Modulating Tumor Cell Functions by Tunable Nanopatterned Ligand Presentation
Abstract
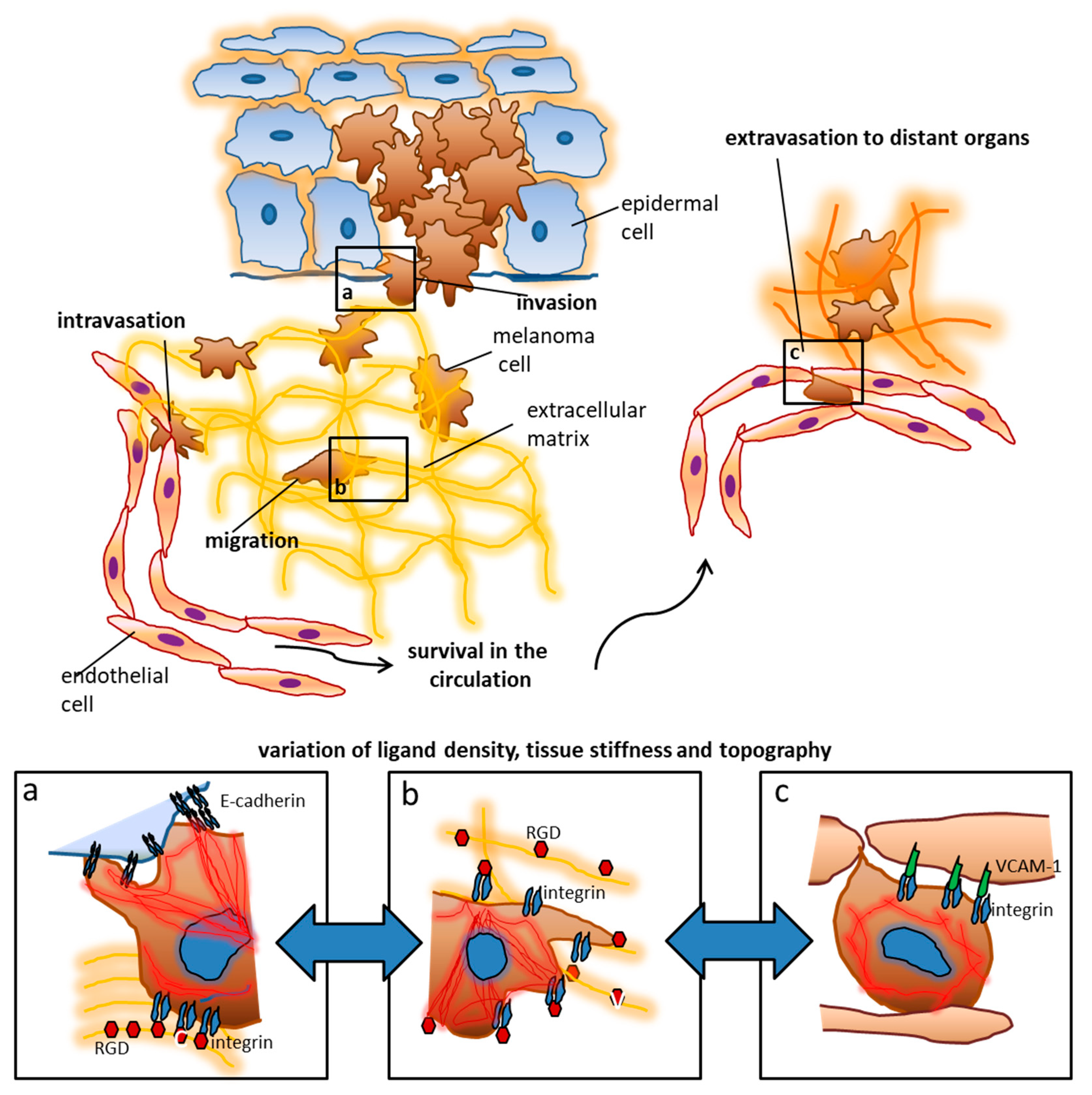
1. Setting the Stage: Environmental Signals Modulate Cellular Functions
2. Biophysical Properties of the Extracellular Matrix—More Than Just a Scaffold
3. YAP and TAZ—Master Regulators of Mechanosensing
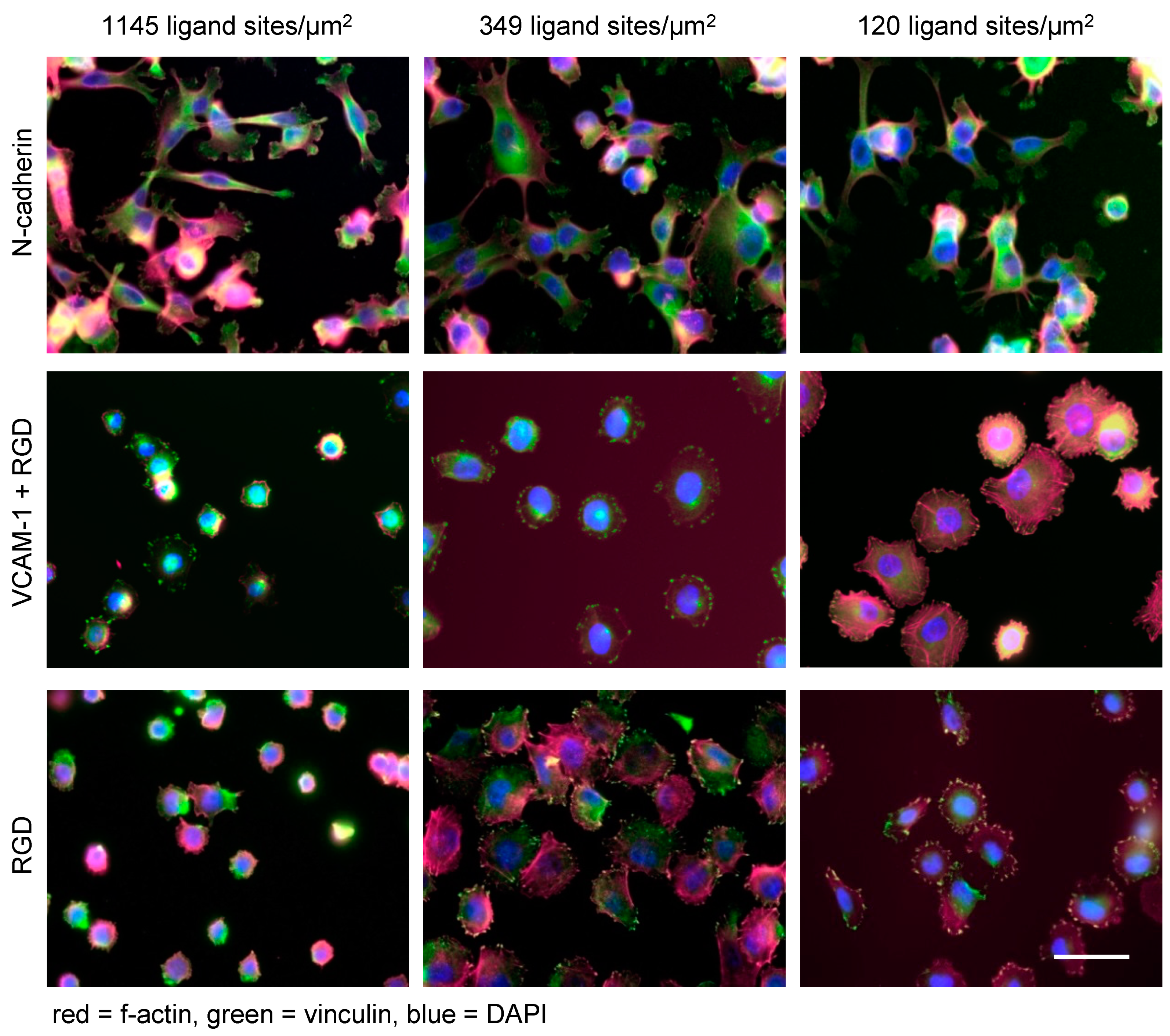
4. Nanoscale Ligand Control in Experimental Models
5. Modulation of Tumor Cell Functions by Nanostructured Ligands
6. Limitations of Current Nanomodel Systems
7. Biophysical Cues Modulate Cell Death and Survival
Author Contributions
Funding
Conflicts of Interest
References
- Gumbiner, B.M. Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell 1996, 84, 345–357. [Google Scholar] [CrossRef]
- Maheshwari, G.; Brown, G.; Lauffenburger, D.A.; Wells, A.; Griffith, L.G. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 2000, 113, 1677–1686. [Google Scholar] [PubMed]
- Boudreau, N.J.; Jones, P.L. Extracellular matrix and integrin signalling: The shape of things to come. Biochem. J. 1999, 339, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Somers, W.S.; Tang, J.; Shaw, G.D.; Camphausen, R.T. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell 2000, 103, 467–479. [Google Scholar] [CrossRef]
- Alon, R.; Ley, K. Cells on the run: Shear-regulated integrin activation in leukocyte rolling and arrest on endothelial cells. Curr. Opin. Cell. Biol. 2008, 20, 525–532. [Google Scholar] [CrossRef]
- Ley, K.; Laudanna, C.; Cybulsky, M.I.; Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007, 7, 678–689. [Google Scholar] [CrossRef]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef]
- Ju, R.J.; Stehbens, S.J.; Haass, N.K. The Role of Melanoma Cell-Stroma Interaction in Cell Motility, Invasion, and Metastasis. Front. Med. 2018, 5, 307. [Google Scholar] [CrossRef]
- Cordes, N. Integrin-mediated cell-matrix interactions for prosurvival and antiapoptotic signaling after genotoxic injury. Cancer Lett. 2006, 242, 11–19. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Kuphal, S.; Bauer, R.; Bosserhoff, A.K. Integrin signaling in malignant melanoma. Cancer Metastasis Rev. 2005, 24, 195–222. [Google Scholar] [CrossRef] [PubMed]
