A Bibliometric Review of Deep Learning Approaches in Skin Cancer Research
Abstract
1. Introduction
2. Material and Methods
2.1. Goal and Research Question
- RQ1:
- What is the trend in the number of publications on the use of deep learning in skin cancer classification from year to year?
- RQ2:
- Which countries contribute the most publications on deep learning for skin cancer classification, and how is this distributed?
- RQ3:
- Who are the most productive authors in publishing articles on the use of deep learning for skin cancer classification?
- RQ4:
- Which journals publish the most articles on deep learning for skin cancer classification?
- RQ5:
- Which countries have the most cited publications in research on deep learning for skin cancer classification?
- RQ6:
- Which organizations or institutions are the most productive in publishing research on deep learning for skin cancer classification?
- RQ7:
- What are the most cited articles in research on deep learning for skin cancer?
- RQ8:
- What are the most frequently used keywords in research on deep learning in skin cancer classification, and how have their usage patterns or trends evolved over time?
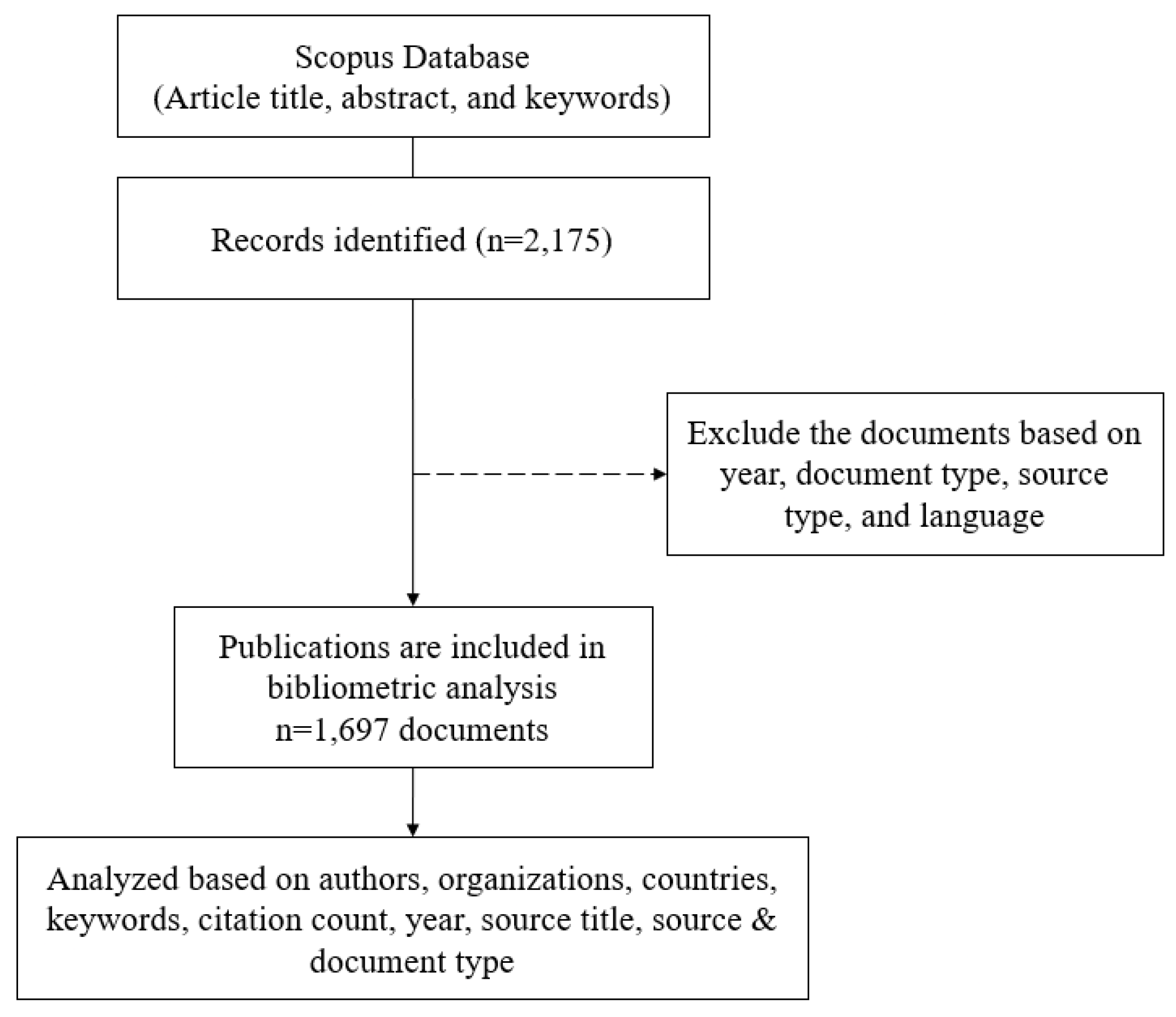
