Metadata Stewardship in Nanosafety Research: Community-Driven Organisation of Metadata Schemas to Support FAIR Nanoscience Data
Abstract
:1. Introduction
1.1. FAIRification: Science, Library Science or Both?
- capturing a snapshot of current NM data and metadata curation practices and issues,
- the development of recommendations for moving the nanoinformatics community toward increasingly standardised curation practices, and
- the facilitation of collaborations between researchers, product developers, and others working with NMs that establish and utilise common datasets for cross-boundary work (e.g., application of data from academic institutions to NM product development in industry).
- To be findable in a scientific context:
- ○
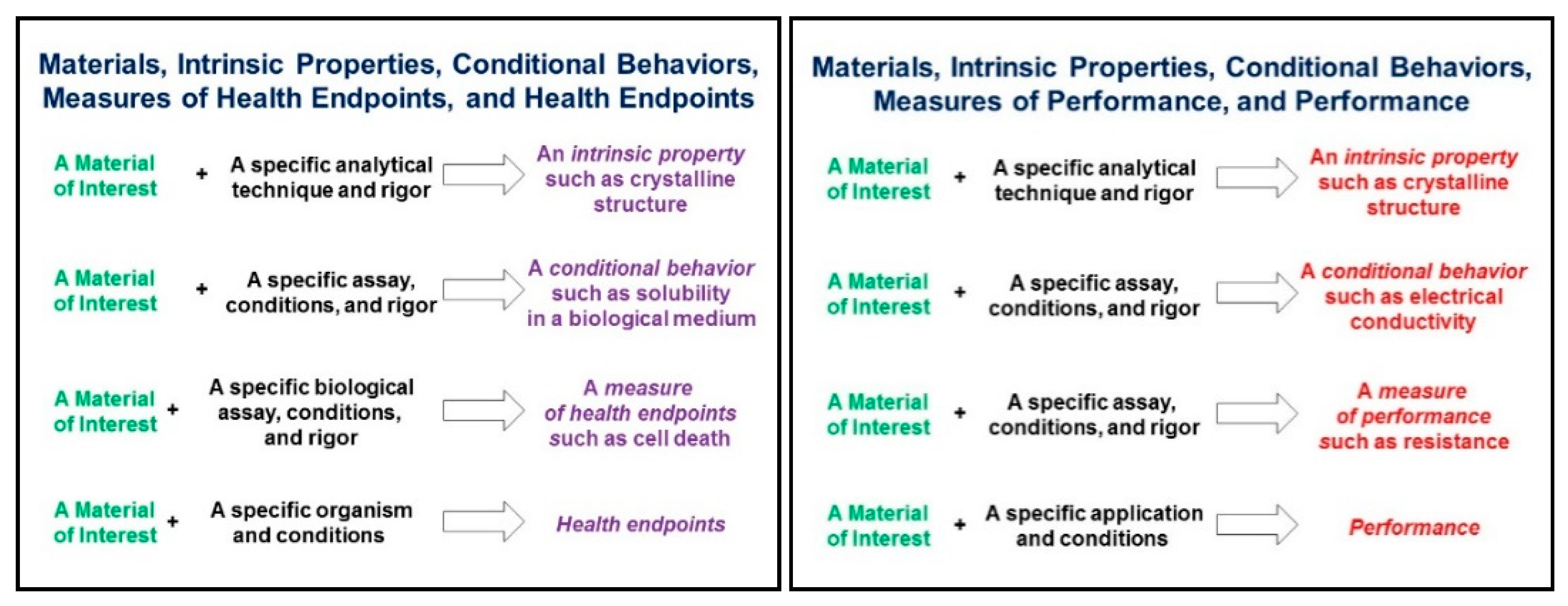
- SF1: Use standard, unambiguous identifiers for characterising your samples, test systems and experimental details, presenting as much information as possible. As per the proposals of the GO FAIR Chemistry Implementation Network [38], coordination and some formalisation is needed to promote interoperability between different types. For example, chemistry-aware identifiers like IUPAC names, PubChem compound identifiers, InChIs and the recently proposed NInChIs (nano-InChIs, see Lynch et al. in this special issue) provide orthogonal information to compound/substance names and CAS RN, and the use of both is preferred.
- To be accessible:
- ○
- SA1: Annotate metadata and data and especially metadata/data schemas with standardised ontologies to make them computer accessible. It would be desirable to create a “dictionary” of terms regularly used and thus to use persistent ontological IDs for the metadata and data produced.
- ○
- SA2: Make sure that the metadata can be accessed from the same resource as the data. If the data warehouse is not flexible enough to provide the scientific metadata through its metadata access functionalities, provide it in a standardised way (either standard file formats like ISA or supplemented by a clear access protocol—see recommendations for further details).
- ○
- SA3: Provide your protocols in FAIR resources, in addition to the materials and methods section of the corresponding paper. Remember that your data might be used in another context and exact descriptions are needed. Linking protocols to data via electronic laboratory notebooks is one approach to achieve this.
- ○
- SA4: Document small deviations from the original/standardised protocol with your metadata/data. For example, if different samples are using different DMSO concentrations for controlling the production and properties of nanomaterials, this should be reported per sample in contrast to the protocol where only the DMSO range will be given.
- To be interoperable:
- ○
- SI1 links to descriptions of the test methods, protocols and quality control measures: Provide direct res to give the user the chance to evaluate data interoperability. In this way, additional information, which cannot all be covered by the metadata can be easily accessed.
- ○
- SI2: Report protocol metadata in a structured and annotated way to allow harmonisation and interlinking of data. Even if duplication of information in the protocol and the metadata is sometimes needed or even preferred, guarantee consistency between both.
- To be reusable:
- ○
- SR1: Do not limit the reported metadata to fulfil only the requirements of the study for which the data was produced. Sections 6 and 7 provide examples on the usage of data in a different computational context than the experimental initially intended.
- ○
- SR2: Establish a feedback loop between data creators, analysts and customers to continuously improve the metadata completeness and quality. Keep in mind that scientific progress can lead to new use cases and go beyond “standards” defined at a specific point of time.
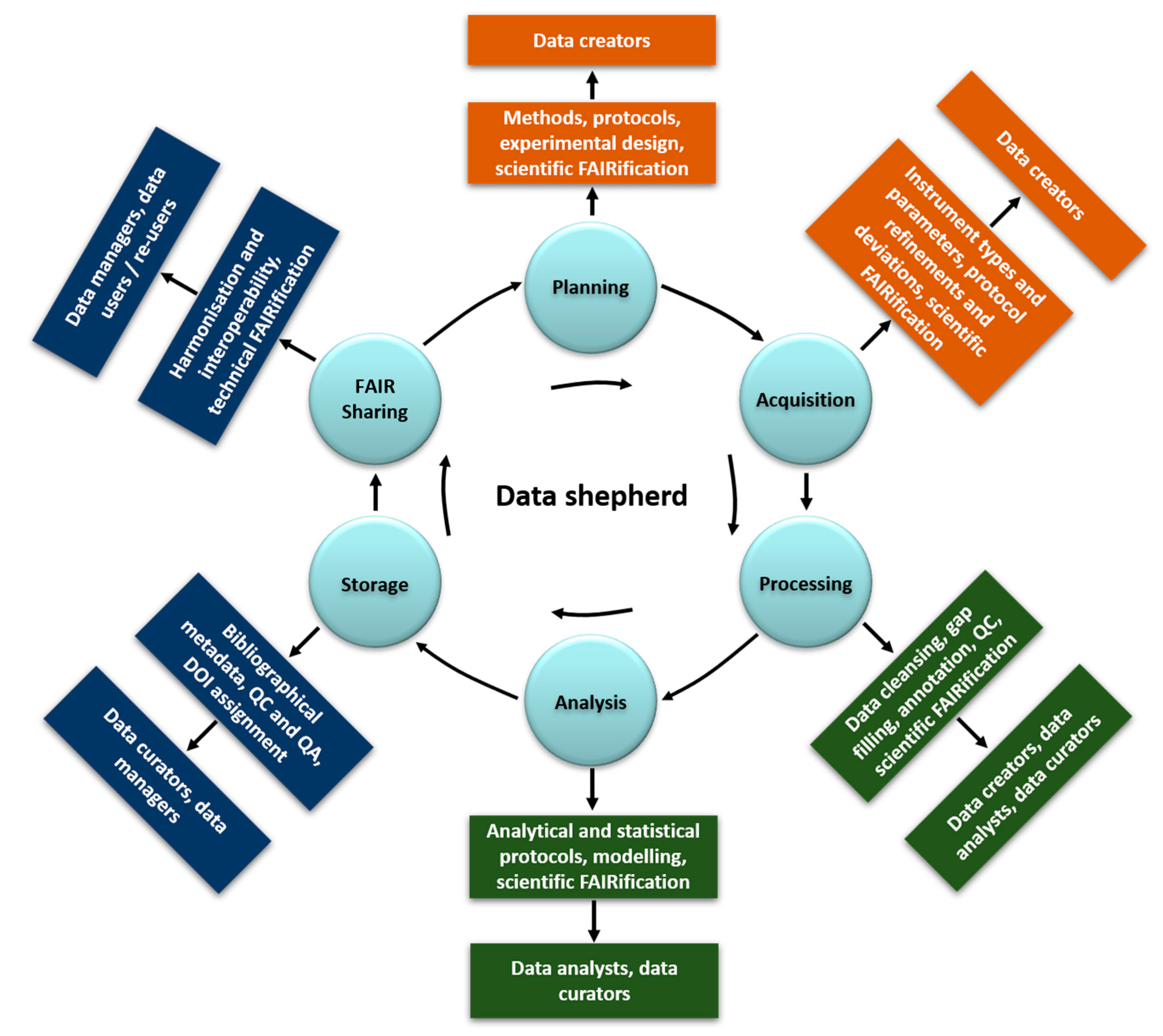
1.2. Roles and Responsibilities in (Meta) Data Collection, Curation and Accessibility, Their Dependencies and the Need for Data Shepherds
- Data customers: requestors, accessors, users, and re-users of the needed or produced data (evaluation of the scientific and technical FAIRification step by testing for the final goal of usability and reusability in real applications)
- Data creator: the experimentalists planning and generating the data (planning, acquisition, and manipulation in the data lifecycle, scientific FAIRification steps in Figure 2)
- Data analyst: data handling, manipulation, analysis including modelling (processing and analysis, scientific FAIRification steps in Figure 2)
- Data curator: data and metadata capturing and quality and completeness control (data manipulation and storage in Figure 2)
- Data manager: data digitisation in a structured and harmonised format. Metadata implementation and link to data (storage and technical FAIRification steps)
- Data shepherd: a new role strongly encouraged here, which is defined in detail below, who operates throughout the data lifecycle.