- Cosgarea, I.; Ritter, C.; Becker, J.C.; Schadendorf, D.; Ugurel, S. Update on the clinical use of kinase inhibitors in melanoma. J. Dtsch. Dermatol. Ges. 2017, 15, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Wilden, S.M.; Lang, B.M.; Mohr, P.; Grabbe, S. Immune checkpoint inhibitors: A milestone in the treatment of melanoma. J. Dtsch. Dermatol. Ges. 2016, 14, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Ugurel, S.; Röhmel, J.; Ascierto, P.A.; Flaherty, K.T.; Grob, J.J.; Hauschild, A.; Larkin, J.; Long, G.V.; Lorigan, P.; McArthur, G.A.; et al. Survival of patients with advanced metastatic melanoma: The impact of novel therapies-update 2017. Eur. J. Cancer 2017, 83, 247–257. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef]
- Valenick, L.V.; Schwarzbauer, J.E. Ligand density and integrin repertoire regulate cellular response to LPA. Matrix Biol. 2006, 25, 223–231. [Google Scholar] [CrossRef]
- Wang, J.F.; Zhang, X.F.; Groopman, J.E. Stimulation of beta 1 integrin induces tyrosine phosphorylation of vascular endothelial growth factor receptor-3 and modulates cell migration. J. Biol. Chem. 2001, 276, 41950–41957. [Google Scholar] [CrossRef]
- Carter, A. Integrins as target: First phase III trial launches, but questions remain. J. Natl. Cancer Inst. 2010, 102, 675–677. [Google Scholar] [CrossRef]
- Eskens, F.A.; Dumez, H.; Hoekstra, R.; Perschl, A.; Brindley, C.; Böttcher, S.; Wynendaele, W.; Drevs, J.; Verweij, J.; van Oosterom, A.T. Phase I and pharmacokinetic study of continuous twice weekly intravenous administration of Cilengitide (EMD 121974), a novel inhibitor of the integrins alphavbeta3 and alphavbeta5 in patients with advanced solid tumours. Eur. J. Cancer 2003, 39, 917–926. [Google Scholar] [CrossRef]
- O’Donnell, P.H.; Undevia, S.D.; Stadler, W.M.; Karrison, T.M.; Nicholas, M.K.; Janisch, L.; Ratain, M.J. A phase I study of continuous infusion cilengitide in patients with solid tumors. Investig. New Drugs 2012, 30, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.; Williams, G.; Gour, B.J.; Blaschuk, O.W.; Doherty, P. A novel family of cyclic peptide antagonists suggests that N-cadherin specificity is determined by amino acids that flank the HAV motif. J. Biol. Chem. 2000, 275, 4007–4012. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.; Mojares, E.; Del Rio Hernandez, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, C.; Marganski, W.A.; Kim, S.; Brown, C.T.; Gunderia, V.; Dembo, M.; Wong, J.Y. Influence of type I collagen surface density on fibroblast spreading, motility, and contractility. Biophys. J. 2003, 85, 3329–3335. [Google Scholar] [CrossRef]
- Shebanova, O.; Hammer, D.A. Biochemical and mechanical extracellular matrix properties dictate mammary epithelial cell motility and assembly. Biotechnol. J. 2012, 7, 397–408. [Google Scholar] [CrossRef]
