholar] [CrossRef] [PubMed]
- Trabulo, S.; Cardoso, A.L.; Mano, M.; De Lima, M.C. Cell-Penetrating Peptides-Mechanisms of Cellular Uptake and Generation of Delivery Systems. Pharmaceuticals 2010, 3, 961–993. [Google Scholar] [CrossRef] [PubMed]
- Bechara, C.; Sagan, S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef]
- Seong, S.Y.; Matzinger, P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 2004, 4, 469–478. [Google Scholar] [CrossRef]
- Moyano, D.F.; Goldsmith, M.; Solfiell, D.J.; Landesman-Milo, D.; Miranda, O.R.; Peer, D.; Rotello, V.M. Nanoparticle hydrophobicity dictates immune response. J. Am. Chem. Soc. 2012, 134, 3965–3967. [Google Scholar] [CrossRef]
- Wang, D.; Sun, B.; Feng, M.; Feng, H.; Gong, W.; Liu, Q.; Ge, S. Role of scavenger receptors in dendritic cell function. Hum. Immunol. 2015, 76, 442–446. [Google Scholar] [CrossRef]
- Heather, C.D.; Verchere, C.B. IAPP and type 1 diabetes: Implications for immunity, metabolism and islet transplants. J. Mol. Endocrinol. 2018, 60, R57–R75. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Hao, W.; Wolf, L.; Kiliaan, A.J.; Penke, B.; Rübe, C.E.; Walter, J.; Heneka, M.T.; Hartmann, T.; et al. TLR2 Is a Primary Receptor for Alzheimer’s Amyloid β Peptide To Trigger Neuroinflammatory Activation. J. Immunol. 2012, 188, 1098. [Google Scholar] [CrossRef]
- Westwell-Roper, C.; Denroche, H.C.; Ehses, J.A.; Verchere, C.B. Differential Activation of Innate Immune Pathways by Distinct Islet Amyloid Polypeptide (IAPP) Aggregates. J. Biol. Chem. 2016, 291, 8908–8917. [Google Scholar] [CrossRef]
- Davis, B.K.; Wen, H.; Ting, J.P.Y. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011, 29, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sunagar, R.; Gosselin, E. Bacterial Protein Toll-Like-Receptor Agonists: A Novel Perspective on Vaccine Adjuvants. Front. Immunol. 2019, 10, 1144. [Google Scholar] [CrossRef] [PubMed]
- Janabi, M.; Yamashita, S.; Hirano, K.; Sakai, N.; Hiraoka, H.; Matsumoto, K.; Zhang, Z.; Nozaki, S.; Matsuzawa, Y. Oxidized LDL-induced NF-kappa B activation and subsequent expression of proinflammatory genes are defective in monocyte-derived macrophages from CD36-deficient patients. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1953–1960. [Google Scholar] [CrossRef] [PubMed]
- Platt, N.; Gordon, S. Is the class A macrophage scavenger receptor (SR-A) multifunctional? The mouse’s tale. J. Clin. Invest. 2001, 108, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, P.; Hall, J.B.; McLeland, C.B.; Dobrovolskaia, M.A.; McNeil, S.E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Deliv. Rev. 2009, 61, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Colino, J.; Duke, L.; Snapper, C.M. Autologous albumin enhances the humoral immune response to capsular polysaccharide covalently coattached to bacteria-sized latex beads. Eur. J. Immunol. 2014, 44, 1433–1443. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Aggarwal, P.; Hall, J.B.; McNeil, S.E. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol. Pharm. 2008, 5, 487–495. [Google Scholar] [CrossRef]
- Oyewumi, M.O.; Kumar, A.; Cui, Z. Nano-microparticles as immune adjuvants: Correlating particle sizes and the resultant immune responses. Expert Rev. Vaccines 2010, 9, 1095–1107. [Google Scholar] [CrossRef]
- Mallapragada, S.K.; Narasimhan, B. Immunomodulatory biomaterials. Int. J. Pharm. 2008, 364, 265–271. [Google Scholar] [CrossRef]
- Jain, S.; Yap, W.T.; Irvine, D.J. Synthesis of protein-loaded hydrogel particles in an aqueous two-phase system for coincident antigen and CpG oligonucleotide delivery to antigen-presenting cells. Biomacromolecules 2005, 6, 2590–2600. [Google Scholar] [CrossRef]
- Aguilar, J.C.; Rodriguez, E.G. Vaccine adjuvants revisited. Vaccine 2007, 25, 3752–3762. [Google Scholar] [CrossRef] [PubMed]
- Cella, M.; Engering, A.; Pinet, V.; Pieters, J.; Lanzavecchia, A. Inflammatory stimuli induce accumulation of MHC class II complexes on dendritic cells. Nature 1997, 388, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Delamarre, L.; Holcombe, H.; Mellman, I. Presentation of exogenous antigens on major histocompatibility complex (MHC) class I and MHC class II molecules is differentially regulated during dendritic cell maturation. J. Exp. Med. 2003, 198, 111–122. [Google Scholar] [CrossRef] [PubMed]