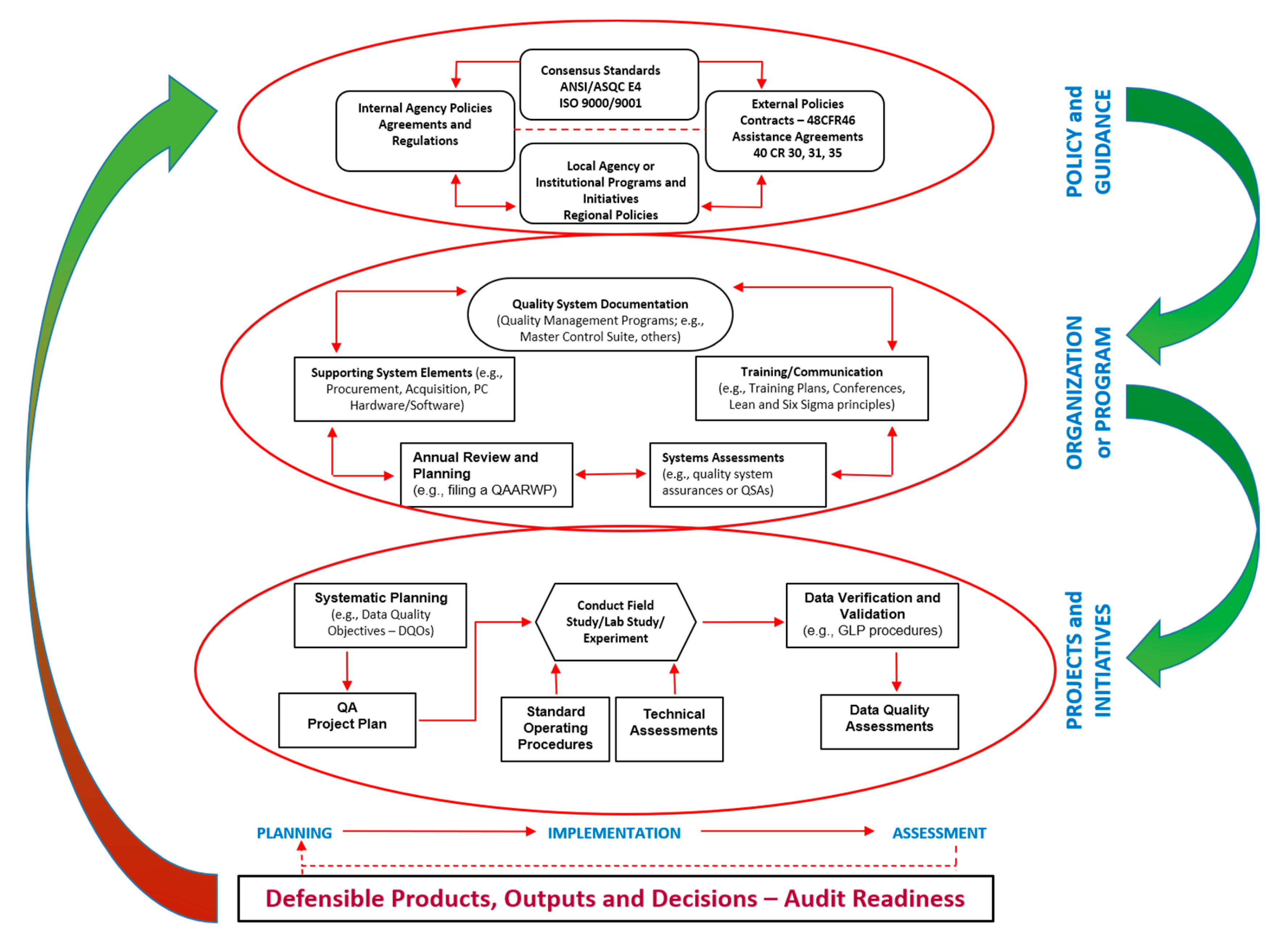
1.3. Quality Management Concepts and Systems—Relevant to NMs and Nanosafety Assessment
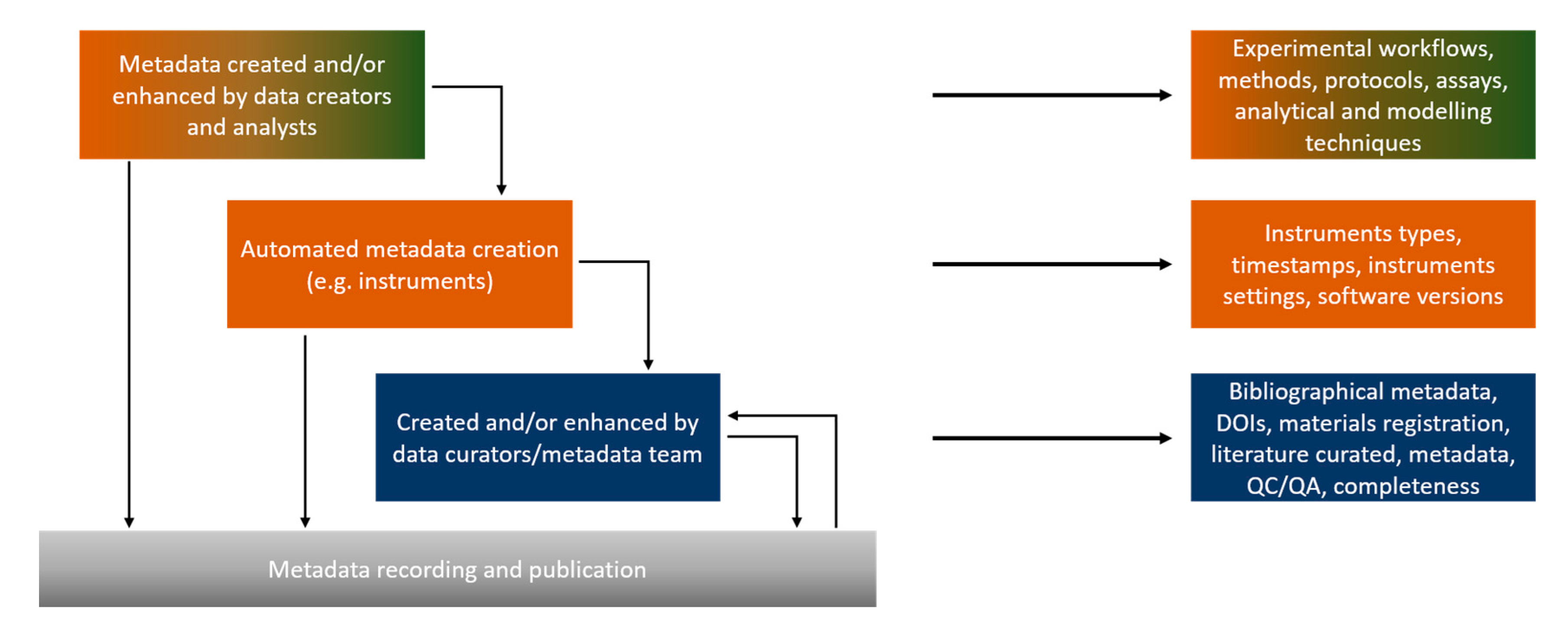
1.4. Metadata Types, Schemas, Standards and Semantic Annotation
- Reference metadata: metadata describing the contents and the quality of the statistical data [55].
- Structural metadata: metadata acting as identifiers and descriptors of the data [56], so include ontological aspects such as assay and NM type, instruments etc.
- Statistical metadata: scientific metadata about statistical data [57].
- Bibliographic metadata, which include the necessary administrative information for the presented data, i.e., the dataset owner and contact details, the license, the publication status etc.
- Technical metadata: information on the data file types, the size of data, dates of creation and/or modification, types of compression and more, mostly related with databases and interoperability.
2. Materials and Methods
2.1. Metadata Databases Questionnaire to Build Community-Driven Consensus on Metadata for Nanosafety
- Project databases: NanoCommons Knowledge base, ACEnano, IOM (representing a number of FP7 and H2020 projects including MARINA and PATROLS);
- Institutional or “Centre” databases: Nanomaterial-Biological Interactions (NBI) Knowledge base, Safe and Sustainable Nanotechnology (S2NANO), RIVM-ECOTOX and the Center for the Environmental Interaction of Nanomaterials NanoInformatics Knowledge Commons (CEINT-NIKC).
2.2. Case Studies to Demonstrate the Value and Relevance of Metadata in Nanosafety Data Harmonisation
2.2.1. Minimum Information Reporting on Nanoparticle (NP) Agglomeration as Source for In Vitro Delivered Dose Variations Critical to Human Hazard Assessment
Objectives of This Case Study
- To determine the impact of a few different agglomeration scenarios (primary particle vs. well-defined agglomerate vs. three different mixtures thereof) of two types of NPs (TiO2 and SiO2) on biological in vitro endpoints.
- To collect (meta)data regarding particle agglomeration, which are relevant for in vitro experimentation using adherent cellular models.
- To define a minimal set of information (data and metadata) most relevant for NP agglomeration to facilitate interpretation of DD in in vitro bioassays.
