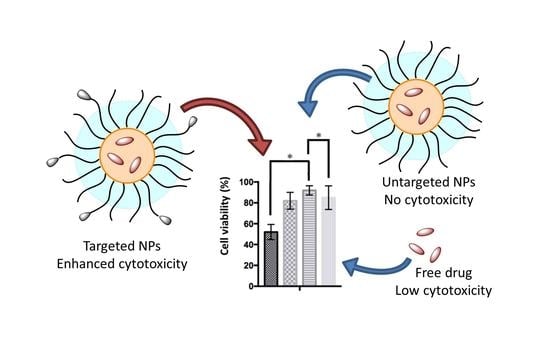
Multifunctional, CD44v6-Targeted ORMOSIL Nanoparticles Enhance Drugs Toxicity in Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. General
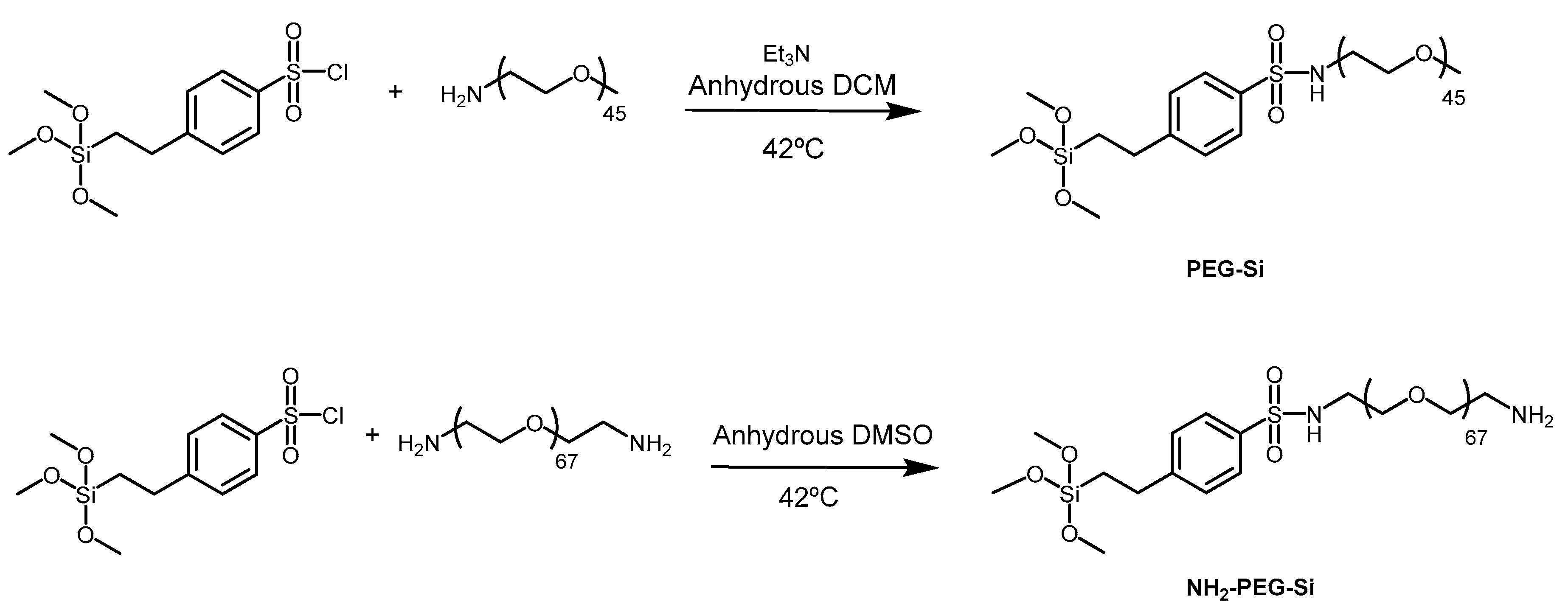
2.1.1. Synthesis of PEG2000-OMe-Si (2)
2.1.2. Synthesis of PEG3000-NH2-Si (3)
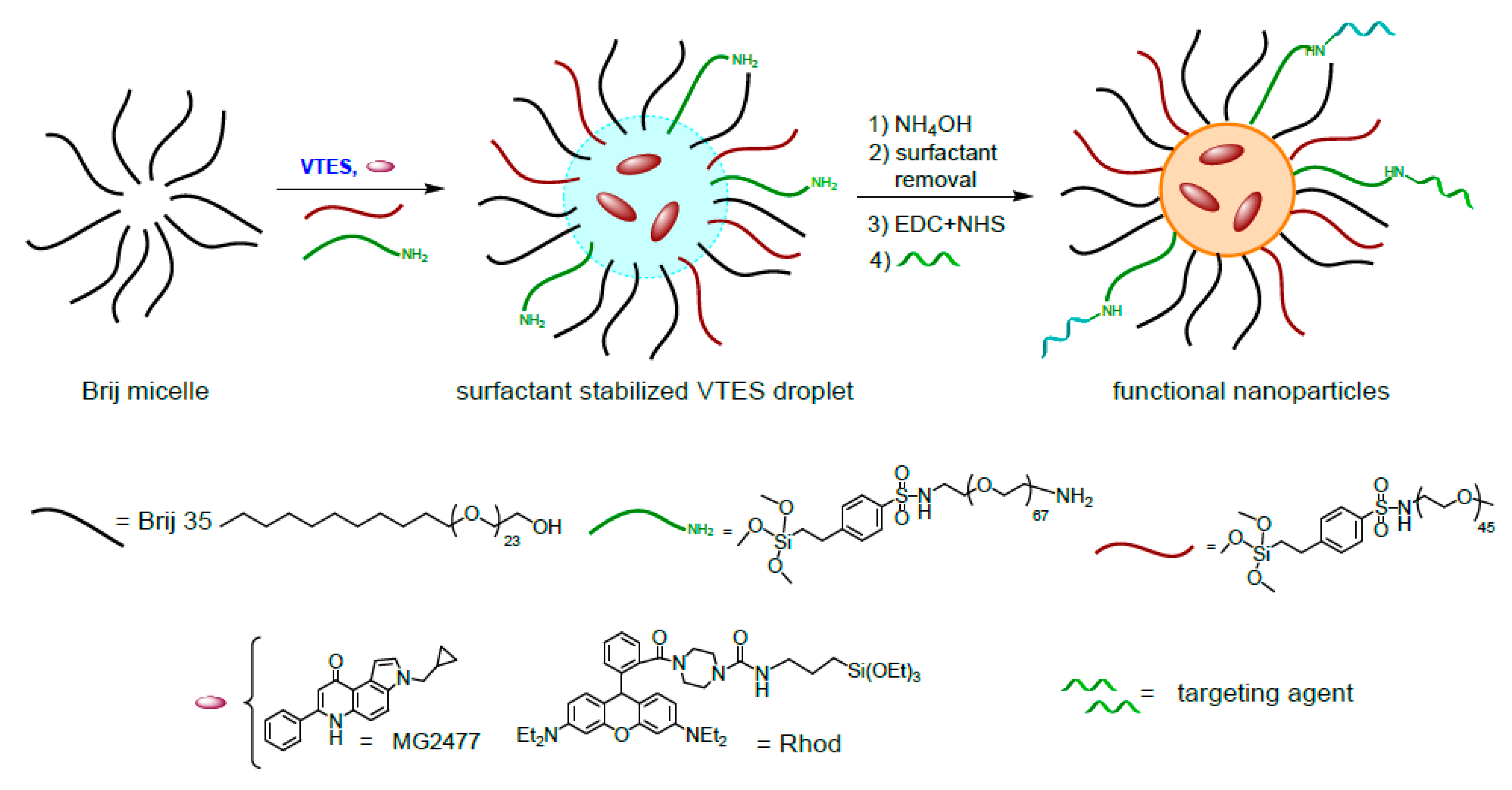
2.1.3. Synthesis and Purification of Rhod-NPs and NH2-Rho-NPs
2.1.4. Synthesis and Purification of MG2477-NPs and NH2-MG2477-NPs
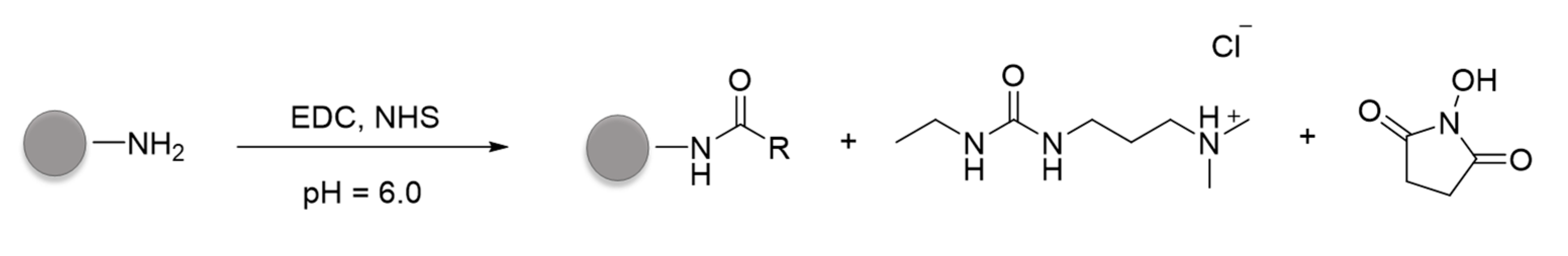
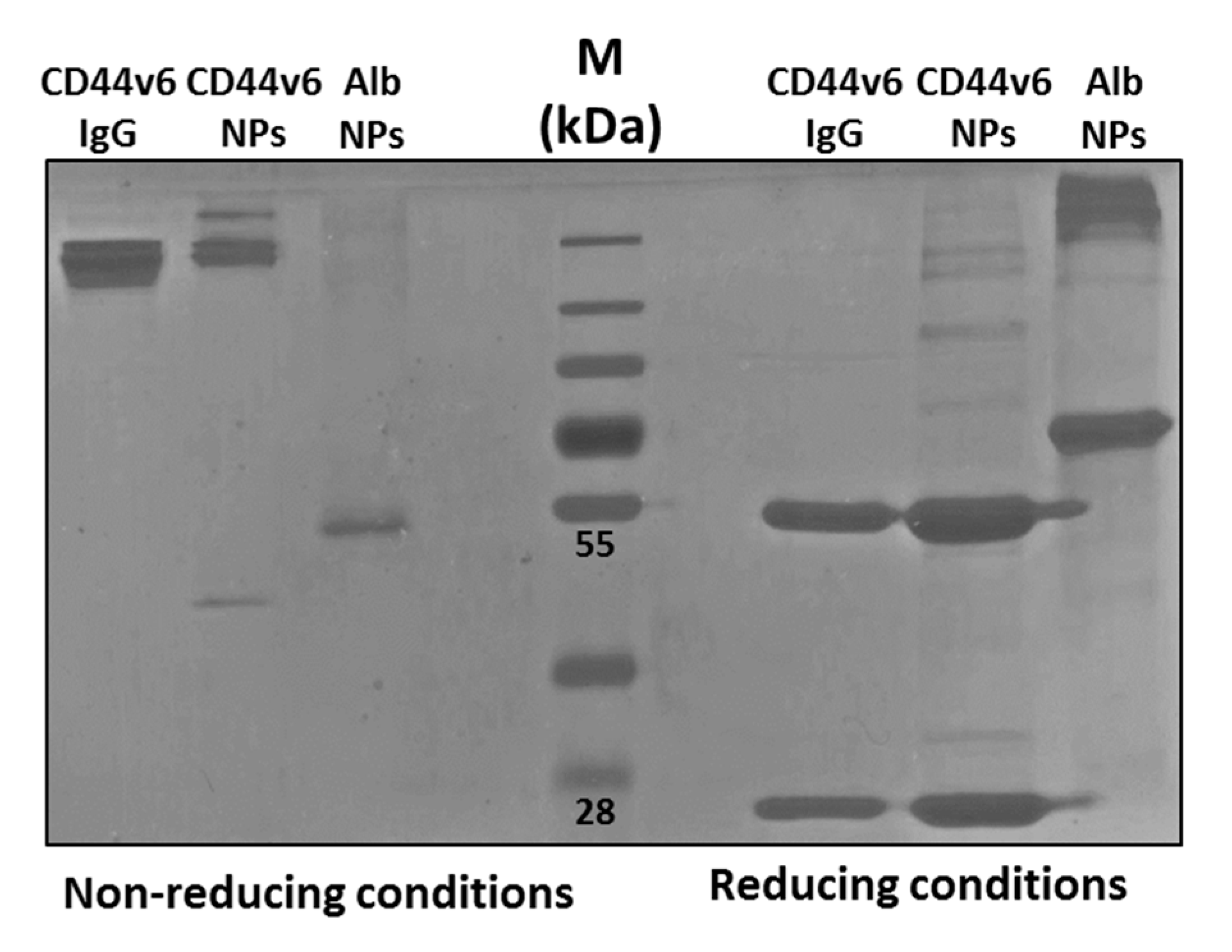
2.1.5. Conjugation of Anti-CD44v6 Antibody to NPs (Ab-CD44v6-NPs)
2.1.6. Conjugation of HA to NPs
2.1.7. Quantification of Antibody Conjugated to Ab-CD44v6-NPs
2.2. Cellular Uptake and Cytotoxicity
2.2.1. Cell Cultures
2.2.2. Production and Purification of Anti-CD44v6 Antibody
2.2.3. Transfection of Cell Cultures
2.2.4. Binding and Internalization Experiments
2.2.5. Confocal Experiments
2.2.6. MTT Cell Viability Assay
2.2.7. Competition Assay
3. Results and Discussion
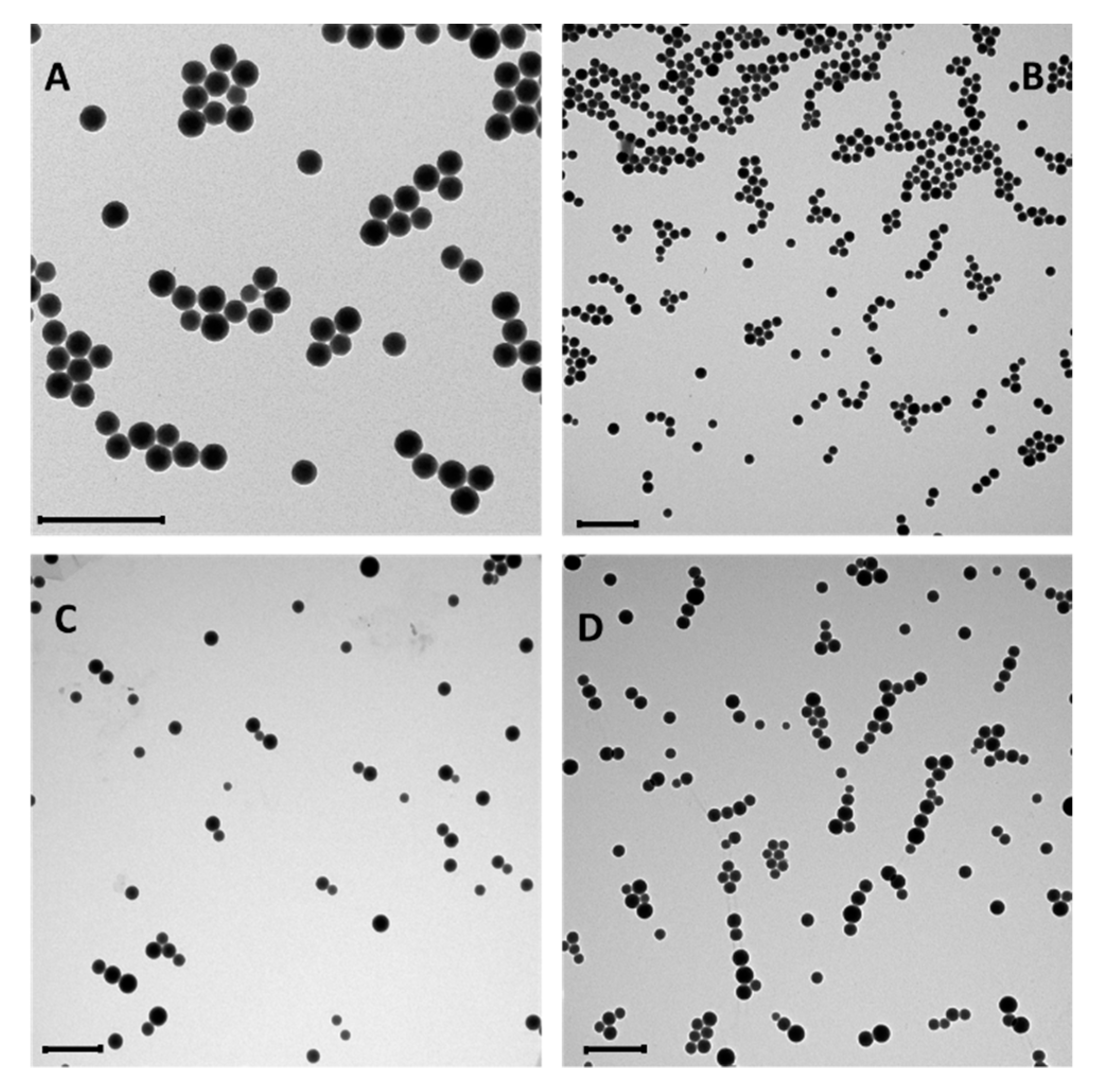
3.1. Synthesis and Conjugation of the Nanoparticles
3.2. Characterization of the Conjugated Nanoparticles
3.3. Nanoparticles Interaction with Cells
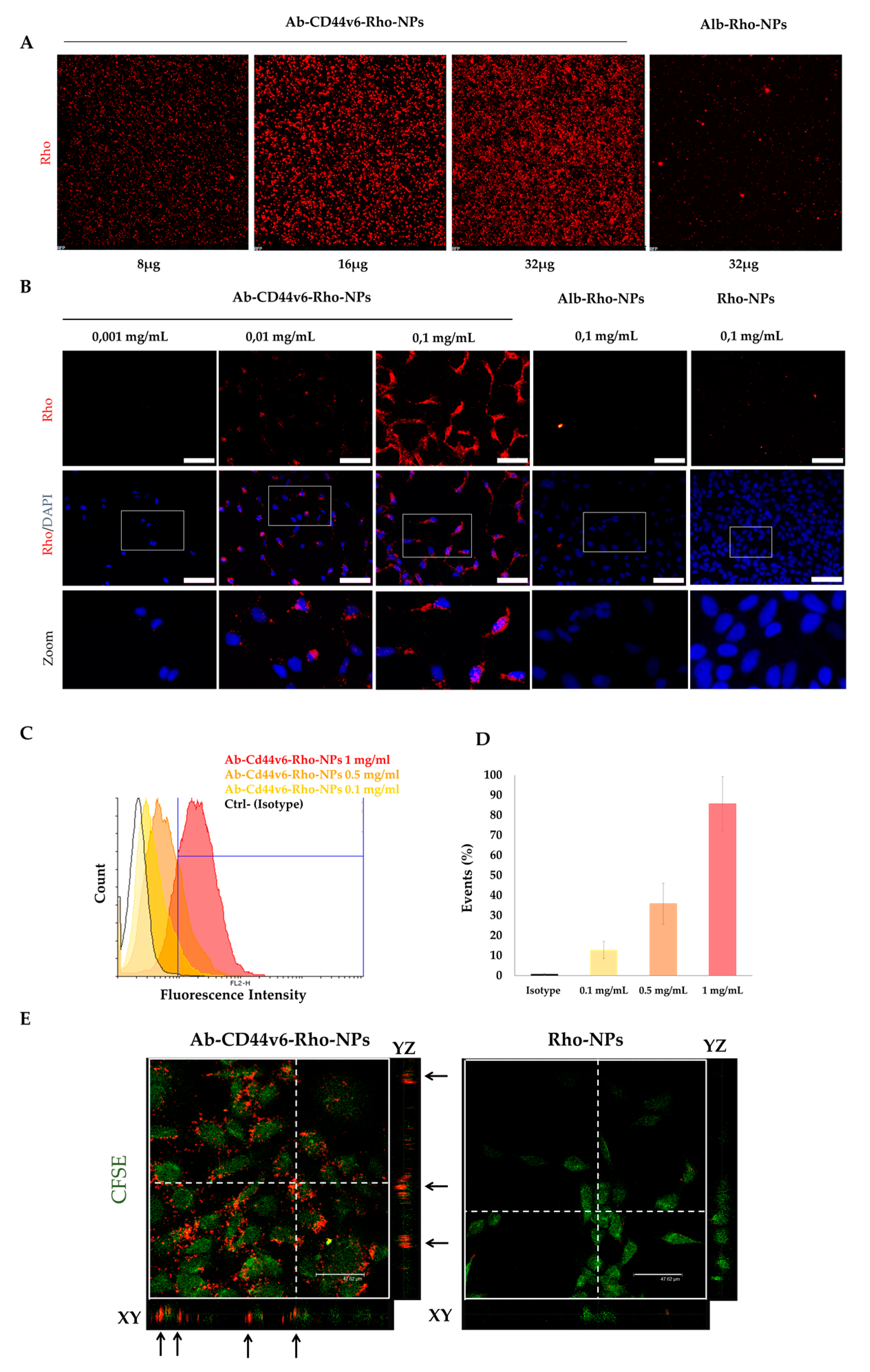
3.3.1. Cell Attachment and Internalization of Ab-CD44v6-Rho-NPs
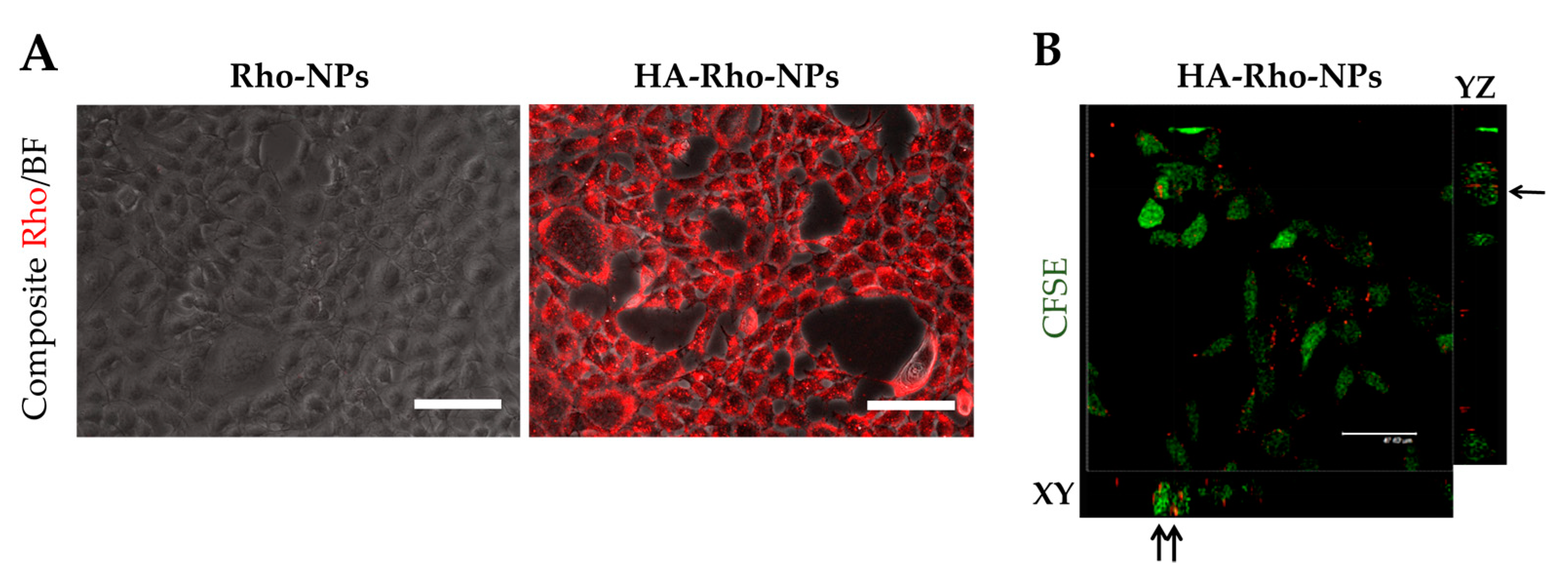
3.3.2. Cell Attachment and Internalization of HA-Rho-NPs
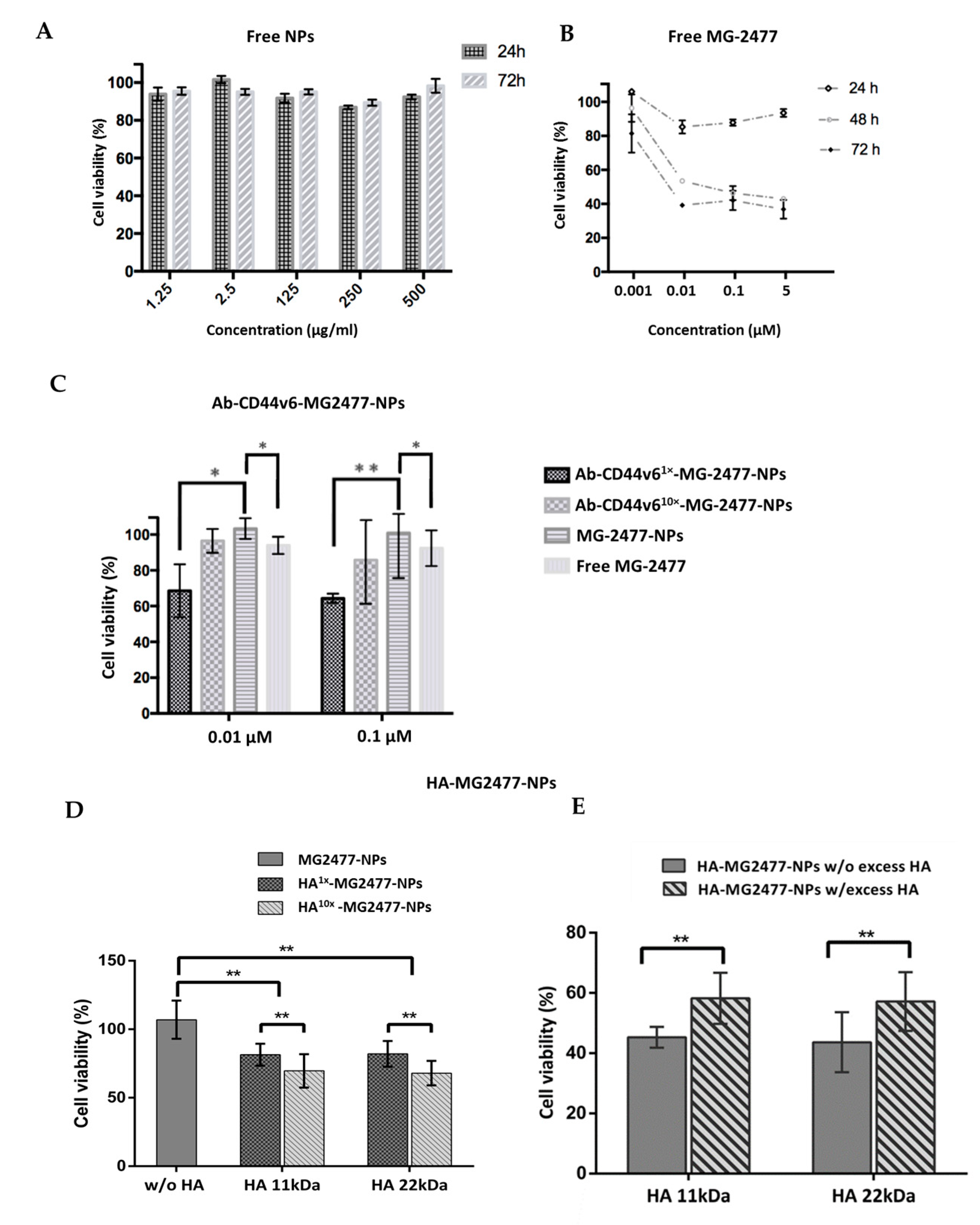
3.3.3. Delivery of the Quinolone Derivative MG2477
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vos, T.; Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abate, K.H.; Abd-Allah, F.; Abdulle, A.M.; Abebo, T.A.; Abera, S.F.; Aboyans, V.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [Google Scholar] [CrossRef]
- Feng, S.S.; Zhao, L.; Zhang, Z.; Bhakta, G.; Win, K.Y.; Dong, Y.; Chien, S. Chemotherapeutic engineering: Vitamin E TPGS-emulsified nanoparticles of biodegradable polymers realized sustainable paclitaxel chemotherapy for 168 h in vivo. Chem. Eng. Sci. 2007, 62, 6641–6648. [Google Scholar] [CrossRef]
- Pelaz, B.; Alexiou, C.; Alvarez-Puebla, R.A.; Alves, F.; Andrews, A.M.; Ashraf, S.; Balogh, L.P.; Ballerini, L.; Bestetti, A.; Brendel, C.; et al. Diverse Applications of Nanomedicine. ACS Nano 2017, 11, 2313–2381. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Q.; Han, N.; Bai, L.; Li, J.; Liu, J.; Che, E.; Hu, L.; Zhang, Q.; Jiang, T.; et al. Mesoporous silica nanoparticles in drug delivery and biomedical applications. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 313–327. [Google Scholar] [CrossRef]