- Kumar, S.; Das, A.; Sen, S. Extracellular matrix density promotes EMT by weakening cell-cell adhesions. Mol. Biosyst. 2014, 10, 838–850. [Google Scholar] [CrossRef]
- Li, L.; Klim, J.R.; Derda, R.; Courtney, A.H.; Kiessling, L.L. Spatial control of cell fate using synthetic surfaces to potentiate TGF-beta signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11745–11750. [Google Scholar] [CrossRef]
- Smith, Q.; Stukalin, E.; Kusuma, S.; Gerecht, S.; Sun, S.X. Stochasticity and Spatial Interaction Govern Stem Cell Differentiation Dynamics. Sci. Rep. 2015, 5, 12617. [Google Scholar] [CrossRef]
- Young, J.L.; Holle, A.W.; Spatz, J.P. Nanoscale and mechanical properties of the physiological cell-ECM microenvironment. Exp. Cell Res. 2016, 343, 3–6. [Google Scholar] [CrossRef]
- Cimmino, C.; Rossano, L.; Netti, P.A.; Ventre, M. Spatio-Temporal Control of Cell Adhesion: Toward Programmable Platforms to Manipulate Cell Functions and Fate. Front. Bioeng. Biotechnol. 2018, 6, 190. [Google Scholar] [CrossRef]
- Rapier, R.; Huq, J.; Vishnubhotla, R.; Bulic, M.; Perrault, C.M.; Metlushko, V.; Cho, M.; Tay, R.T.; Glover, S.C. The extracellular matrix microtopography drives critical changes in cellular motility and Rho A activity in colon cancer cells. Cancer Cell Int. 2010, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Gu, L.; Darnell, M.; Klumpers, D.; Bencherif, S.A.; Weaver, J.C.; Huebsch, N.; Mooney, D.J. Substrate stress relaxation regulates cell spreading. Nat. Commun. 2015, 6, 6364. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Models Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Butcher, D.T.; Alliston, T.; Weaver, V.M. A tense situation: Forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108–122. [Google Scholar] [CrossRef]
- Miskolczi, Z.; Smith, M.P.; Rowling, E.J.; Ferguson, J.; Barriuso, J.; Wellbrock, C. Collagen abundance controls melanoma phenotypes through lineage-specific microenvironment sensing. Oncogene 2018, 37, 3166–3182. [Google Scholar] [CrossRef]
- Barcus, C.E.; Keely, P.J.; Eliceiri, K.W.; Schuler, L.A. Stiff collagen matrices increase tumorigenic prolactin signaling in breast cancer cells. J. Biol. Chem. 2013, 288, 12722–12732. [Google Scholar] [CrossRef]
- Hartmann, N.; Giese, N.A.; Giese, T.; Poschke, I.; Offringa, R.; Werner, J.; Ryschich, E. Prevailing role of contact guidance in intrastromal T-cell trapping in human pancreatic cancer. Clin. Cancer Res. 2014, 20, 3422–3433. [Google Scholar] [CrossRef]
- Abrams, G.A.; Schaus, S.S.; Goodman, S.L.; Nealey, P.F.; Murphy, C.J. Nanoscale topography of the corneal epithelial basement membrane and Descemet’s membrane of the human. Cornea 2000, 19, 57–64. [Google Scholar] [CrossRef]