In Silico Modelling of NP Agglomeration
2.2.2. NMs Dissolution: Achieving Consensus on Terminology and Metadata Usage
3. Results
3.1. Community-Driven Consensus on Metadata for Nanosafety
3.1.1. Data Coverage
3.1.2. Metadata and Data Templates
3.1.3. New Challenges Arising from Nanoinformatics
3.1.4. Data Management, FAIRification and Reusability
3.1.5. Metadata on Test Methods and Protocols
3.1.6. Quality Assurance and Quality Control
3.1.7. Metadata Awareness and Training
3.2. Case Studies to Demonstrate the Value and Relevance of Metadata in Nanosafety Data Harmonisation
3.2.1. Essential Information Reporting on NP Agglomeration as a Source of In Vitro Delivered Dose Variations Critical to Human Hazard Assessment
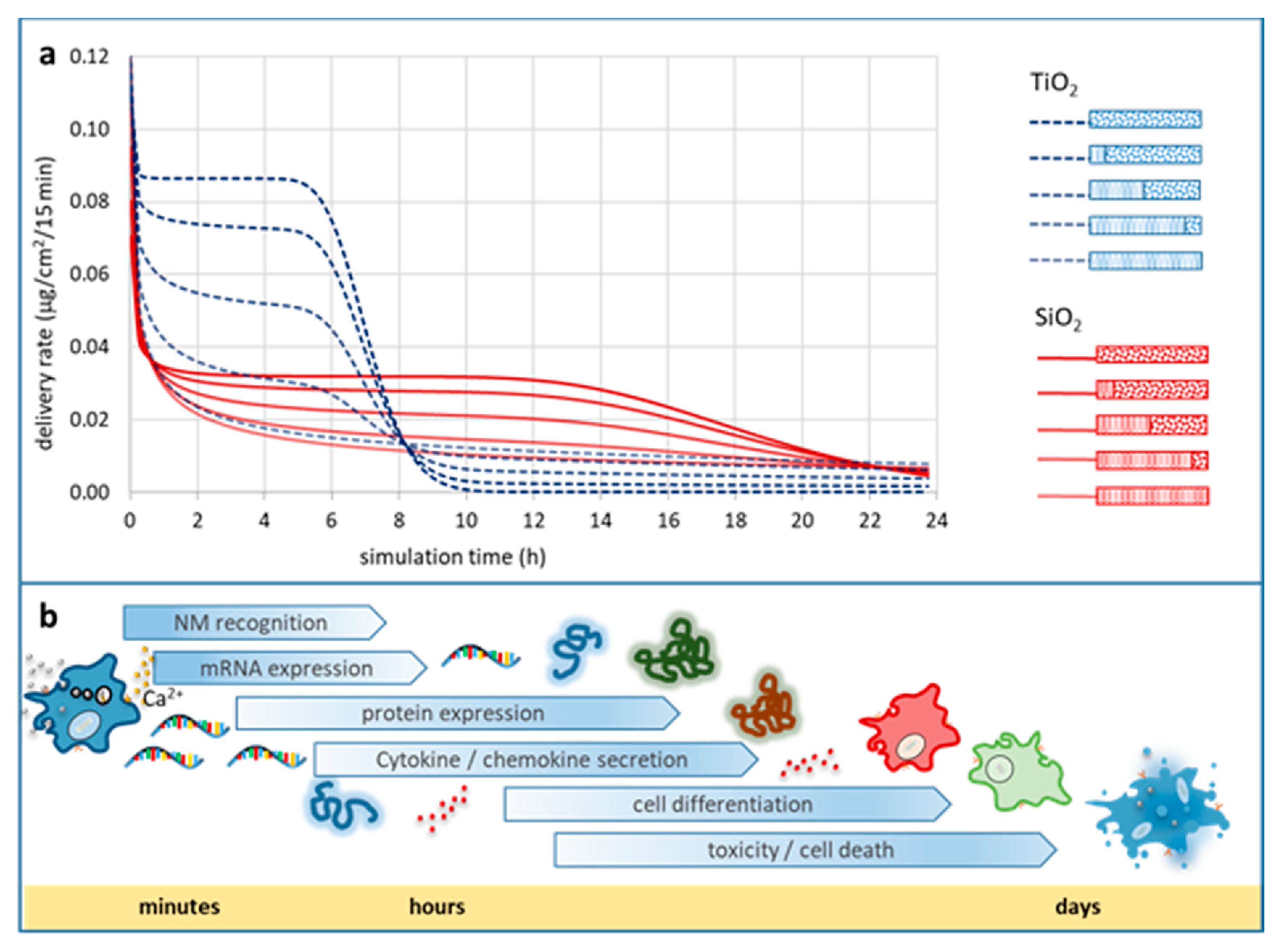
Impact of Particle Agglomeration on the Time Scales of Biologic Responses
Metadata on Particle Agglomeration Relevant for In Vitro Bioassays
Essential Information Set for Bioassays Enabling Better Data FAIRness and Reproducibility
- At one end of the continuum are (meta)data, which cannot be determined with reasonable effort within an experimental setup (e.g., shape, primary particle density). Experimental confirmation, however, could be replaced by well-reasoned suggestions for these without introducing a wide margin of error to the dataset and the experimental outcomes.
- At the other end of the continuum are (meta)data with a high impact on the confidence in the results from the respective bioassay, as they render the experimental outcomes sensitive to high variations (e.g., packing factor, effective density of agglomerates, agglomerate stability). Therefore, a thorough physicochemical characterisation is essential for these NP properties, and metadata have to include all information needed to facilitate for e.g., assessment of the in vitro DD or for in vivo-to-in vitro dose bridging studies.
3.2.2. NMs Dissolution: Achieving Consensus on Terminology and Metadata Usage
- Terms and definitions: There is consensus that the terms dissolution, solubility and leaching, are pertinent to nanosafety and warrant specific definitions for this field. Respondents either accepted the suggested definitions or offered improvements, but did not propose additional terms. There was less acceptance with the suggested visualisation of a NP, though it may yet be a useful tool in prompting a fuller description of the particle/nanoform under study. The visualisation was for a core-shell NP with a nanoscale surface coating. Further labelling and explanation are needed. Two colleagues viewed the dissolution definition as vague relative to that for melting.
- Suggested unit of measurement: There is near consensus that it would be desirable to have an accepted unit of measurement with caveats on normalising to particle surface area (flux) when measurements are usually reported as solution concentration. Those interested in therapy and toxicity focus more on the release of drug active or toxicant (dissolution rate in the questionnaire) and less so on poorly soluble carrier materials. Flux is more acceptable when the whole of the particle is of interest. The most relevant time scale will vary by experiment.
- Catalogue of competing reactions: There is consensus that there are competing reactions to be considered in the experimental design (no one challenged the concept and additional phenomena were suggested); reasonable agreement that the investigator should express their results in the form of a chemical reaction. The range of comments is similar to the distinctions made above for dissolution rate and dissolution flux.
- Induction time effects: There is consensus that the concept is pertinent to experimental design and nanoform architecture (NP structure). As with competing reactions, no one challenged the concept and several proposed additional sources of induction effects.
- Catalogue of media: Significant commentary from respondents that the media listed in the survey are prominent, but the number of suggestions for additional media implies that a general approach on listing metadata should be pursued over listing media. There was some agreement on listing media according to pathways, but the purpose for selecting a medium was paramount.
- Catalogue of current standardised methods: Significant commentary as a number of respondents were not aware of the extent of this listing, especially those with a nanosafety background not being aware of the test methods used in drug development.
- Catalogue of calculational models used for data interpretation: The listed models are pertinent, but significant commentary indicates that they are not used widely in the nanosafety community and still tied to the investigator’s disciplinary field rather than generalised.
- Five respondents mentioned silver as the material with the most complete data set and two proposed zinc oxide (Question 10c).
- Terms and Definitions: distinction between solubility and dissolution similar to the definitions used in the questionnaire. Many terms are used to express nuances: dissolution; ion-leaching; shedding of ions; released ions; dynamic (non-equilibrium); quasi-dynamic system; non-equilibrium; solubility; equilibrium; static (equilibrium); static-solubility; solubility limit; and supersaturation.
- Units of measurement: ng/cm2/h and %/day; stoichiometry used for chemical reaction.
- Competing reactions: not discussed as such. Terms used: binding events; secondary NPs; re-precipitation; recrystallisation; bioprocessing; transformation; modulation of biopersistence; and complex re-speciation.
- Induction time effects: not discussed as such, but indicated through terms that reflect the trajectory from under-saturation to supersaturation, such as: slowing onset of dissolution; saturation-related events; Ostwald ripening; and concentration limited. The length of time to be associated with a specific induction effect is not yet established.
- Media: not on the questionnaire’s list. Terms used are: physiological buffer; phagolysomal simulant fluid; receptor medium; dissolution buffers; eluates; simulant fluids; pH 4.5.
- Methods: flow-through method was on the questionnaire’s list; flow-by method is not. Terms include: flow-by abiotic and flow-by dialysis; and flow-through abiotic dissolution.
- Models: first order kinetics using stoichiometry to relate barium ion concentration in the existing medium to solid mass.
3.3. Conclusions on the Presented Case Studies
- Whether common metadata for both experimental and in silico workflows can be identified that can fully explain a phenomenon;
- Whether consensus can be reached between experts from different scientific fields on a particular subject regarding the necessary metadata and approaches.
4. Discussion on Metadata Challenges and Recommendations for the Nano-Community
4.1. Metadata Related Challenges for the Nano-Community Need Ongoing Attention and Mitigation
4.2. (Meta) Data Generation and Capture Need to Be Implemented into DMPs along with the Use of Modern (Meta) Data Capturing Tools (ELNs)
4.3. Relevant Databases Need to Make Metadata Submission Mandatory and Implement QA Processes
4.4. Community Alignment Is Needed with Respect to Ontological Development
4.5. Community Consensus Is Needed to Promote Scientific and Technical Data FAIRness
4.6. Change in Scientific Mindset and Re-Education Is Needed to Support Implementation of the Scientific FAIR Principles
4.7. Community Collaboration and Introduction of Data Shepherds Are Needed to Accelerate Progress
4.8. User-Friendly Tools Need to Be Developed and Implement
4.9. Recognise That Creating and Adhering to Community Standards Is Not Easy
“Standards are like toothbrushes—everyone has one, but no one wants to use someone else’s.”
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- European Commission: Commission Recommendation on a Code of Conduct for Responsible Nanosciences and Nanotechnologies Research and Council Conclusions on Responsible Nanosciences and Nanotechnologies Research. Available online: http://ec.europa.eu/research/science-society/document_library/pdf_06/nanocode-apr09_en.pdf (accessed on 17 March 2020).
- Organisation for Economic Co-operation and Development: Responsible Development of Nanotechnology. Summary Results from a Survey Activity DSTI/STP/NANO(2013)9/FINAL. Available online: http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=dsti/stp/nano(2013)9/final&doclanguage=en (accessed on 17 March 2020).