- West, M.A.; Wallin, R.P.; Matthews, S.P.; Svensson, H.G.; Zaru, R.; Ljunggren, H.G.; Prescott, A.R.; Watts, C. Enhanced dendritic cell antigen capture via toll-like receptor-induced actin remodeling. Science 2004, 305, 1153–1157. [Google Scholar] [CrossRef]
- Fischer, N.O.; Rasley, A.; Corzett, M.; Hwang, M.H.; Hoeprich, P.D.; Blanchette, C.D. Colocalized delivery of adjuvant and antigen using nanolipoprotein particles enhances the immune response to recombinant antigens. J. Am. Chem. Soc. 2013, 135, 2044–2047. [Google Scholar] [CrossRef]
- Tacken, P.J.; Zeelenberg, I.S.; Cruz, L.J.; van Hout-Kuijer, M.A.; van de Glind, G.; Fokkink, R.G.; Lambeck, A.J.; Figdor, C.G. Targeted delivery of TLR ligands to human and mouse dendritic cells strongly enhances adjuvanticity. Blood 2011, 118, 6836–6844. [Google Scholar] [CrossRef]
- Moon, J.J.; Suh, H.; Li, A.V.; Ockenhouse, C.F.; Yadava, A.; Irvine, D.J. Enhancing humoral responses to a malaria antigen with nanoparticle vaccines that expand Tfh cells and promote germinal center induction. Proc. Natl. Acad. Sci. USA 2012, 109, 1080–1085. [Google Scholar] [CrossRef]
- Lee, E.B.; Jeon, H.M.; Kim, C.U.; Park, S.M.; Cho, G.; Kim, H.J.; Kim, Y.; Kim, D.J.; Kim, Y.S.; Lee, H.; et al. Attachment of flagellin enhances the immunostimulatory activity of a hemagglutinin-ferritin nano-cage. Nanomed. Nanotechnol. Biol. Med. 2019, 17, 223–235. [Google Scholar] [CrossRef]



| Candidate | Scaffold | Assembling Motifs | Morphology | Diameter (nm) | Target | Epitope | Linker | Route | Immune Response | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| N-3M2e | Nucleo-protein N | Coiled-like | 10 to 11-mer rings | 15 | Influenza A | (M2e)3 | EL | IN * | ↑ IgG (IgG1 and IgG2a) and IgA titers | [73] |
| Pfs25:IMX: Pfs28 | IMX313 | Coiled-coil | Heptameric spider-like rings | 10 | Malaria P. falciparum | Pfs25 * Pfs28 * | Snoop and Spy Tag/Catcher | IM * | ↑ IgG titers | [74] |
| P4c-Mal | SAP | Coiled-coil | Icosahedral nanoparticle | 25 | Malaria P. berghei | (DPPPPNPN)2D | GSG | IP * | ↑ IgG (IgG1, IgG2a, IgG2) titers 100% PI *, Th1/Th2 response | [75] |
| D123-ferritin | Ferritin | Coiled-coil | Octahedral cage | 20 | EBV * | gp350 | (SG3)2 | IM * | ↑ IgG titers, PI | [76] |
| eOD-GT6 eOD-GT8 | LS * | Coiled-coil | Icosahedral cage | 25–30 | HIV-1 | eOD * of gp120 | GS linker | IP * | ↑ IgG and IgM titers | [77,78] |
| CpG-gp-E2 | E2 | Coiled-coil | Dodecahedral cage | 30 | Cancer melanoma | gp100 CpG | Chemical crosslinkers | SC * | Activation of Ag-specific * TC cells, tumor growth delay | [79] |
| (NANP)3-Q11 | Q11 | Cross-β-sheet | Unbranched fibrils | 15 | Malaria P. falciparum | 3 x NANP from CSP | SGSG | SC * | ↑ IgG (T-cell-mediated) | [80] |
| E2EP3-I10 | I10 | Cross-β-sheet | Unbranched fibrils | 6–8 | Chikungunya virus | E2EP3 | GGGG | SC * | ↑ IgG (IgG1, IgG2a, IgG2b and IgG3) | [69] |
| PEP-C/PAD-C | Coil29 | Coiled-coil | Unbranched fibers | 6–10 | Glyoblastoma | PEPvIII, PADRE | SGSG | IP * | Efficient internalization by APCs, ↑ IgG (IgG1, IgG2a and IgG2b) titers | [56] |
| hFliC | FliC | Coiled-coil | Filaments | 35 | Dengue virus | DENV2 E | (GGGS)2 | IN * IP * | ↑ IgG (IgG1), IgA titers ↑ IgG, IgM titers, TD * and TI * responses | [81] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zottig, X.; Côté-Cyr, M.; Arpin, D.; Archambault, D.; Bourgault, S. Protein Supramolecular Structures: From Self-Assembly to Nanovaccine Design. Nanomaterials 2020, 10, 1008. https://doi.org/10.3390/nano10051008
Zottig X, Côté-Cyr M, Arpin D, Archambault D, Bourgault S. Protein Supramolecular Structures: From Self-Assembly to Nanovaccine Design. Nanomaterials. 2020; 10(5):1008. https://doi.org/10.3390/nano10051008
Chicago/Turabian StyleZottig, Ximena, Mélanie Côté-Cyr, Dominic Arpin, Denis Archambault, and Steve Bourgault. 2020. "Protein Supramolecular Structures: From Self-Assembly to Nanovaccine Design" Nanomaterials 10, no. 5: 1008. https://doi.org/10.3390/nano10051008
APA StyleZottig, X., Côté-Cyr, M., Arpin, D., Archambault, D., & Bourgault, S. (2020). Protein Supramolecular Structures: From Self-Assembly to Nanovaccine Design. Nanomaterials, 10(5), 1008. https://doi.org/10.3390/nano10051008