- Kraning-Rush, C.M.; Reinhart-King, C.A. Controlling matrix stiffness and topography for the study of tumor cell migration. Cell Adh. Migr. 2012, 6, 274–279. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Welf, E.S.; Naik, U.P.; Ogunnaike, B.A. A Spatial Model for Integrin Clustering as a Result of Feedback between Integrin Activation and Integrin Binding. Biophys. J. 2012, 103, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Kanchanawong, P.; Shtengel, G.; Pasapera, A.M.; Ramko, E.B.; Davidson, M.W.; Hess, H.F.; Waterman, C.M. Nanoscale architecture of integrin-based cell adhesions. Nature 2010, 468, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Panciera, T.; Azzolin, L.; Cordenonsi, M.; Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell. Biol. 2017, 18, 758–770. [Google Scholar] [CrossRef]
- Oria, R.; Wiegand, T.; Escribano, J.; Elosegui-Artola, A.; Uriarte, J.J.; Moreno-Pulido, C.; Platzman, I.; Delcanale, P.; Albertazzi, L.; Navajas, D.; et al. Force loading explains spatial sensing of ligands by cells. Nature 2017, 552, 219–224. [Google Scholar] [CrossRef]
- Plouffe, S.W.; Lin, K.C.; Moore, J.L., 3rd; Tan, F.E.; Ma, S.; Ye, Z.; Qiu, Y.; Ren, B.; Guan, K.L. The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. J. Biol. Chem. 2018, 293, 11230–11240. [Google Scholar] [CrossRef]
- Sun, M.; Spill, F.; Zaman, M.H. A Computational Model of YAP/TAZ Mechanosensing. Biophys. J. 2016, 110, 2540–2550. [Google Scholar] [CrossRef]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP and TAZ: A signalling hub of the tumour microenvironment. Nat. Rev. Cancer 2019, 19, 454–464. [Google Scholar] [CrossRef]
- Kim, M.H.; Kim, J.; Hong, H.; Lee, S.H.; Lee, J.K.; Jung, E.; Kim, J. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. Embo J. 2016, 35, 462–478. [Google Scholar] [CrossRef]
- Haemmerle, M.; Taylor, M.L.; Gutschner, T.; Pradeep, S.; Cho, M.S.; Sheng, J.; Lyons, Y.M.; Nagaraja, A.S.; Dood, R.L.; Wen, Y.; et al. Platelets reduce anoikis and promote metastasis by activating YAP1 signaling. Nat. Commun. 2017, 8, 310. [Google Scholar] [CrossRef] [PubMed]
- Hinterdorfer, P.; Garcia-Parajo, M.F.; Dufrene, Y.F. Single-molecule imaging of cell surfaces using near-field nanoscopy. Acc. Chem. Res. 2012, 45, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Cavalcanti-Adam, E.A.; Glass, R.; Blümmel, J.; Eck, W.; Kantlehner, M.; Kessler, H.; Spatz, J.P. Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem 2004, 5, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Dzamba, B.J.; Peters, D.M. Arrangement of cellular fibronectin in noncollagenous fibrils in human fibroblast cultures. J. Cell Sci. 1991, 100, 605–612. [Google Scholar]