- Tavano, R.; Segat, D.; Reddi, E.; Kos, J.; Rojnik, M.; Kocbek, P.; Iratni, S.; Scheglmann, D.; Colucci, M.; Rio-Echevarria, I.M.; et al. Procoagulant properties of bare and highly PEGylated vinyl-modified silica nanoparticles. Nanomedicine 2010, 5, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Rojnik, M.; Kocbek, P.; Moret, F.; Compagnin, C.; Celotti, L.; Bovis, M.J.; Woodhams, J.H.; Macrobert, A.J.; Scheglmann, D.; Helfrich, W.; et al. In vitro and in vivo characterization of temoporfin-loaded PEGylated PLGA nanoparticles for use in photodynamic therapy. Nanomedicine 2012, 7, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Gozuacik, D.; Yagci-Acar, H.F.; Akkoc, Y.; Kosar, A.; Dogan-Ekici, A.I.; Ekici, S. Anticancer use of nanoparticles as nucleic acid carriers. J. Biomed. Nanotechnol. 2014, 10, 1751–1783. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.; Hamidi, M. Passive and active targeting in cancer therapy by liposomes and lipid nanoparticles. Drug Metab. Pers. Ther. 2019, 34. [Google Scholar] [CrossRef] [PubMed]
- Huynh, E.; Zhengm, G. Cancer nanomedicine: Addressing the dark side of the enhanced permeability and retention effect. Nanomedicine 2015, 10, 1993–1995. [Google Scholar] [CrossRef] [PubMed]
- Vander Linden, C.; Corbet, C. Therapeutic Targeting of Cancer Stem Cells: Integrating and Exploiting the Acidic Niche. Front. Oncol. 2019, 19, 159. [Google Scholar] [CrossRef]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ishida, T.; Okada, Y.; Ise, S.; Harashima, H.; Kiwada, H. Effect of transferrin receptor-targeted liposomal doxorubicin in P-glycoprotein-mediated drug resistant tumor cells. Int. J. Pharm. 2007, 329, 94–102. [Google Scholar] [CrossRef]
- Iinuma, H.; Maruyama, K.; Okinaga, K.; Sasaki, K.; Sekine, T.; Ishida, O.; Ogiwara, N.; Johkura, K.; Yonemura, Y. Intracellular targeting therapy of cisplatin-encapsulated transferrin-polyethylene glycol liposome on peritoneal dissemination of gastric cancer. Int. J. Cancer 2002, 99, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Kirpotin, D.B.; Drummond, D.C.; Shao, Y.; Refaat Shalaby, M.; Hong, K.; Nielsen, U.B.; Marks, J.D.; Benz, C.C.; Park, J.W. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006, 66, 6732–6740. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, N.A.; Gomes-da-Silva, L.C.; Moura, V.; Simões, S.; Moreira, J.N. Simultaneous active intracellular delivery of doxorubicin and C6-ceramide shifts the additive/antagonistic drug interaction of non-encapsulated combination. J. Control. Release 2014, 196, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Zalba, S.; Seynhaeve, A.L.B.; Debets, R.; Ten Hagen, T.L.M. Liposomes targeted to MHC-restricted antigen improve drug delivery and antimelanoma response. Int. J. Nanomed. 2019, 14, 2069–2089. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Hiramoto, S.; Amano, Y.; Emoto, S.; Yamaguchi, H.; Ishigami, H.; Kitayama, J.; Ito, T. Intraperitoneal Delivery of Cisplatin via a Hyaluronan-Based Nanogel/in Situ Cross-Linkable Hydrogel Hybrid System for Peritoneal Dissemination of Gastric Cancer. Mol. Pharm. 2017, 14, 3105–3113. [Google Scholar] [CrossRef]
- Hoang, B.; Ekdawim, S.N.; Reilly, R.M.; Allen, C. Active targeting of block copolymer micelles with trastuzumab Fab fragments and nuclear localization signal leads to increased tumor uptake and nuclear localization in HER2-overexpressing xenografts. Mol. Pharm. 2013, 10, 4229–4241. [Google Scholar] [CrossRef]
- Alama, N.; Koul, M.; Mintoo, M.J.; Khare, V.; Gupta, R.; Rawat, N.; Sharma, P.R.; Singh, S.K.; Mondhe, D.M.; Gupta, P.N. Development and characterization of hyaluronic acid modified PLGA based nanoparticles for improved efficacy of cisplatin in solid tumor. Biomed. Pharmacother. 2017, 95, 856–864. [Google Scholar] [CrossRef]
- Danhier, F.; Ansorena, E.; Silva, J.M.; Coco, R.; Le Breton, A.; Préat, V. PLGA-based nanoparticles: An overview of biomedical applications. J. Control. Release 2012, 161, 505–522. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Bjerkvig, R.; Tysnes, B.B.; Aboody, K.S.; Najbauer, J.; Terzis, A.J. Opinion: The origin of the cancer stem cell: Current controversies and new insights. Nat. Rev. Cancer 2005, 11, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, A.; Deshpande, K.; Arfuso, F.; Newsholme, P.; Dharmarajan, A. Cancer stem cell metabolism: A potential target for cancer therapy. Mol. Cancer 2016, 15, 69. [Google Scholar] [CrossRef]
- Ganesh, S.; Iyer, A.K.; Morrissey, D.V.; Amiji, M.M. Hyaluronic acid based self-assembling nanosystems for CD44 target mediated siRNA delivery to solid tumors. Biomaterials 2013, 34, 3489–3502. [Google Scholar] [CrossRef]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef]