- Izak-Nau, E.; Huk, A.; Reidy, B.; Uggerud, H.; Vadset, M.; Eiden, S.; Voetz, M.; Himly, M.; Duschl, A.; Dusinska, M.; et al. Impact of storage conditions and storage time on silver nanoparticles’ physicochemical properties and implications for their biological effects. RSC Adv. 2015, 5, 84172–84185. [Google Scholar] [CrossRef] [Green Version]
- Lynch, I.; Lee, R.G. In Support of the Inclusion of Data on Nanomaterials Transformations and Environmental Interactions into Existing Regulatory Frameworks. In Managing Risk in Nanotechnology: Topics in Governance, Assurance and Transfer; Murphy, F., McAlea, E.M., Mullins, M., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 145–169. [Google Scholar] [CrossRef]
- Mitrano, D.M.; Motellier, S.; Clavaguera, S.; Nowack, B. Review of nanomaterial aging and transformations through the life cycle of nano-enhanced products. Environ. Int. 2015, 77, 132–147. [Google Scholar] [CrossRef] [PubMed]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical−Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Lowry, G.V.; Gregory, K.B.; Apte, S.C.; Lead, J.R. Transformations of Nanomaterials in the Environment. Environ. Sci. Technol. 2012, 46, 6893–6899. [Google Scholar] [CrossRef]
- Avramescu, M.-L.; Rasmussen, P.E.; Chénier, M.; Gardner, H.D. Influence of pH, particle size and crystal form on dissolution behaviour of engineered nanomaterials. Environ. Sci. Pollut. Res. 2017, 24, 1553–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, S.F.; Sørensen, S.N.; Skjolding, L.M.; Hartmann, N.B.; Baun, A. Revising REACH guidance on information requirements and chemical safety assessment for engineered nanomaterials for aquatic ecotoxicity endpoints: Recommendations from the EnvNano project. Environ. Sci. Eur. 2017, 29, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Chemicals Agency: Get Ready for New REACH Requirements for Nanomaterials. Available online: https://echa.europa.eu/-/get-ready-for-new-reach-requirements-for-nanomaterials (accessed on 17 March 2020).
- United States Environmental Protection Agency: Control of Nanoscale Materials under the Toxic Substances Control Act. Available online: https://www.epa.gov/reviewing-new-chemicals-under-toxic-substances-control-act-tsca/control-nanoscale-materials-under (accessed on 17 March 2020).
- Steinhäuser, K.G.; Sayre, P.G.; Nowack, B. Reliability of methods and data for regulatory assessment of nanomaterial risks. NanoImpact 2018, 10, 68–69. [Google Scholar] [CrossRef]
- Organisation for Economic Co-Operation and Development: Series on the Safety of Manufactured Nanomaterials: Alternative Testing Strategies in Risk Assessment of Manufactured Nanomaterials: Current State of Knowledge and Research needs to Advance their Use (ENV/JM/MONO(2016)63). Available online: http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=ENV/JM/MONO(2016)63&doclanguage=en (accessed on 17 March 2020).
- Organisation for Economic Co-Operation and Development: Series on the Safety of Manufactured Nanomaterials: Strategies, Techniques and Sampling Protocols for Determining the Concentrations of Manufactured Nanomaterials in Air at the Workplace (ENV/JM/MONO(2017)30). Available online: http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono(2017)30&doclanguage=en (accessed on 17 March 2020).
- Stone, V.; Johnston, H.J.; Balharry, D.; Gernand, J.M.; Gulumian, M. Approaches to Develop Alternative Testing Strategies to Inform Human Health Risk Assessment of Nanomaterials. Risk Anal. 2016, 36, 1538–1550. [Google Scholar] [CrossRef]
- Burden, N.; Aschberger, K.; Chaudhry, Q.; Clift, M.J.D.; Doak, S.H.; Fowler, P.; Johnston, H.; Landsiedel, R.; Rowland, J.; Stone, V. The 3Rs as a framework to support a 21st century approach for nanosafety assessment. Nano Today 2017, 12, 10–13. [Google Scholar] [CrossRef]
- Berggren, E.; White, A.; Ouedraogo, G.; Paini, A.; Richarz, A.N.; Bois, F.Y.; Exner, T.; Leite, S.; Grunsven, L.A.V.; Worth, A.; et al. Ab initio chemical safety assessment: A workflow based on exposure considerations and non-animal methods. Comput. Toxicol. 2017, 4, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Rogiers, V.; Benfenati, E.; Bernauer, U.; Bodin, L.; Carmichael, P.; Chaudhry, Q.; Coenraads, P.J.; Cronin, M.T.D.; Dent, M.; Dusinska, M.; et al. The way forward for assessing the human health safety of cosmetics in the EU—Workshop proceedings. Toxicology 2020, 436, 152421. [Google Scholar] [CrossRef] [PubMed]
- Drasler, B.; Sayre, P.; Steinhäuser, K.G.; Petri-Fink, A.; Rothen-Rutishauser, B. In vitro approaches to assess the hazard of nanomaterials. NanoImpact 2017, 8, 99–116. [Google Scholar] [CrossRef]
- Oberdörster, G.; Kuhlbusch, T.A.J. In vivo effects: Methodologies and biokinetics of inhaled nanomaterials. NanoImpact 2018, 10, 38–60. [Google Scholar] [CrossRef]
- Cohen, Y.; Rallo, R.; Liu, R.; Liu, H.H. In Silico Analysis of Nanomaterials Hazard and Risk. ACC Chem. Res. 2013, 46, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Varsou, D.-D.; Afantitis, A.; Tsoumanis, A.; Melagraki, G.; Sarimveis, H.; Valsami-Jones, E.; Lynch, I. A safe-by-design tool for functionalised nanomaterials through the Enalos Nanoinformatics Cloud platform. Nanoscale Adv. 2019, 1, 706–718. [Google Scholar] [CrossRef] [Green Version]
- Afantitis, A.; Melagraki, G.; Tsoumanis, A.; Valsami-Jones, E.; Lynch, I. A nanoinformatics decision support tool for the virtual screening of gold nanoparticle cellular association using protein corona fingerprints. Nanotoxicology 2018, 12, 1148–1165. [Google Scholar] [CrossRef] [PubMed]
- Melagraki, G.; Afantitis, A. A Risk Assessment Tool for the Virtual Screening of Metal Oxide Nanoparticles through Enalos InSilicoNano Platform. Curr. Top. Med. Chem. 2015, 15, 1827–1836. [Google Scholar] [CrossRef]
- OECD. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models. Available online: https://www.oecd.org/env/guidance-document-on-the-validation-of-quantitative-structure-activity-relationship-q-sar-models-9789264085442-en.htm (accessed on 7 October 2020).
- Powers, C.M.; Mills, K.A.; Morris, S.A.; Klaessig, F.; Gaheen, S.; Lewinski, N.; Hendren, C.O. Nanocuration workflows: Establishing best practices for identifying, inputting, and sharing data to inform decisions on nanomaterials. Beilstein J. Nanotechnol. 2015, 6, 1860–1871. [Google Scholar] [CrossRef] [Green Version]
- Hendren, C.O.; Powers, C.M.; Hoover, M.D.; Harper, S.L. The Nanomaterial Data Curation Initiative: A collaborative approach to assessing, evaluating, and advancing the state of the field. Beilstein J. Nanotechnol. 2015, 6, 1752–1762. [Google Scholar] [CrossRef]
- Marchese Robinson, R.L.; Lynch, I.; Peijnenburg, W.; Rumble, J.; Klaessig, F.; Marquardt, C.; Rauscher, H.; Puzyn, T.; Purian, R.; Åberg, C.; et al. How should the completeness and quality of curated nanomaterial data be evaluated? Nanoscale 2016, 8, 9919–9943. [Google Scholar] [CrossRef]
- Karcher, S.; Willighagen, E.L.; Rumble, J.; Ehrhart, F.; Evelo, C.T.; Fritts, M.; Gaheen, S.; Harper, S.L.; Hoover, M.D.; Jeliazkova, N.; et al. Integration among databases and data sets to support productive nanotechnology: Challenges and recommendations. NanoImpact 2018, 9, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Hoover, M.D.; Cash, L.J.; Feitshans, I.L.; Hendren, C.O.; Harper, S.L. Chapter 5-A Nanoinformatics Approach to Safety, Health, Well-Being, and Productivity. In Nanotechnology Environmental Health and Safety (Third Edition); Hull, M.S., Bowman, D.M., Eds.; William Andrew Publishing: Boston, MA, USA, 2018; pp. 83–117. [Google Scholar] [CrossRef]
- Thomas, D.G.; Gaheen, S.; Harper, S.L.; Fritts, M.; Klaessig, F.; Hahn-Dantona, E.; Paik, D.; Pan, S.; Stafford, G.A.; Freund, E.T.; et al. ISA-TAB-Nano: A Specification for Sharing Nanomaterial Research Data in Spreadsheet-based Format. BMC Biotechnol. 2013, 13, 2. [Google Scholar] [CrossRef] [Green Version]
- National Cancer Institute - ISA-TAB-Nano. Available online: https://wiki.nci.nih.gov/display/ICR/ISA-TAB-Nano#ISA-TAB-Nano-title (accessed on 18 May 2020).
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, S.K.; Porter, A.L.; Youtie, J.; Shapira, P. Capturing new developments in an emerging technology: An updated search strategy for identifying nanotechnology research outputs. Scientometrics 2013, 95, 351–370. [Google Scholar] [CrossRef]
- Hjørland, B. Theories are Knowledge Organizing Systems (KOS). Knowl. Organ. 2015, 42, 113–128. [Google Scholar] [CrossRef]
- Hjørland, B. Data (with Big Data and Database Semantics). Knowl. Organ. 2018, 45, 685–708. [Google Scholar] [CrossRef]
- Jørn Nielsen, H.; Hjørland, B. Curating research data: The potential roles of libraries and information professionals. J. Doc. 2014, 70, 221–240. [Google Scholar] [CrossRef]
- Coles, S.J.; Frey, J.G.; Willighagen, E.L.; Chalk, S.J. Taking FAIR on the ChIN: The Chemistry Implementation Network. Data Intell. 2020, 2, 131–138. [Google Scholar] [CrossRef]
- Iglesia, D.d.l.; Harper, S.; Hoover, M.D.; Klaessig, F.; Lippell, P.; Maddux, B.; Morse, J.; Nel, A.; Rajan, K.; Reznik-Zellen, R.; et al. Nanoinformatics 2020 Roadmap; National Nanomanufacturing Network: Amherst, MA, USA, 2011. [Google Scholar]
- Haase, A.; Klaessig, F. EU-US Roadmap NanoInformatics 2030; EU NanoSafety Cluster. 2018. Available online: https://zenodo.org/record/1486012#.X4gxnu2-uUl (accessed on 27 March 2020).