- Angelin, A.; Weigel, S.; Garrecht, R.; Meyer, R.; Bauer, J.; Kumar, R.K.; Hirtz, M.; Niemeyer, C.M. Multiscale Origami Structures as Interface for Cells. Angew. Chem. Int. Ed. Engl. 2015, 54, 15813–15817. [Google Scholar] [CrossRef]
- Leggett, G.J. Light-directed nanosynthesis: Near-field optical approaches to integration of the top-down and bottom-up fabrication paradigms. Nanoscale 2012, 4, 1840–1855. [Google Scholar] [CrossRef]
- Le, D.H.T.; Commandeur, U.; Steinmetz, N.F. Presentation and Delivery of Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand via Elongated Plant Viral Nanoparticle Enhances Antitumor Efficacy. ACS Nano 2019, 13, 2501–2510. [Google Scholar] [CrossRef]
- Kruss, S.; Erpenbeck, L.; Amschler, K.; Mundinger, T.A.; Boehm, H.; Helms, H.J.; Friede, T.; Andrews, R.K.; Schön, M.P.; Spatz, J.P. Adhesion maturation of neutrophils on nanoscopically presented platelet glycoprotein Ibalpha. ACS Nano 2013, 7, 9984–9996. [Google Scholar] [CrossRef]
- Lohmüller, T.; Aydin, D.; Schwieder, M.; Morhard, C.; Louban, I.; Pacholski, C.; Spatz, J.P. Nanopatterning by block copolymer micelle nanolithography and bioinspired applications. Biointerphases 2011, 6, MR1–MR12. [Google Scholar] [CrossRef]
- Arnold, M.; Hirschfeld-Warneken, V.C.; Lohmüller, T.; Heil, P.; Blümmel, J.; Cavalcanti-Adam, E.A.; Lopez-Garcia, M.; Walther, P.; Kessler, H.; Geiger, B.; et al. Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing. Nano Lett. 2008, 8, 2063–2069. [Google Scholar] [CrossRef]
- Huang, J.; Grater, S.V.; Corbellini, F.; Rinck, S.; Bock, E.; Kemkemer, R.; Kessler, H.; Ding, J.; Spatz, J.P. Impact of order and disorder in RGD nanopatterns on cell adhesion. Nano Lett. 2009, 9, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Bati-Ayaz, G.; Can, A.; Pesen-Okvur, D. Cellular distribution of invadopodia is regulated by nanometer scale surface protein patterns. Eur. J. Cell Biol. 2017, 96, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Horzum, U.; Ozdil, B.; Pesen-Okvur, D. Differentiation of Normal and Cancer Cell Adhesion on Custom Designed Protein Nanopatterns. Nano Lett. 2015, 15, 5393–5403. [Google Scholar] [CrossRef] [PubMed]
- Takkunen, M.; Hukkanen, M.; Liljestrom, M.; Grenman, R.; Virtanen, I. Podosome-like structures of non-invasive carcinoma cells are replaced in epithelial-mesenchymal transition by actin comet-embedded invadopodia. J. Cell Mol. Med. 2010, 14, 1569–1593. [Google Scholar] [CrossRef] [PubMed]