- Toole, B.P. Hyaluronan: From extracellular glue to pericellular cue. Nat. Rev. Cancer 2004, 4, 528–539. [Google Scholar] [CrossRef]
- Auvinen, P.; Tammi, R.; Tammi, M.; Johansson, R.; Kosma, V.M. Expression of CD44s, CD44v3 and CD44v6 in benign and malignant breast lesions: Correlation and colocalization with hyaluronan. Histopathology 2005, 47, 420–428. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef]
- Yan, Y.; Zuo, X.; Wie, D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl. Med. 2015, 9, 1033–1043. [Google Scholar] [CrossRef]
- Wang, L.; Zuo, X.; Xie, K.; Wie, D. The Role of CD44 and Cancer Stem Cells. Cancer Stem Cells 2017, 1692, 31–42. [Google Scholar]
- Günthert, U.; Hofmann, M.; Rudy, W.; Reber, S.; Zöller, M.; Haussmann, I.; Matzku, S.; Wenzel, A.; Ponta, H.; Herrlich, P. A new variant of glycoproteinCD44 confers metastatic potential to rat carcinoma cells. Cell 1991, 65, 13–24. [Google Scholar] [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Kodama, H.; Murata, S.; Ishida, M.; Yamamoto, H.; Yamaguchi, T.; Kaida, S.; Miyake, T.; Takebayashi, K.; Kushima, R.; Tani, M. Prognostic impact of CD44-positive cancer stem-like cells at the invasive front of gastric cancer. Br. J. Cancer 2017, 116, 186–194. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef]
- Toole, B.P. Hyaluronan-CD44 Interactions in Cancer: Paradoxes and Possibilities. Clin Cancer Res. 2009, 15, 7462–7468. [Google Scholar] [CrossRef]
- Motohara, T.; Fujimoto, K.; Tayama, S.; Narantuya, D.; Sakaguchi, I.; Tashiro, H.; Katabuchi, H. CD44 Variant 6 as a Predictive Biomarker for Distant Metastasis in Patients with Epithelial Ovarian Cancer. Obst. Gynecol. 2016, 127, 1003–1011. [Google Scholar] [CrossRef]
- Jin, Y.-J.; Ubonvan, T.; Kim, D.D. Hyaluronic Acid in Drug Delivery Systems. J. Pharm. Investig. 2010, 40, 33–43. [Google Scholar] [CrossRef]
- Eliaz, R.E.; Szoka Jr, F.C. Liposome-encapsulated Doxorubicin Targeted to CD44: A Strategy to Kill CD44-overexpressing Tumor Cells. Cancer Res. 2001, 61, 2592–2601. [Google Scholar]
- Lee, H.; Lee, K.; Park, T.G. Hyaluronic Acid−Paclitaxel Conjugate Micelles: Synthesis, Characterization, and Antitumor Activity. Bioconjug. Chem. 2008, 19, 1319–1325. [Google Scholar] [CrossRef]