- Hoover, M.D.; Myers, D.S.; Cash, L.J.; Guilmette, R.A.; Kreyling, W.G.; Oberdörster, G.; Smith, R.; Cassata, J.R.; Boecker, B.B.; Grissom, M.P. Application of an Informatics-Based Decision-Making Framework and Process to the Assessment of Radiation Safety in Nanotechnology. Health Phys. 2015, 108, 179–194. [Google Scholar] [CrossRef]
- Woodall, G.M.; Hoover, M.D.; Williams, R.; Benedict, K.; Harper, M.; Soo, J.-C.; Jarabek, A.M.; Stewart, M.J.; Brown, J.S.; Hulla, J.E.; et al. Interpreting Mobile and Handheld Air Sensor Readings in Relation to Air Quality Standards and Health Effect Reference Values: Tackling the Challenges. Atmosphere (Basel) 2017, 8, 182. [Google Scholar] [CrossRef] [Green Version]
- Iavicoli, I.; Leso, V.; Ricciardi, W.; Hodson, L.L.; Hoover, M.D. Opportunities and challenges of nanotechnology in the green economy. Environ. Health 2014, 13, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koshovets, O.B.; Ganichev, N.A. Nanotechnology and the new technological revolution: Expectations and reality. Stud. Russ. Econ. Dev. 2017, 28, 391–397. [Google Scholar] [CrossRef]
- European Commission: Open Data. Available online: https://ec.europa.eu/digital-single-market/en/open-data (accessed on 17 March 2020).
- European Commission: H2020 Online Manual: Open Access and Data Management. Available online: https://ec.europa.eu/research/participants/docs/h2020-funding-guide/cross-cutting-issues/open-access-dissemination_en.htm (accessed on 17 March 2020).
- European Commission: H2020 Online Manual: Data Management. Available online: https://ec.europa.eu/research/participants/docs/h2020-funding-guide/cross-cutting-issues/open-access-data-management/data-management_en.htm (accessed on 17 March 2020).
- Skobelev, D.; Zaytseva, T.; Kozlov, A.; Perepelitsa, V.; Makarova, A. Laboratory information management systems in the work of the analytic laboratory. Meas. Tech. 2011, 53, 1182–1189. [Google Scholar] [CrossRef]
- Gibbon, G.A. A brief history of LIMS. J. Lab. Autom. 1996, 32, 1–5. [Google Scholar] [CrossRef]
- Elliott, M. Electronic laboratory notebooks–A foundation for scientific knowledge management. Atrium Res. Consult. 2004. [Google Scholar] [CrossRef]
- Morris, D. ELNs: Arch enemies or best of friends. Drug Discov. World 2009, 24–28. [Google Scholar]
- EPA Requirements for Quality Management Plans, EPA QA/R-2, March 2001, Washington, DC, USA. Available online: https://www.epa.gov/sites/production/files/2016-06/documents/r2-final.pdf (accessed on 2 June 2020).
- International Organization for Standardization: ISO/IEC JTC1 SC32 Working Group: Metadata Standards. Available online: http://metadata-standards.org/ (accessed on 17 March 2020).
- Organisation for Economic Co-Operation and Development: Glossary of Statistical Terms: Metadata. Available online: https://stats.oecd.org/glossary/detail.asp?ID=5136 (accessed on 17 March 2020).
- Organisation for Economic Co-Operation and Development: Glossary of Statistical Terms: Reference Metadata. Available online: https://stats.oecd.org/glossary/detail.asp?ID=7076 (accessed on 17 March 2020).
- Organisation for Economic Co-Operation and Development: Glossary of Statistical Terms: Structural Metadata. Available online: https://stats.oecd.org/glossary/detail.asp?ID=2579 (accessed on 17 March 2020).
- Organisation for Economic Co-Operation and Development: Glossary of Statistical Terms: Statistical Metadata. Available online: https://stats.oecd.org/glossary/detail.asp?ID=1647 (accessed on 17 March 2020).
- Willighagen, E. European Registry of Materials. Available online: https://github.com/NanoCommons/identifiers (accessed on 17 March 2020).
- European Chemicals Agency: Guidance on Information Requirements and Chemical Safety Assessment: Chapter R.4: Evaluation of Available Information. Available online: https://echa.europa.eu/documents/10162/13632/information_requirements_r4_en.pdf (accessed on 17 March 2020).
- Klimisch, H.J.; Andreae, M.; Tillmann, U. A Systematic Approach for Evaluating the Quality of Experimental Toxicological and Ecotoxicological Data. Regul. Toxicol. Pharmacol. 1997, 25, 1–5. [Google Scholar] [CrossRef] [Green Version]
- OECD. Organisation for Economic Co-Operation and Development: OECD Guidelines for the Testing of Chemicals. Available online: https://www.oecd-ilibrary.org/environment/oecd-guidelines-for-the-testing-of-chemicals_72d77764-en (accessed on 18 May 2020).
- Becker, R.A.; Janus, E.R.; White, R.D.; Kruszewski, F.H.; Brackett, R.E. Good Laboratory Practices and Safety Assessments. Environ. Health Perspect. 2009, 117, A482–A483. [Google Scholar] [CrossRef] [Green Version]
- Leonelli, S. Data-Centric Biology: A Philosophical Study; University of Chicago Press: Chicago, IL, USA, 2016. [Google Scholar]
- Willis, C.; Greenberg, J.; White, H. Analysis and synthesis of metadata goals for scientific data. J. Am. Soc. Inform. Sci. 2012, 63, 1505–1520. [Google Scholar] [CrossRef] [Green Version]
- Rumble, J.; Broome, J.; Hodson, S. Building an international consensus on multi-disciplinary metadata standards: A codata case history in nanotechnology. Data Sci. J. 2019, 18. [Google Scholar] [CrossRef] [Green Version]
- Gilchrist, A. Thesauri, taxonomies and ontologies—An etymological note. J. Doc. 2003, 59, 7–18. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.; Pappu, R.; Baker, N. NPO: Ontology for Cancer Nanotechnology Research. Nat. Preced. 2009. [Google Scholar] [CrossRef]
- Hastings, J.; Jeliazkova, N.; Owen, G.; Tsiliki, G.; Munteanu, C.R.; Steinbeck, C.; Willighagen, E. eNanoMapper: Harnessing ontologies to enable data integration for nanomaterial risk assessment. J. Biomed. Semant. 2015, 6, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeliazkova, N.; Chomenidis, C.; Doganis, P.; Fadeel, B.; Grafström, R.; Hardy, B.; Hastings, J.; Hegi, M.; Jeliazkov, V.; Kochev, N. The eNanoMapper database for nanomaterial safety information. Beilstein J. Nanotechnol. 2015, 6, 1609–1634. [Google Scholar] [CrossRef] [Green Version]
- Stoumann, J.; Larsen, M.; Falk, J.; Randrup, A.G.; Schultz, E.A.; Nielsen, S.K. Preliminary Analysis: Introduction of FAIR Data in Denmark; Danish Agency for Science and Higher Education: Copenhagen, Denmark, 2018; Available online: https://ufm.dk/en/publications/2018/filer/preliminary-analysis-introduction-of-fair-data-in-denmark_oxford-research-og-hbs.pdf (accessed on 18 May 2020).
- Carrara, W.; Chan, W.; Fischer, S.; Steenbergen, E.V. European Commission: Creating Value through Open Data: Study on the Impact of Re-Use of Public Data Resources. Available online: https://www.europeandataportal.eu/sites/default/files/edp_creating_value_through_open_data_0.pdf (accessed on 18 May 2020).
- Himly, M.; Geppert, M.; Hofer, S.; Hofstätter, N.; Horejs-Höck, J.; Duschl, A. When Would Immunologists Consider a Nanomaterial to be Safe? Recommendations for Planning Studies on Nanosafety. Small 2020, 16, 1907483. [Google Scholar] [CrossRef] [Green Version]
- Teeguarden, J.G.; Hinderliter, P.M.; Orr, G.; Thrall, B.D.; Pounds, J.G. Particokinetics In Vitro: Dosimetry Considerations for In Vitro Nanoparticle Toxicity Assessments. Toxicol. Sci. 2007, 95, 300–312. [Google Scholar] [CrossRef]
- DeLoid, G.M.; Cohen, J.M.; Pyrgiotakis, G.; Pirela, S.V.; Pal, A.; Liu, J.; Srebric, J.; Demokritou, P. Advanced computational modeling for in vitro nanomaterial dosimetry. Part. Fibre Toxicol. 2015, 12, 32. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.M.; DeLoid, G.M.; Demokritou, P. A critical review of in vitro dosimetry for engineered nanomaterials. Nanomedicine 2015, 10, 3015–3032. [Google Scholar] [CrossRef]
- DeLoid, G.M.; Cohen, J.M.; Pyrgiotakis, G.; Demokritou, P. Preparation, characterization, and in vitro dosimetry of dispersed, engineered nanomaterials. Nat. Protoc. 2017, 12, 355–371. [Google Scholar] [CrossRef]
- Hinderliter, P.M.; Minard, K.R.; Orr, G.; Chrisler, W.B.; Thrall, B.D.; Pounds, J.G.; Teeguarden, J.G. ISDD: A computational model of particle sedimentation, diffusion and target cell dosimetry for in vitro toxicity studies. Part. Fibre Toxicol. 2010, 7, 36. [Google Scholar] [CrossRef] [Green Version]
- SCCS. Scientific Committee on Consumer Safety: Opinion on Solubility of Synthetic Amorphous Silica (SAS), (SCCS/1606/19). Available online: https://ec.europa.eu/health/sites/health/files/scientific_committees/consumer_safety/docs/sccs_o_228.pdf (accessed on 18 May 2020).