2.2. Data Collection
2.3. Data Exclusion
2.4. Data Analysis
3. Results
3.1. RQ1: Publication Trends
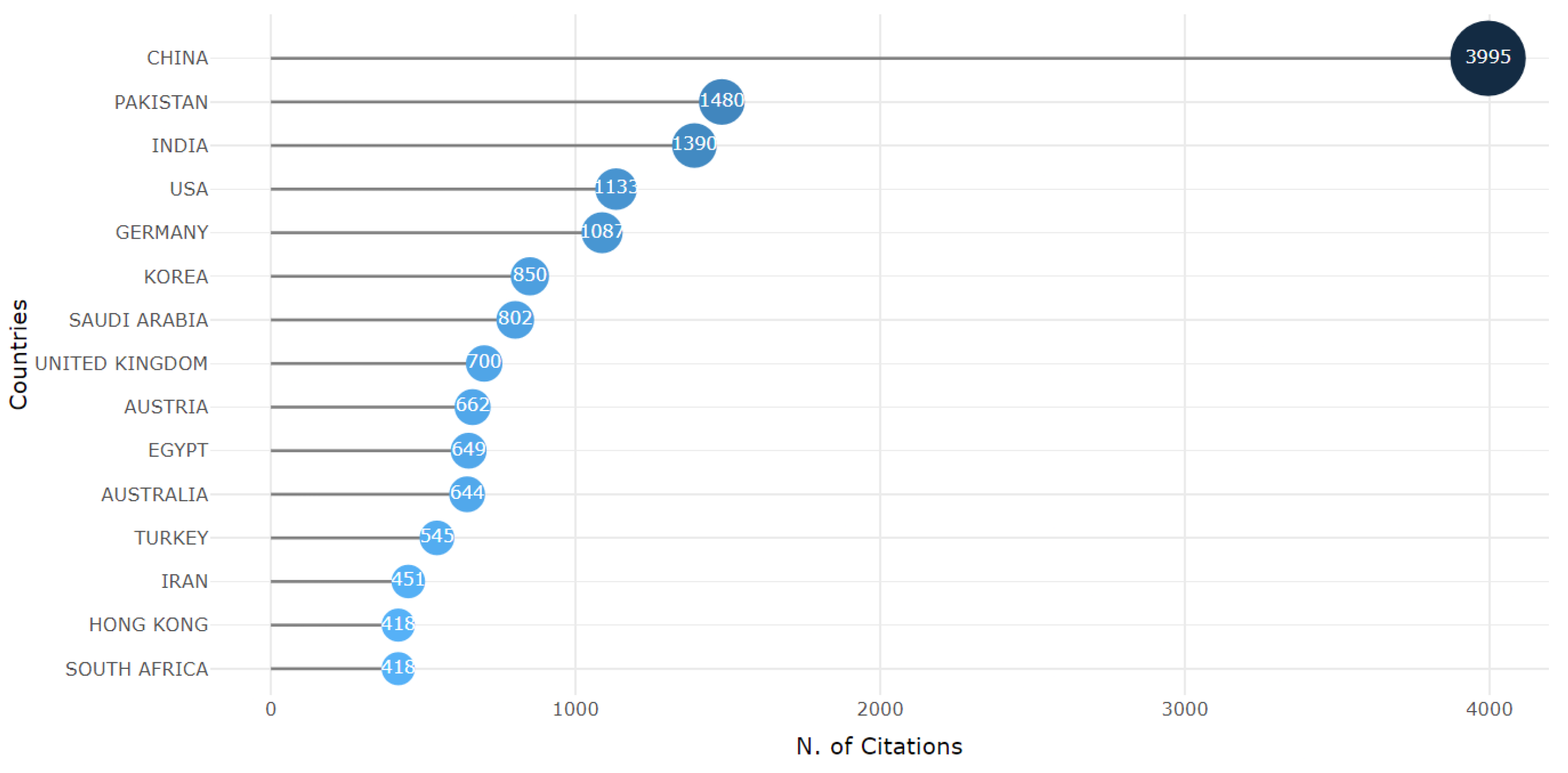
3.2. RQ2: Countries Distribution
3.3. RQ3: The Most Productive Authors
3.4. RQ4: The Most Productive Journals
3.5. RQ5: The Most Cited Countries
3.6. RQ6: The Most Productive Organizations
3.7. RQ7: The Most Cited Articles
3.7.1. Advancements in Semi-Supervised Learning for Medical Image Segmentation
3.7.2. State-of-the-Art Performance in Segmentation and Classification
3.7.3. Ensemble and Attention-Based Methods Enhancing Accuracy
3.7.4. Impact Beyond Skin Lesions
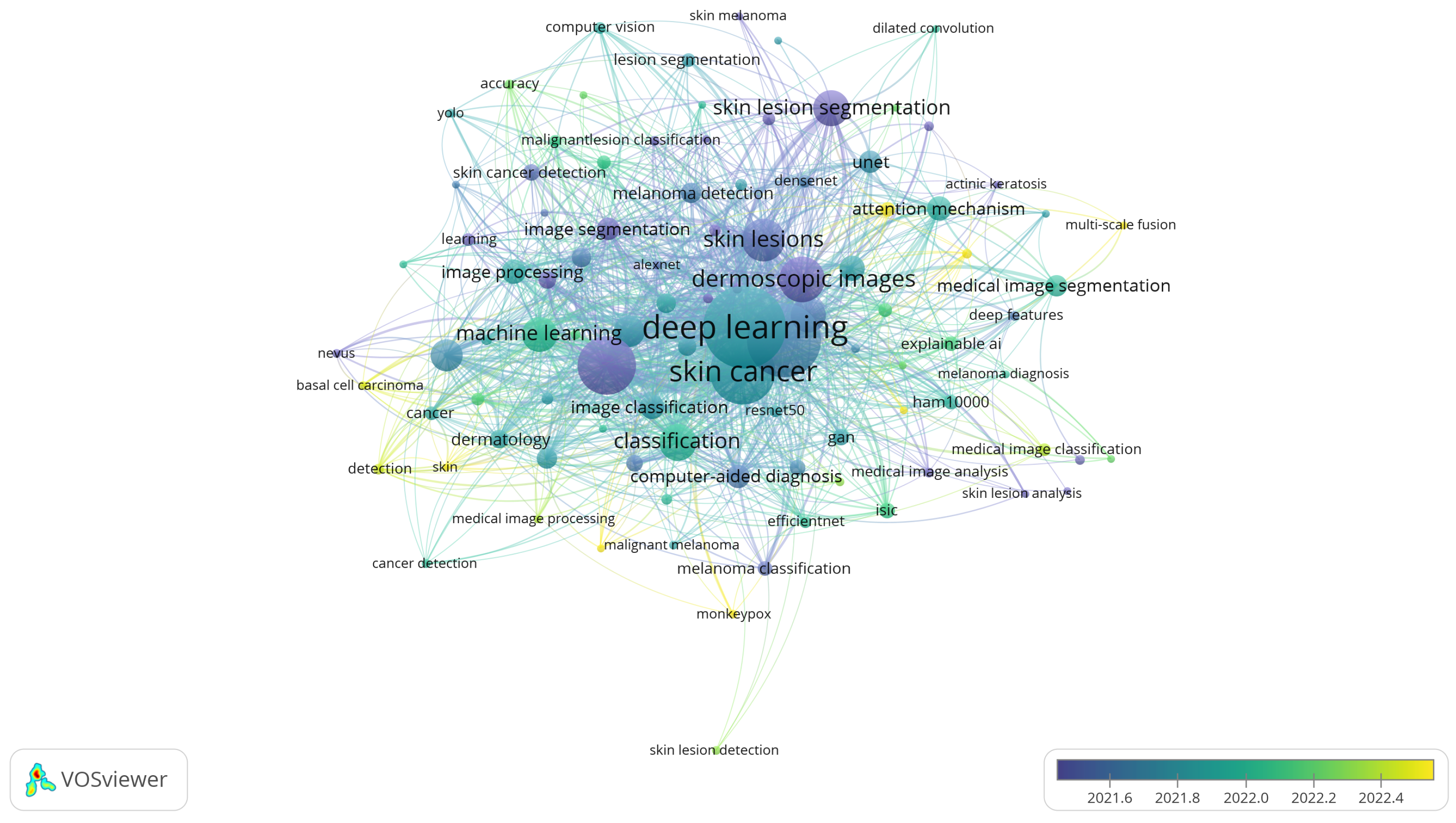
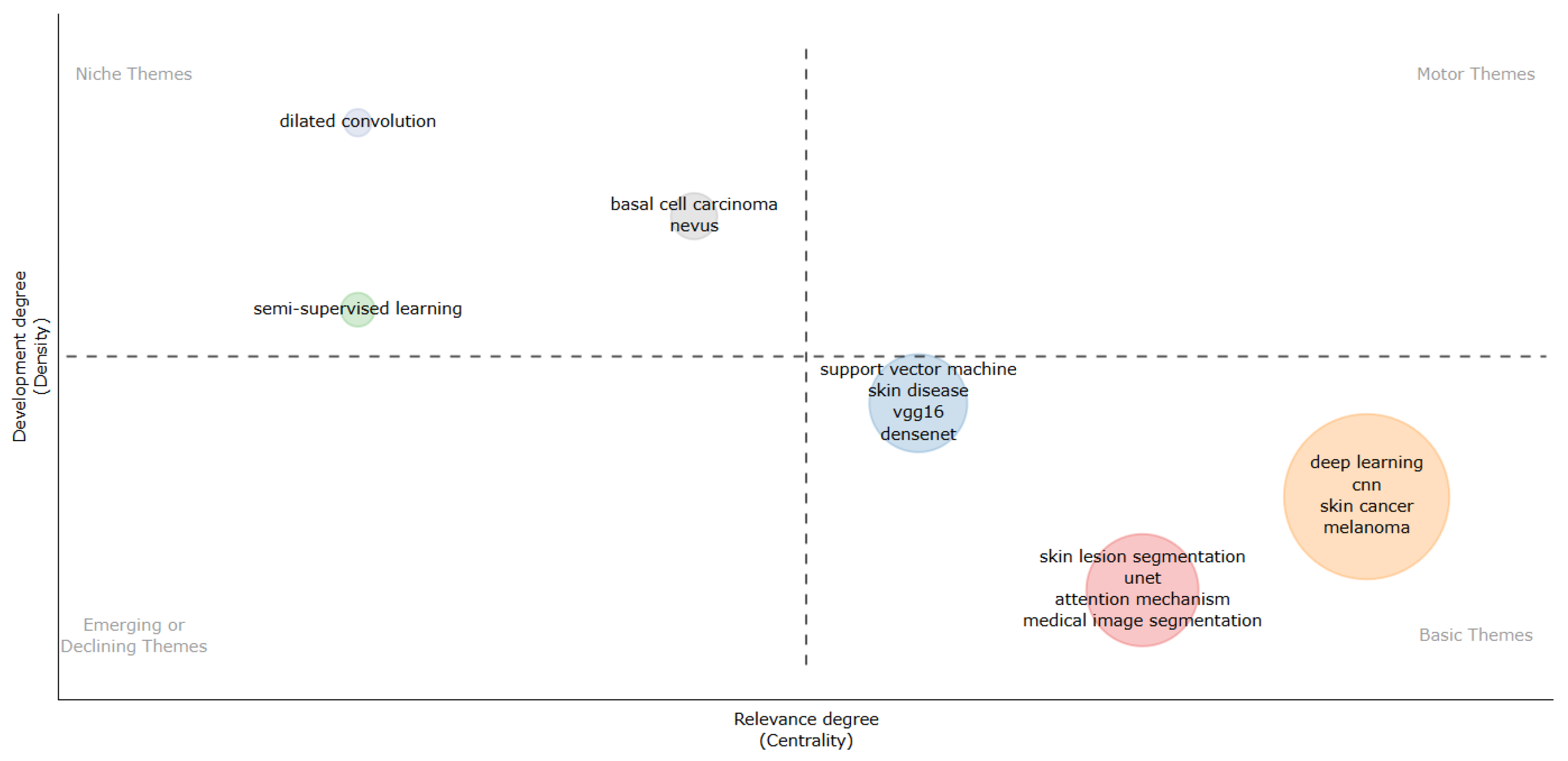
3.8. RQ8: Keywords and Research Trends
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Zhang, X.; Zhou, S.; Li, B.; Wang, Y.; Lu, K.; Liu, W.; Wang, Z. Automatic segmentation of pericardial adipose tissue from cardiac MR images via semi-supervised method with difference-guided consistency. Med. Phys. 2025, 52, 1679–1692. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, L.; Li, G.; Lin, W.; Wang, L. A foundation model for enhancing magnetic resonance images and downstream segmentation, registration and diagnostic tasks. Nat. Biomed. Eng. 2024. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, Y.; Seidlitz, J.; Bethlehem, R.A.I.; Alexander-Bloch, A.; Dorfschmidt, L.; Li, G.; Elison, J.T.; Lin, W.; Wang, L. A lifespan-generalizable skull-stripping model for magnetic resonance images that leverages prior knowledge from brain atlases. Nat. Biomed. Eng. 2025. [Google Scholar] [CrossRef] [PubMed]
- Mahmutoglu, M.A.; Preetha, C.J.; Meredig, H.; Tonn, J.C.; Weller, M.; Wick, W.; Bendszus, M.; Brugnara, G.; Vollmuth, P. Deep Learning–based Identification of Brain MRI Sequences Using a Model Trained on Large Multicentric Study Cohorts. Radiol. Artif. Intell. 2024, 6, e230095. [Google Scholar] [CrossRef]
- Sharma, A.K.; Tiwari, S.; Aggarwal, G.; Goenka, N.; Kumar, A.; Chakrabarti, P.; Chakrabarti, T.; Gono, R.; Leonowicz, Z.; Jasiński, M. Dermatologist-Level Classification of Skin Cancer Using Cascaded Ensembling of Convolutional Neural Network and Handcrafted Features Based Deep Neural Network. IEEE Access 2022, 10, 17920–17932. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Utikal, J.S.; Grabe, N.; Schadendorf, D.; Klode, J.; Berking, C.; Steeb, T.; Enk, A.H.; Von Kalle, C. Skin Cancer Classification Using Convolutional Neural Networks: Systematic Review. J. Med. Internet Res. 2018, 20, e11936. [Google Scholar] [CrossRef]