- Mironov, V.; Kasyanov, V.; Shu, X.Z.; Eisenberg, C.; Eisenberg, L.; Gonda, S.; Trusk, T.; Markwald, R.R.; Prestwich, G.D. Fabrication of tubular tissue constructs by centrifugal casting of cells suspended in an in situ crosslinkable hyaluronan-gelatin hydrogel. Biomaterials 2005, 26, 7628–7635. [Google Scholar] [CrossRef] [PubMed]
- Karousou, E.; Misra, S.; Ghatak, S.; Dobra, K.; Götte, M.; Vigetti, D.; Passi, A.; Karamanos, N.K.; Skandalis, S.S. Roles and targeting of the HAS/hyaluronan/CD44 molecular system in cancer. Matrix Biol. 2017, 59, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Ekici, S.; Cerwinka, W.H.; Duncan, R.; Gomez, P.; Civantos, F.; Soloway, M.S.; Lokeshwar, V.B. Comparison of the prognostic potential of hyaluronic acid, hyaluronidase (HYAL-1), CD44v6 and microvessel density for prostate cancer. Int. J. Cancer 2004, 112, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Girbl, T.; Hinterseer, E.; Grössinger, E.M.; Asslaber, D.; Oberascher, K.; Weiss, L.; Hauser-Kronberger, C.; Neureiter, D.; Kerschbaum, H.; Naor, D.; et al. CD40-mediated activation of chronic lymphocytic leukemia cells promotes their CD44-dependent adhesion to hyaluronan and restricts CCL21-induced motility. Cancer Res. 2013, 73, 561–570. [Google Scholar] [CrossRef]
- Wang, Z.; Zöller, M. Exosomes, metastases, and the miracle of cancer stem cell markers. Cancer Metastasis Rev. 2019, 38, 259–295. [Google Scholar] [CrossRef]
- Vallet-Regí, M.; Colilla, M.; Izquierdo-Barba, I.; Manzano, M. Mesoporous Silica Nanoparticles for Drug Delivery: Current Insights. Molecules 2018, 23, 47. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, W.; Hong, C.; Pan, C. Silica Nanotubes Decorated by pH-Responsive Diblock Copolymers for Controlled Drug Release. ACS Appl. Mater. Interfaces 2015, 7, 3618–3625. [Google Scholar] [CrossRef]
- Alao Amolegbe, S.; Hirano, Y.; Oluwatope Adebayo, J.; George Ademowo, O.; Abidemi Balogun, E.; Ayoola Obaleye, J.; Ursine Krettli, A.; Yu, C.; Hayami, S. Mesoporous silica nanocarriers encapsulated antimalarials with high therapeutic performance. Sci. Rep. 2018, 8, 3078. [Google Scholar] [CrossRef]
- Echevarria, I.M.; Selvestrel, F.; Segat, D.; Guarino, G.; Tavano, R.; Causin, V.; Reddi, E.; Papini, E.; Mancin, F. Highly PEGylated silica nanoparticles: “ready to use” stealth functional nanocarriers. J. Mater. Chem. 2010, 10, 2780–2787. [Google Scholar] [CrossRef]