- Arts, J.H.E.; Hadi, M.; Irfan, M.-A.; Keene, A.M.; Kreiling, R.; Lyon, D.; Maier, M.; Michel, K.; Petry, T.; Sauer, U.G.; et al. A decision-making framework for the grouping and testing of nanomaterials (DF4nanoGrouping). Regul. Toxicol. Pharmacol. 2015, 71, S1–S27. [Google Scholar] [CrossRef] [Green Version]
- Klaessig, F.C. Dissolution as a paradigm in regulating nanomaterials. Environ. Sci. Nano 2018, 5, 1070–1077. [Google Scholar] [CrossRef]
- MARINA—Managing the Risks of Enginnered Nanomaterials. Available online: https://www.safenano.org/research/marina/ (accessed on 28 May 2020).
- SUN—Sustainable Nanotechnologies Project. Available online: http://www.sun-fp7.eu/ (accessed on 28 May 2020).
- NanoSolutions. Available online: https://nanosolutionsfp7.com/ (accessed on 28 May 2020).
- PATROLS—Advanced Tools for NanoSafety Testing. Available online: https://www.patrols-h2020.eu/ (accessed on 28 May 2020).
- Gracious. Available online: https://www.h2020gracious.eu/ (accessed on 28 May 2020).
- NanoMILE—Engineered Nanomaterial Mechanisms of Interactions with Living Systems and the Environment: A Universal Framework for Safe Nanotechnology. Available online: http://nanomile.eu-vri.eu/ (accessed on 28 May 2020).
- NanoFASE—Nanomaterial Fate and Speciation in the Environment. Available online: http://nanofase.eu/ (accessed on 28 May 2020).
- NanoFARM. Available online: https://research.ce.cmu.edu/nanofarm/ (accessed on 28 May 2020).
- SmartNanoTox—Smart Tools for Gauging Nano Hazards. Available online: http://www.smartnanotox.eu/ (accessed on 28 May 2020).
- Pon, R.K.; Buttler, D.J. METADATA REGISTRY, ISO/IEC 11179. Available online: http://metadata-standards.org/11179/ (accessed on 18 May 2020).
- Dublin Core Metadata Initiative: DCMI Metadata Terms. Available online: https://www.dublincore.org/specifications/dublin-core/dcmi-terms/ (accessed on 17 March 2020).
- The DOI System: ISO 26324. Available online: https://www.doi.org/ (accessed on 17 March 2020).
- Wieczorek, J.; Bloom, D.; Guralnick, R.; Blum, S.; Döring, M.; Giovanni, R.; Robertson, T.; Vieglais, D. Darwin Core: An evolving community-developed biodiversity data standard. PLoS ONE 2012, 7, e29715. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J. Understanding Metadata and Metadata Schemes. Cat. Classif Q. 2005, 40, 17–36. [Google Scholar] [CrossRef]
- Taylor, C.F.; Field, D.; Sansone, S.-A.; Aerts, J.; Apweiler, R.; Ashburner, M.; Ball, C.A.; Binz, P.-A.; Bogue, M.; Booth, T.; et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: The MIBBI project. Nat. Biotech. 2008, 26, 889–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faria, M.; Björnmalm, M.; Thurecht, K.J.; Kent, S.J.; Parton, R.G.; Kavallaris, M.; Johnston, A.P.R.; Gooding, J.J.; Corrie, S.R.; Boyd, B.J.; et al. Minimum information reporting in bio–nano experimental literature. Nat. Nanotechnol. 2018, 13, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Chetwynd, A.J.; Wheeler, K.E.; Lynch, I. Best practice in reporting corona studies: Minimum information about Nanomaterial Biocorona Experiments (MINBE). Nano Today 2019, 28, 100758. [Google Scholar] [CrossRef]
- Olsen, L. Global Change Master Directory: Directory Interchange Format (DIF) Writer’s Guide. Available online: https://gcmd.nasa.gov/r/u/difguide/ (accessed on 18 May 2020).
- Rumble, J.; Freiman, S.; Teague, C. Materials on the Nanoscale - Uniform Description System Version 2.0. Chem. Internat. 2016, 38, 25. [Google Scholar] [CrossRef]
- E3144—Standard Guide for Reporting the Physical and Chemical Characteristics of Nano-Objects. Available online: https://www.astm.org/Standards/E3144.htm (accessed on 2 June 2020).
- E3206—Standard Guide for Reporting the Physical and Chemical Characteristics of a Collection of Nano-Objects. Available online: https://www.astm.org/Standards/E3206.htm (accessed on 2 June 2020).
- E3172—Standard Guide for Reporting Production Information and Data for Nano-Objects. Available online: https://www.astm.org/Standards/E3172.htm (accessed on 2 June 2020).
- Center for the Environmental Implication of Nanotechnology: NanoInformatics Knowledge Commons (NIKC). Available online: https://ceint.duke.edu/research/nikc (accessed on 19 March 2020).
- Waltemath, D.; Adams, R.; Beard, D.A.; Bergmann, F.T.; Bhalla, U.S.; Britten, R.; Chelliah, V.; Cooling, M.T.; Cooper, J.; Crampin, E.J.; et al. Minimum Information About a Simulation Experiment (MIASE). PLoS Comp. Biol. 2011, 7, e1001122. [Google Scholar] [CrossRef]
- Baas, A.F.D. What Makes A Material Function?—Let Me Compute the Ways: Modelling in H2020 LEIT-NMBP Programme Materials and Nanotechnology Projects—Study; Directorate-General for Research and Innovation (European Commission): Brussels, Belgium, 2017. [Google Scholar] [CrossRef]
- Novère, N.L.; Finney, A.; Hucka, M.; Bhalla, U.S.; Campagne, F.; Collado-Vides, J.; Crampin, E.J.; Halstead, M.; Klipp, E.; Mendes, P.; et al. Minimum information requested in the annotation of biochemical models (MIRIAM). Nat. Biotechnol. 2005, 23, 1509–1515. [Google Scholar] [CrossRef] [Green Version]
- Ghedini, E. MODA, A Common Ground for Modelling Data Generalization: Introduction, Use Case and Possible Improvements. Available online: https://emmc.info/moda-workflow-templates/ (accessed on 18 May 2020).
- European Materials Modelling Council: European Materials Modelling Ontology Version 1.0. Available online: https://github.com/emmo-repo/EMMO (accessed on 27 April 2020).
- European Chemicals Agency: Guidance on Information Requirements and Chemical Safety Assessment. Chapter R.6: QSARs and Grouping of Chemicals. Available online: https://echa.europa.eu/documents/10162/13632/information_requirements_r6_en.pdf/77f49f81-b76d-40ab-8513-4f3a533b6ac9 (accessed on 18 May 2020).
- Trinh, T.X.; Choi, J.-S.; Jeon, H.; Byun, H.-G.; Yoon, T.-H.; Kim, J. Quasi-SMILES-Based Nano-Quantitative Structure–Activity Relationship Model to Predict the Cytotoxicity of Multiwalled Carbon Nanotubes to Human Lung Cells. Chem. Res. Toxicol. 2018, 31, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, P. Cytotoxicity of quantum dots: Use of quasiSMILES in development of reliable models with index of ideality of correlation and the consensus modelling. J. Hazard. Mater. 2020, 402, 123777. [Google Scholar] [CrossRef]
- Ahmadi, S. Mathematical modeling of cytotoxicity of metal oxide nanoparticles using the index of ideality correlation criteria. Chemosphere 2020, 242, 125192. [Google Scholar] [CrossRef] [PubMed]
- Qi, R.; Pan, Y.; Cao, J.; Jia, Z.; Jiang, J. The cytotoxicity of nanomaterials: Modeling multiple human cells uptake of functionalized magneto-fluorescent nanoparticles via nano-QSAR. Chemosphere 2020, 249, 126175. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, P. Quantitative structure toxicity analysis of ionic liquids toward acetylcholinesterase enzyme using novel QSTR models with index of ideality of correlation and correlation contradiction index. J. Mol. Liq. 2020, 318, 114055. [Google Scholar] [CrossRef]
- Choi, J.-S.; Trinh, T.X.; Yoon, T.-H.; Kim, J.; Byun, H.-G. Quasi-QSAR for predicting the cell viability of human lung and skin cells exposed to different metal oxide nanomaterials. Chemosphere 2019, 217, 243–249. [Google Scholar] [CrossRef]
- Papadiamantis, A.G.; Jänes, J.; Voyiatzis, E.; Sikk, L.; Burk, J.; Burk, P.; Tsoumanis, A.; Ha, M.K.; Yoon, T.H.; Valsami-Jones, E.; et al. Predicting cytotoxicity of metal oxide nanoparticles using Isalos Analytics platform. Nanomaterials 2020, 10, 2017. [Google Scholar] [CrossRef]
- Jafari, K.; Fatemi, M.H. Application of nano-quantitative structure–property relationship paradigm to develop predictive models for thermal conductivity of metal oxide-based ethylene glycol nanofluids. J. Therm. Anal. Calorim. 2020. [Google Scholar] [CrossRef]
- Jafari, K.; Fatemi, M.H. A new approach to model isobaric heat capacity and density of some nitride-based nanofluids using Monte Carlo method. Adv. Powder Technol. 2020, 31, 3018–3027. [Google Scholar] [CrossRef]
- JRC. JRC QSAR Model Database: (Q)SAR Model Reporting Format Inventory. Available online: https://sourceforge.net/projects/qmrf/ (accessed on 13 October 2020).
- JRC. QMRF Template. Available online: https://github.com/ntua-unit-of-control-and-informatics/QSAR-Models/blob/master/QMRF%20template.md (accessed on 13 October 2020).