- Schvartzman, M.; Palma, M.; Sable, J.; Abramson, J.; Hu, X.; Sheetz, M.P.; Wind, S.J. Nanolithographic control of the spatial organization of cellular adhesion receptors at the single-molecule level. Nano Lett. 2011, 11, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Muller, J.; Depoil, D.; Mayya, V.; Sheetz, M.P.; Dustin, M.L.; Wind, S.J. Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nat. Nanotechnol. 2018, 13, 610–617. [Google Scholar] [CrossRef]
- Clement, N.; Patriarche, G.; Smaali, K.; Vaurette, F.; Nishiguchi, K.; Troadec, D.; Fujiwara, A.; Vuillaume, D. Large array of sub-10-nm single-grain Au nanodots for use in nanotechnology. Small 2011, 7, 2607–2613. [Google Scholar] [CrossRef]
- Trasobares, J.; Vaurette, F.; Francois, M.; Romijn, H.; Codron, J.L.; Vuillaume, D.; Theron, D.; Clement, N. High speed e-beam lithography for gold nanoarray fabrication and use in nanotechnology. Beilstein J. Nanotechnol. 2014, 5, 1918–1925. [Google Scholar] [CrossRef]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Castro, C.E.; Kilchherr, F.; Kim, D.N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. [Google Scholar] [CrossRef]
- Linko, V.; Dietz, H. The enabled state of DNA nanotechnology. Curr. Opin. Biotechnol. 2013, 24, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Patel, K.; Perez-Garrido, S.; Marshall, J.F.; Palma, M. DNA Origami Nanoarrays for Multivalent Investigations of Cancer Cell Spreading with Nanoscale Spatial Resolution and Single-Molecule Control. ACS Nano 2019, 13, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell. Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Marlar, S.; Abdellatef, S.A.; Nakanishi, J. Reduced adhesive ligand density in engineered extracellular matrices induces an epithelial-mesenchymal-like transition. Acta Biomater. 2016, 39, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Amschler, K.; Beyazpinar, I.; Erpenbeck, L.; Kruss, S.; Spatz, J.P.; Schön, M.P. Morphological Plasticity of Human Melanoma Cells Is Determined by Nanoscopic Patterns of E- and N-Cadherin Interactions. J. Investig. Dermatol. 2019, 139, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Amschler, K.; Erpenbeck, L.; Kruss, S.; Schön, M.P. Nanoscale integrin ligand patterns determine melanoma cell behavior. ACS Nano 2014, 8, 9113–9125. [Google Scholar] [CrossRef]