- Selvestrel, F.; Moret, F.; Segat, D.; Woodhams, J.H.; Fracasso, G.; Echevarria, I.M.; Baù, L.; Rastrelli, F.; Compagning, C.; Reddi, E.; et al. Targeted delivery of photosensitizers: Efficacy and selectivity issues revealed by multifunctional ORMOSIL nanovectors in cellular systems. Nanoscale 2013, 5, 6106–6116. [Google Scholar] [CrossRef]
- Gonzalez-García, T.; Fernández, S.; Lubian, E.; Mancin, F.; Ferrero, M. Preparation of ORMOSIL nanoparticles conjugated with vitamin D3 analogues and their biological evaluation. RSC Adv. 2016, 6, 31840–31849. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kang, S.; Ahn, M.; Jang, H.; Min, D.-H. Development of dual-pore coexisting branched silica nanoparticles for efficient gene–chemo cancer therapy. Small 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-A.; Chen, W.; Zhang, L.; Wu, H.H.; Zink, J.I. A Responsive Mesoporous Silica Nanoparticle Platform for Magnetic Resonance Imaging-Guided High-Intensity Focused UltrasoundStimulated Cargo Delivery with Controllable Location, Time, and Dose. J. Am. Chem. Soc. 2019, 141, 17670–17684. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Glackin, C.A.; Horwitz, M.A.; Zink, J.I. Nanomachines and Other Caps on Mesoporous Silica Nanoparticles for Drug Delivery. Acc. Chem. Res. 2019, 52, 1531–1542. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Cheng, C.-A.; Cosco, E.D.; Ramakrishnan, S.; Lingg, J.G.P.; Bruns, O.T.; Zink, J.I.; Sletten, E.M. Shortwave Infrared Imaging with J-Aggregates Stabilized in Hollow Mesoporous Silica Nanoparticles. J. Am. Chem. Soc. 2019, 141, 12475–12480. [Google Scholar] [CrossRef] [PubMed]
- Ciriminna, R.; Sciortino, M.; Alonzo, G.; de Schrijver, A.; Pagliaro, M. From Molecules to Systems: Sol−Gel Microencapsulation in Silica-Based Materials. Chem. Rev. 2011, 111, 765–789. [Google Scholar] [CrossRef] [PubMed]
- Roy, I.; Ohulchanskyy, T.Y.; Pudavar, H.E.; Bergey, E.J.; Oseroff, A.R.; Morgan, J.; Dougherty, T.J.; Prasad, P.N. Ceramic-Based Nanoparticles Entrapping Water-Insoluble Photosensitizing Anticancer Drugs: A Novel Drug−Carrier System for Photodynamic Therapy. J. Am. Chem. Soc. 2003, 125, 7860–7865. [Google Scholar] [CrossRef]
- Tavano, R.; Gabrielli, L.; Lubian, E.; Fedeli, C.; Visentin, S.; Polverino De Laureto, P.; Arrigoni, G.; Geffner-Smith, A.; Chen, F.; Simberg, D.; et al. C1q-Mediated Complement Activation and C3 Opsonization Trigger Recognition of Stealth Poly(2-methyl-2-oxazoline)-Coated Silica Nanoparticles by Human Phagocytes. ACS Nano 2018, 12, 5834–5847. [Google Scholar] [CrossRef]