- Varsou, D.-D.; Afantitis, A.; Tsoumanis, A.; Papadiamantis, A.; Valsami-Jones, E.; Lynch, I.; Melagraki, G. Zeta-Potential Read-Across Model Utilizing Nanodescriptors Extracted via the NanoXtract Image Analysis Tool Available on the Enalos Nanoinformatics Cloud Platform. Small 2020, 16, 1906588. [Google Scholar] [CrossRef]
- Karatzas, P.; Melagraki, G.; Ellis, L.-J.A.; Lynch, I.; Varsou, D.-D.; Afantitis, A.; Tsoumanis, A.; Doganis, P.; Sarimveis, H. Development of deep learning models for predicting the effects of exposure to engineered nanomaterials on Daphnia Magna. Small 2020. In press. [Google Scholar] [CrossRef]
- Linkert, M.; Rueden, C.T.; Allan, C.; Burel, J.-M.; Moore, W.; Patterson, A.; Loranger, B.; Moore, J.; Neves, C.; MacDonald, D.; et al. Metadata matters: Access to image data in the real world. J. Cell Biol. 2010, 189, 777–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilal, M.; Oh, E.; Liu, R.; Breger, J.C.; Medintz, I.L.; Cohen, Y. Bayesian Network Resource for Meta-Analysis: Cellular Toxicity of Quantum Dots. Small 2019, 15, 1900510. [Google Scholar] [CrossRef]
- Gernand, J.M.; Casman, E.A. The toxicity of carbon nanotubes. Risk Anal. 2014, 34. [Google Scholar]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Oh, E.; Liu, R.; Nel, A.; Gemill, K.B.; Bilal, M.; Cohen, Y.; Medintz, I.L. Meta-analysis of cellular toxicity for cadmium-containing quantum dots. Nat. Nanotechnol. 2016, 11, 479–486. [Google Scholar] [CrossRef]
- Ha, M.K.; Trinh, T.X.; Choi, J.S.; Maulina, D.; Byun, H.G.; Yoon, T.H. Toxicity Classification of Oxide Nanomaterials: Effects of Data Gap Filling and PChem Score-based Screening Approaches. Sci. Rep. 2018, 8, 3141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.K.; Seo, M.; Shin, S.E.; Kim, K.-Y.; Park, J.-W.; No, K.T. Meta-analysis of Daphnia magna nanotoxicity experiments in accordance with test guidelines. Environ. Sci. Nano 2018, 5, 765–775. [Google Scholar] [CrossRef]
- Labouta, H.I.; Asgarian, N.; Rinker, K.; Cramb, D.T. Meta-Analysis of Nanoparticle Cytotoxicity via Data-Mining the Literature. ACS Nano 2019, 13, 1583–1594. [Google Scholar] [CrossRef]
- Wilhelm, S.; Tavares, A.J.; Chan, W.C.W. Reply to “Evaluation of nanomedicines: Stick to the basics”. Nat. Rev. Mater. 2016, 1, 16074. [Google Scholar] [CrossRef]
- Papadiamantis, A.; Farcal, L.; Willighagen, E.; Lynch, I.; Exner, T. D10.1 Initial Draft of Data Management Plan (Open Data Pilot) V 2.0; NanoCommons: Birmingham, UK, 2019. [Google Scholar] [CrossRef]
- Bunakov, V.; Griffin, T.; Matthews, B.; Cozzini, S. Metadata for Experiments in Nanoscience Foundries; Springer: Cham, Switzerland, 2017; pp. 248–262. [Google Scholar]
- Biomax. Browse Available Ontologies. Available online: https://ssl.biomax.de/nanocommons/bioxm_portal/bin/view/BioXM/Ontologies (accessed on 27 May 2020).
- Kodali, V.; Littke, M.H.; Tilton, S.C.; Teeguarden, J.G.; Shi, L.; Frevert, C.W.; Wang, W.; Pounds, J.G.; Thrall, B.D. Dysregulation of Macrophage Activation Profiles by Engineered Nanoparticles. ACS Nano 2013, 7, 6997–7010. [Google Scholar] [CrossRef]
- Rodriguez-Lorenzo, L.; Rothen-Rutishauser, B.; Petri-Fink, A.; Balog, S. Nanoparticle Polydispersity Can Strongly Affect In Vitro Dose. Part. Part. Syst. Char. 2015, 32, 321–333. [Google Scholar] [CrossRef]
- Hirota, K.; Terada, H. Endocytosis of particle formulations by macrophages and its application to clinical treatment. Molecular regulation of endocytosis. InTech 2012, 1, 1. [Google Scholar] [CrossRef] [Green Version]
- Park, M.V.D.Z.; Neigh, A.M.; Vermeulen, J.P.; de la Fonteyne, L.J.J.; Verharen, H.W.; Briedé, J.J.; van Loveren, H.; de Jong, W.H. The effect of particle size on the cytotoxicity, inflammation, developmental toxicity and genotoxicity of silver nanoparticles. Biomaterials 2011, 32, 9810–9817. [Google Scholar] [CrossRef] [PubMed]
- Waters, K.M.; Masiello, L.M.; Zangar, R.C.; Tarasevich, B.J.; Karin, N.J.; Quesenberry, R.D.; Bandyopadhyay, S.; Teeguarden, J.G.; Pounds, J.G.; Thrall, B.D. Macrophage Responses to Silica Nanoparticles are Highly Conserved Across Particle Sizes. Toxicol. Sci. 2008, 107, 553–569. [Google Scholar] [CrossRef] [Green Version]
- Keller, J.G.; Graham, U.M.; Koltermann-Jülly, J.; Gelein, R.; Ma-Hock, L.; Landsiedel, R.; Wiemann, M.; Oberdörster, G.; Elder, A.; Wohlleben, W. Predicting dissolution and transformation of inhaled nanoparticles in the lung using abiotic flow cells: The case of barium sulfate. Sci. Rep. 2020, 10, 458. [Google Scholar] [CrossRef] [PubMed]
- Dair, B.J.; Saylor, D.M.; Cargal, T.E.; French, G.R.; Kennedy, K.M.; Casas, R.S.; Guyer, J.E.; Warren, J.A.; Kim, C.-S.; Pollack, S.K. The Effect of Substrate Material on Silver Nanoparticle Antimicrobial Efficacy. J. Nanosci. Nanotechnol. 2010, 10, 8456–8462. [Google Scholar] [CrossRef]
- Pettibone, J.M.; Adamcakova-Dodd, A.; Thorne, P.S.; O’Shaughnessy, P.T.; Weydert, J.A.; Grassian, V.H. Inflammatory response of mice following inhalation exposure to iron and copper nanoparticles. Nanotoxicology 2008, 2, 189–204. [Google Scholar] [CrossRef]
- Koltermann-Jülly, J.; Keller, J.G.; Vennemann, A.; Werle, K.; Müller, P.; Ma-Hock, L.; Landsiedel, R.; Wiemann, M.; Wohlleben, W. Abiotic dissolution rates of 24 (nano)forms of 6 substances compared to macrophage-assisted dissolution and in vivo pulmonary clearance: Grouping by biodissolution and transformation. NanoImpact 2018, 12, 29–41. [Google Scholar] [CrossRef]
- Guldberg, M.; Christensen, V.R.; Krøis, W.; Sebastian, K. Method for determining in-vitro dissolution rates of man-made vitreous fibres. Glass Sci. Technol. (Frankfurt) 1995, 68, 181–187. [Google Scholar]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans; International Agency for Research on Cancer; World Health Organization. Man-Made Vitreous Fibres; World Health Organization: Lyon, France, 2002. [Google Scholar]
- ISO/TR 19057:2017(en) Nanotechnologies—Use and Application of Acellular In Vitro Tests and Methodologies to Assess Nanomaterial Biodurability. Available online: https://www.iso.org/obp/ui/#iso:std:iso:tr:19057:ed-1:v1:en (accessed on 28 May 2020).
- Marques, M.R.; Loebenberg, R.; Almukainzi, M. Simulated biological fluids with possible application in dissolution testing. Dissolution Technol. 2011, 18, 15–28. [Google Scholar] [CrossRef]
- Mercer, T.T. On the Role of Particle Size in the Dissolution of Lung Burdens. Health Phys. 1967, 13. [Google Scholar] [CrossRef]
- Schmidt, J.; Vogelsberger, W. Dissolution Kinetics of Titanium Dioxide Nanoparticles: The Observation of an Unusual Kinetic Size Effect. J. Phys. Chem. B 2006, 110, 3955–3963. [Google Scholar] [CrossRef] [PubMed]
- Utembe, W.; Potgieter, K.; Stefaniak, A.B.; Gulumian, M. Dissolution and biodurability: Important parameters needed for risk assessment of nanomaterials. Part. Fibre Toxicol. 2015, 12, 11. [Google Scholar] [CrossRef] [Green Version]
- Baker, M. Reproducibility crisis? Nature 2016, 533, 353–366. [Google Scholar]
- Hutson, M. Artificial intelligence faces reproducibility crisis. Science 2018, 359, 725. [Google Scholar] [CrossRef]
- Stark, P.B. Before reproducibility must come preproducibility. Nature 2018, 557, 613–614. [Google Scholar] [CrossRef]
- Jacobsen, A.; Azevedo, R.d.M.; Juty, N.; Batista, D.; Coles, S.; Cornet, R.; Courtot, M.; Crosas, M.; Dumontier, M.; Evelo, C.T.; et al. FAIR Principles: Interpretations and Implementation Considerations. Data Intell. 2020, 2, 10–29. [Google Scholar] [CrossRef]
- Wiemann, S.; Mehrle, A.; Hahne, F.; Hermjakob, H.; Apweiler, R.; Arlt, D.; Bechtel, S.; Bielke, W.; Birmingham, A.; Smith, Q.; et al. MIACA—Minimum Information about a Cellular Assay: Standardized Description of Cell-Based Functional Assay Projects. Available online: http://europepmc.org/abstract/CTX/C6970 (accessed on 18 May 2020).
- Mehrle, A.; Wiemann, S.; Rosenfelder, H. MIACA—Minimum Information about a Cellular Assay. Available online: https://sourceforge.net/projects/miaca/files/ (accessed on 15 May 2020).
- Yan, X.; Sedykh, A.; Wang, W.; Yan, B.; Zhu, H. Construction of a web-based nanomaterial database by big data curation and modeling friendly nanostructure annotations. Nat. Commun. 2020, 11, 2519. [Google Scholar] [CrossRef] [PubMed]


 100:0,
100:0,  80:20,
80:20,  50:50,
50:50,  20:80,
20:80,  0:100; medium: water; dish depth: 0.25 cm; administered dose: 10 µg/mL.
0:100; medium: water; dish depth: 0.25 cm; administered dose: 10 µg/mL.
 100:0,
100:0,  80:20,
80:20,  50:50,
50:50,  20:80,
20:80,  0:100; medium: water; dish depth: 0.25 cm; administered dose: 10 µg/mL.