- Jinnai, S.; Yamazaki, N.; Hirano, Y.; Sugawara, Y.; Ohe, Y.; Hamamoto, R. The Development of a Skin Cancer Classification System for Pigmented Skin Lesions Using Deep Learning. Biomolecules 2020, 10, 1123. [Google Scholar] [CrossRef]
- Ravi, V. Attention Cost-Sensitive Deep Learning-Based Approach for Skin Cancer Detection and Classification. Cancers 2022, 14, 5872. [Google Scholar] [CrossRef]
- Obayya, M.; Arasi, M.A.; Almalki, N.S.; Alotaibi, S.S.; Al Sadig, M.; Sayed, A. Internet of Things-Assisted Smart Skin Cancer Detection Using Metaheuristics with Deep Learning Model. Cancers 2023, 15, 5016. [Google Scholar] [CrossRef]
- Gouda, W.; Sama, N.U.; Al-Waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183. [Google Scholar] [CrossRef]
- Alam, T.M.; Shaukat, K.; Khan, W.A.; Hameed, I.A.; Almuqren, L.A.; Raza, M.A.; Aslam, M.; Luo, S. An Efficient Deep Learning-Based Skin Cancer Classifier for an Imbalanced Dataset. Diagnostics 2022, 12, 2115. [Google Scholar] [CrossRef]
- Chaturvedi, S.S.; Tembhurne, J.V.; Diwan, T. A multi-class skin Cancer classification using deep convolutional neural networks. Multimed. Tools Appl. 2020, 79, 28477–28498. [Google Scholar] [CrossRef]