- Roy, I.; Ohulchanskyy, T.Y.; Bharali, D.J.; Pudavar, H.E.; Mistretta, R.A.; Kaur, N.; Prasad, P.N. Optical tracking of organically modified silica nanoparticles as DNA carriers: A nonviral, nanomedicine approach for gene delivery. Proc. Natl. Acad. Sci. USA 2005, 102, 279–284. [Google Scholar] [CrossRef]
- Gasparotto, V.; Castagliuolo, I.; Ferlin, M.G. 3-Substituted 7-Phenyl-Pyrroloquinolinones Show Potent Cytotoxic Activity in Human Cancer Cell Lines. J. Med. Chem. 2007, 50, 5509–5513. [Google Scholar] [CrossRef]
- Viola, G.; Bortolozzi, R.; Hamel, E.; Moro, S.; Brun, P.; Castagliuolo, I.; Ferlin, M.G.; Basso, G. MG2477, a new tubulin inhibitor, induces autophagy through inhibition of the Akt/mTOR pathway and delayed apoptosis in A549 cells. Biochem. Pharmacol. 2012, 83, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Gasparotto, V.; Castagliuolo, I.; Chiarelotto, G.; Pezzi, V.; Montanaro, G.; Brun, P.; Palu, G.; Ferlin, M.G. Synthesis and biological activity of 7-phenyl-6,9-dihydro-3H-pyrrolo[3, 2-f]quinolin-9-ones: A new class of antimitotic agents devoid of aromatase activity. J. Med. Chem. 2006, 49, 1910–1915. [Google Scholar] [CrossRef] [PubMed]
- Compagnin, C.; Baù, L.; Mognato, M.; Celotti, L.; Miotto, G.; Arduini, M.; Moret, F.; Fede, C.; Selvestreli, F.; Rio Echevarria, I.M.; et al. The cellular uptake of meta-tetra(hydroxyphenyl)chlorin entrapped in organically modified silica nanoparticles is mediated by serum proteins. Nanotechnology 2009, 20, 345101. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L. Tissue Sulphidryl Groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Bulaj, G.; Kortemme, T.; Goldenberg, D.P. Ionization−Reactivity Relationships for Cysteine Thiols in Polypeptides. Biochemistry 1998, 37, 8965–8972. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.D.; Kiick, K.L. Tunable degradation of maleimide-thiol adducts in reducing environments. Bioconjug. Chem. 2011, 22, 1946–1953. [Google Scholar] [CrossRef]
- Colombo, M.; Fiandra, L.; Alessio, G.; Mazzucchelli, S.; Nebuloni, M.; De Palma, C.; Kantner, K.; Pelaz, B.; Rotem, R.; Corsi, F.; et al. Tumour homing and therapeutic effect of colloidal nanoparticles depend on the number of attached antibodies. Nat. Commun. 2015, 7, 13818. [Google Scholar] [CrossRef]