- Amschler, K.; Kossmann, E.; Erpenbeck, L.; Kruss, S.; Schill, T.; Schön, M.; Möckel, S.M.C.; Spatz, J.P.; Schön, M.P. Nanoscale Tuning of VCAM-1 Determines VLA-4-Dependent Melanoma Cell Plasticity on RGD Motifs. Mol. Cancer Res. 2018, 16, 528–542. [Google Scholar] [CrossRef]
- Reynolds, A.R.; Hart, I.R.; Watson, A.R.; Welti, J.C.; Silva, R.G.; Robinson, S.D.; Da Violante, G.; Gourlaouen, M.; Salih, M.; Jones, M.C.; et al. Stimulation of tumor growth and angiogenesis by low concentrations of RGD-mimetic integrin inhibitors. Nat. Med. 2009, 15, 392–400. [Google Scholar] [CrossRef]
- Lee, H.; Jang, Y.; Seo, J.; Nam, J.M.; Char, K. Nanoparticle-functionalized polymer platform for controlling metastatic cancer cell adhesion, shape, and motility. ACS Nano 2011, 5, 5444–5456. [Google Scholar] [CrossRef]
- Sonnenschein, C.; Soto, A.M. The aging of the 2000 and 2011 Hallmarks of Cancer reviews: A critique. J. Biosci. 2013, 38, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Garini, Y.; Vermolen, B.J.; Young, I.T. From micro to nano: Recent advances in high-resolution microscopy. Curr. Opin. Biotechnol. 2005, 16, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Hell, S.W. Far-field optical nanoscopy. Science 2007, 316, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.A.; Assoian, R.K. Integrins and cell proliferation: Regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J. Cell Sci. 2001, 114, 2553–2560. [Google Scholar] [PubMed]
- Boisvert-Adamo, K.; Aplin, A.E. Mutant B-RAF mediates resistance to anoikis via Bad and Bim. Oncogene 2008, 27, 3301–3312. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, S.; Pradhan, A.K.; Bhoopathi, P.; Shen, X.N.; August, L.A.; Windle, J.J.; Sarkar, D.; Furnari, F.B.; Cavenee, W.K.; Das, S.K.; et al. MDA-9/Syntenin regulates protective autophagy in anoikis-resistant glioma stem cells. Proc. Natl. Acad. Sci. USA 2018, 115, 5768–5773. [Google Scholar] [CrossRef]
- Zhu, G.; Xu, P.; Guo, S.; Yi, X.; Wang, H.; Yang, Y.; Liu, L.; Shi, Q.; Gao, T.; Li, C. Metastatic melanoma cells rely on Sestrin2 to acquire anoikis resistance via detoxifying intracellular ROS. J. Investig. Dermatol. 2019. [Google Scholar] [CrossRef]
- Stupack, D.G.; Cheresh, D.A. Get a ligand, get a life: Integrins, signaling and cell survival. J. Cell Sci. 2002, 115, 3729–3738. [Google Scholar] [CrossRef]
- Juliano, R. Cooperation between soluble factors and integrin-mediated cell anchorage in the control of cell growth and differentiation. Bioessays 1996, 18, 911–917. [Google Scholar] [CrossRef]