0:100; medium: water; dish depth: 0.25 cm; administered dose: 10 µg/mL.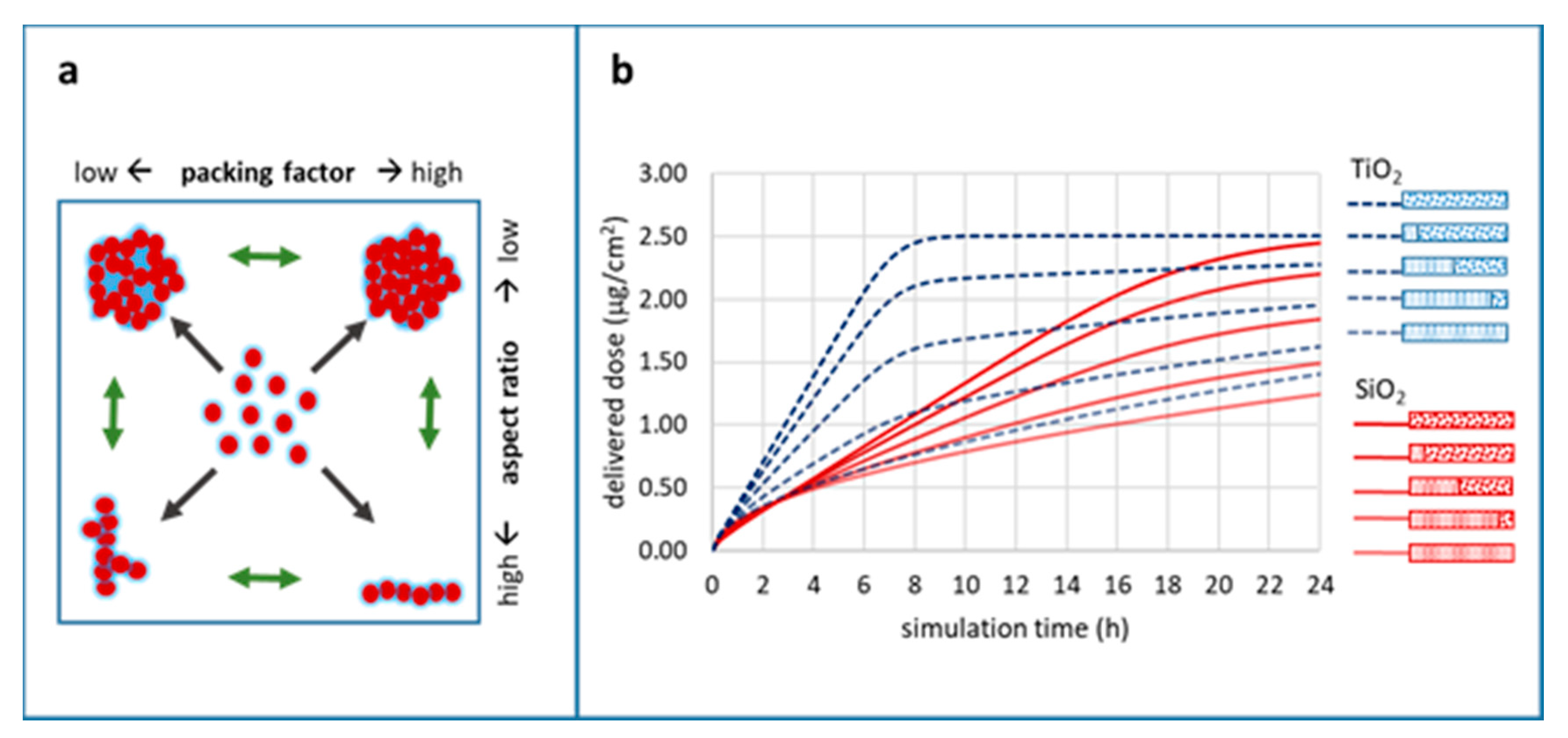
| Set objectives | Design Approach | Collect | Processing | Modelling/Analysis | Validate | Store | Share | Quality Control | Annotation | Determine Relevance | Apply | Confirm Effectiveness | Generalise | Communication and Education | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Creators | X | X | X | X | X | X | X | X | X | X | |||||
| Analysts | X | X | X | X | X | X | X | X | X | X | |||||
| Curators | X | X | X | X | X | X | X | ||||||||
| Managers | X | X | X | X | X | ||||||||||
| Customers | X | X | X | X | X | X | X | ||||||||
| Shepherds | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X |
| Role | Example Term | Definition |
|---|---|---|
| Data creator | Experimental instance | A specific part of an assay or method |
| Data analyst | Training and test instances | A set of specific data entries used for training, testing and validating a predictive model |
| Data curator | NIKC instance | The reported nanomaterial in a system at a specific moment in time |
| Data manager | Database instance | A set of the background processes and memory structure needed by the database software to access the data |
| Data customer | All of the above depending on the specific use case |
| Journal Article | Suggested Data/Metadata/Descriptors | Remarks |
|---|---|---|
| Introduction | Definitions for dissolution, dissolution rate, dissolution profile, and leaching; dissolution stoichiometry; potential for induction effects and competing reactions | Recommended to establish the study’s purpose relative to the literature |
| Materials & Methods | Apparatus relative to standardised test methods; Medium composition; stock dispersion shelf life and solution composition | Explanation that the chosen experimental design achieves the study’s purpose |
| Results—Reporting Units | mg/L/day for the analyte and ng/cm2/hr normalised to the particle surface area; initial and final surface images; and initial dissolution rates & solution compositions; final particle composition | As needed to address the experimental design and to allow for later interoperability and reuse. |
| Discussion | Computational model and characteristic dissolution rate and half-life. | Data analysis and interpretation should be related to study’s purpose |
| References | Sources of terms, apparatus, models | Sufficient to establish a basis for FAIR |
| Data Object | NP Descriptors | (Meta)data | Remarks/Description | Case Study Value |
|---|---|---|---|---|
| Primary particle | Size | Diameter | Diameter of primary particle | 50.0 ± 0 nm |
| Determination method | DLS, NTA, TEM, SEM, … | NTA | ||
| Statistical measure | Mean, mode, median, ... | Mean ± Stdev | ||
| Size qualifier | Hydrodynamic diameter, dried, … | Hydrodynamic diameter | ||
| Shape | Shape of particle (spherical, rod, …) | Spherical | ||
| Aspect ratio | Ratio of sizes in different dimensions | 1 | ||
| Density | Density of primary particle | SiO2: 2.2 g/cm3, TiO2: 4.24 g/cm3 | ||
| Surface charge | Zeta potential of primary particle | −34 mV * | ||
| Porosity | Pore volume fraction | Non porous | ||
| Polydispersity | Polydispersity index, size distribution | Monodisperse | ||
| Dissolution rate | Release rate of molecular monomers and polymers | No significant dissolution | ||
| Synthesis protocol | Protocol of particle synthesis/particle source | Reverse emulsion method: doi:10.1021/la052797 * | ||
| Agglomerate | Size | Diameter | diameter of agglomerate | 250.0 ± 0 nm |
| Determination method | DLS, NTA, TEM, SEM, … | NTA | ||
| Statistical measure | Mean, mode, median, ... | Mean ± Stdev | ||
| Size qualifier | Hydrodynamic diameter, dried, … | Hydrodynamic diameter | ||
| Shape | Shape of agglomerate (spherical, rod, …) | Spherical | ||
| Aspect ratio | Ratio of sizes in different dimensions | 1 | ||
| Packing | Packing factor | Particle fraction of agglomerate volume (intra-agglomerate volume subtracted) | 0.637 | |
| Determination method | Volumetric Centrifugation Method, … | Default value: DOI:10.1186/1743-8977-7-36 | ||
| Effective density | Agglomerate density considering intra-agglomerate fluid | SiO2: 1.76 g/cm3, TiO2: 3.06 g/cm3 | ||
| Polydispersity | Polydispersity index, size distribution | Monodisperse | ||
| Stability | Dissolution, agglomeration or dis-agglomeration over time | Stable | ||
| Experiment | Method | In vivo/in vitro/in silico | In silico | |
| Model | For in silico only: which method is simulated | In vitro, submerged, adherent cells | ||
| Tool | Name | Tool name | ISDD | |
| Version | Software version | Current version | ||
| Reference | Tool/method literature | DOI:10.1186/1743-8977-7-36 | ||
| Medium | Temperature | 310 °K | ||
| Viscosity | 0.00074 Ns/m2 | |||
| Density | 1 g/mL | |||
| Type | Medium type (dH2O, PBS, serum free medium, …) | dH2O * | ||
| Dish depth | Medium height level | 0.25 cm | ||
| Administered dose | Primary particle concentration administered | 10 mg/mL | ||
| Exposure time | Duration of NP exposure/simulation | 24 h * | ||
| Delivered dose | Primary particle mass deposited after exposure time | SiO2: 2.4 mg/cm2 * TiO2: 2.5 mg/cm2 * | ||
| Dispersion protocol | Sample preparation details (sonication, …) | doi:10.3109/17435390.2012.666576 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadiamantis, A.G.; Klaessig, F.C.; Exner, T.E.; Hofer, S.; Hofstaetter, N.; Himly, M.; Williams, M.A.; Doganis, P.; Hoover, M.D.; Afantitis, A.; et al. Metadata Stewardship in Nanosafety Research: Community-Driven Organisation of Metadata Schemas to Support FAIR Nanoscience Data. Nanomaterials 2020, 10, 2033. https://doi.org/10.3390/nano10102033
Papadiamantis AG, Klaessig FC, Exner TE, Hofer S, Hofstaetter N, Himly M, Williams MA, Doganis P, Hoover MD, Afantitis A, et al. Metadata Stewardship in Nanosafety Research: Community-Driven Organisation of Metadata Schemas to Support FAIR Nanoscience Data. Nanomaterials. 2020; 10(10):2033. https://doi.org/10.3390/nano10102033
Chicago/Turabian StylePapadiamantis, Anastasios G., Frederick C. Klaessig, Thomas E. Exner, Sabine Hofer, Norbert Hofstaetter, Martin Himly, Marc A. Williams, Philip Doganis, Mark D. Hoover, Antreas Afantitis, and et al. 2020. "Metadata Stewardship in Nanosafety Research: Community-Driven Organisation of Metadata Schemas to Support FAIR Nanoscience Data" Nanomaterials 10, no. 10: 2033. https://doi.org/10.3390/nano10102033