- Shenton, W.; Davis, S.A.; Mann, S. Directed self-assembly of nanoparticles into macroscopic materials using antibody ± antigen recognition. Adv. Mater. 1995, 11, 449–452. [Google Scholar] [CrossRef]


| Entry | Nanoparticles | Size a (nm) | PDI | Z-Potential a (mV) |
|---|---|---|---|---|
| 1 | NH2-NPs | 96.0 ± 2.34 | 0.019 | −1.00 ± 0.63 |
| 2 | Ab-CD44v61x-NPs | 104 ± 17.3 | 0.539 | −3.62 ± 0.35 |
| 3 | Ab-CD44v610x-NPs | 99.0 ± 18.9 | 0.504 | −1.60 ± 0.52 |
| 4 | NH2-NPs | 140 ± 0.83 | 0.095 | −1.32 ± 0.39 |
| 5 | 11.5 kDa HA1x-NPs | 144 ± 2.68 | 0.034 | −7.07 ± 0.76 |
| 6 | 11.5 kDa HA10x-NPs | 137 ± 2.47 | 0.050 | −8.50 ± 0.15 |
| 7 | 22.5 kDa HA1x-NPs | 128 ± 9.17 | 0.096 | −8.06 ± 0.54 |
| 8 | 22.5 kDa HA10x-NPs | 127 ± 3.04 | 0.076 | −8.73 ± 0.28 |
| Entry | Nanoparticles | Drug Loading a (w/w %) |
|---|---|---|
| 1 | NH2-MG2477-NPs | 0.4 |
| 2 | Ab-CD44v61x-MG2477-NPs | 0.05 |
| 3 | Ab-CD44v610x-MG2477-NPs | 0.05 |
| 4 | 11.5 kDa HA1x-MG2477-NPs | 0.06 |
| 5 | 11.5 kDa HA10x-MG2477-NPs | 0.07 |
| 6 | 22.5 kDa HA1x-MG2477-NPs | 0.05 |
| 7 | 22.5 kDa HA10x-MG2477-NPs | 0.06 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morillas-Becerril, L.; Peta, E.; Gabrielli, L.; Russo, V.; Lubian, E.; Nodari, L.; Ferlin, M.G.; Scrimin, P.; Palù, G.; Barzon, L.; et al. Multifunctional, CD44v6-Targeted ORMOSIL Nanoparticles Enhance Drugs Toxicity in Cancer Cells. Nanomaterials 2020, 10, 298. https://doi.org/10.3390/nano10020298
Morillas-Becerril L, Peta E, Gabrielli L, Russo V, Lubian E, Nodari L, Ferlin MG, Scrimin P, Palù G, Barzon L, et al. Multifunctional, CD44v6-Targeted ORMOSIL Nanoparticles Enhance Drugs Toxicity in Cancer Cells. Nanomaterials. 2020; 10(2):298. https://doi.org/10.3390/nano10020298
Chicago/Turabian StyleMorillas-Becerril, Lucía, Elektra Peta, Luca Gabrielli, Venera Russo, Elisa Lubian, Luca Nodari, Maria Grazia Ferlin, Paolo Scrimin, Giorgio Palù, Luisa Barzon, and et al. 2020. "Multifunctional, CD44v6-Targeted ORMOSIL Nanoparticles Enhance Drugs Toxicity in Cancer Cells" Nanomaterials 10, no. 2: 298. https://doi.org/10.3390/nano10020298
APA StyleMorillas-Becerril, L., Peta, E., Gabrielli, L., Russo, V., Lubian, E., Nodari, L., Ferlin, M. G., Scrimin, P., Palù, G., Barzon, L., Castagliuolo, I., Mancin, F., & Trevisan, M. (2020). Multifunctional, CD44v6-Targeted ORMOSIL Nanoparticles Enhance Drugs Toxicity in Cancer Cells. Nanomaterials, 10(2), 298. https://doi.org/10.3390/nano10020298