- Waring, P.; Mullbacher, A. Cell death induced by the Fas/Fas ligand pathway and its role in pathology. Immunol. Cell Biol. 1999, 77, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Young, J.L.; Monzon, R.I.; Chen, N.; Todorovic, V.; Lau, L.F. Cytotoxicity of TNFalpha is regulated by integrin-mediated matrix signaling. Embo J. 2007, 26, 1257–1267. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Lau, L.F. Deadly liaisons: Fatal attraction between CCN matricellular proteins and the tumor necrosis factor family of cytokines. J. Cell Commun. Signal. 2010, 4, 63–69. [Google Scholar] [CrossRef] [PubMed]

| Method | Cell Type | Ligand Type | Nanoscopic Control | Tumor Progression-Related Outcome | Reference |
|---|---|---|---|---|---|
| BCML (Block-Copolymer Nano-Lithography) | Human melanoma cells | RGD | - Monovalent - global density - spatial distance | Definition of optimum ligand density | [77] |
| Human melanoma cells | VCAM-1 (plus RGD) | - monovalent (plus RGD background) - global density - spatial distance | Antagonistic function of VCAM-1 and RGD upon cell spreading | [78] | |
| Human melanoma cells | N-cadherinE-cadherin | - monovalent - global density - spatial distance | Flexible spreading irrespective of density | [76] | |
| EBL (Electron Beam Lithography) | Human breast cancer cells | Fibronectin (plus Laminin/K-casein background) | - monovalent - gradient | Invadipodia formation | [62] |
| Human breast cancer cells | Fibronectin | - monovalent - gradient | Flexible spreading | [63] | |
| DNA-Origami | Human melanoma cells | - EGF - A20FMDV2 peptide (integrin ligand) | - bivalent - ligand distance - ligand ratio | Cooperative signaling upon spreading | [55] |
| Human breast cancer cells | EGF | - monovalent - ligand number and distribution | EGF ligand architecture determines cellular response | [73] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amschler, K.; Schön, M.P. Modulating Tumor Cell Functions by Tunable Nanopatterned Ligand Presentation. Nanomaterials 2020, 10, 212. https://doi.org/10.3390/nano10020212
Amschler K, Schön MP. Modulating Tumor Cell Functions by Tunable Nanopatterned Ligand Presentation. Nanomaterials. 2020; 10(2):212. https://doi.org/10.3390/nano10020212
Chicago/Turabian StyleAmschler, Katharina, and Michael P. Schön. 2020. "Modulating Tumor Cell Functions by Tunable Nanopatterned Ligand Presentation" Nanomaterials 10, no. 2: 212. https://doi.org/10.3390/nano10020212
APA StyleAmschler, K., & Schön, M. P. (2020). Modulating Tumor Cell Functions by Tunable Nanopatterned Ligand Presentation. Nanomaterials, 10(2), 212. https://doi.org/10.3390/nano10020212