- Gilani, S.Q.; Umair, M.; Naqvi, M.; Marques, O.; Kim, H.C. Adversarial Training Based Domain Adaptation of Skin Cancer Images. Life 2024, 14, 1009. [Google Scholar] [CrossRef] [PubMed]
- Tembhurne, J.V.; Hebbar, N.; Patil, H.Y.; Diwan, T. Skin cancer detection using ensemble of machine learning and deep learning techniques. Multimed. Tools Appl. 2023, 82, 27501–27524. [Google Scholar] [CrossRef]
- Mazhar, T.; Haq, I.; Ditta, A.; Mohsan, S.A.H.; Rehman, F.; Zafar, I.; Gansau, J.A.; Goh, L.P.W. The Role of Machine Learning and Deep Learning Approaches for the Detection of Skin Cancer. Healthcare 2023, 11, 415. [Google Scholar] [CrossRef]
- Furriel, B.C.R.S.; Oliveira, B.D.; Prôa, R.; Paiva, J.Q.; Loureiro, R.M.; Calixto, W.P.; Reis, M.R.C.; Giavina-Bianchi, M. Artificial intelligence for skin cancer detection and classification for clinical environment: A systematic review. Front. Med. 2024, 10, 1305954. [Google Scholar] [CrossRef]
- Brancaccio, G.; Balato, A.; Malvehy, J.; Puig, S.; Argenziano, G.; Kittler, H. Artificial Intelligence in Skin Cancer Diagnosis: A Reality Check. J. Investig. Dermatol. 2024, 144, 492–499. [Google Scholar] [CrossRef]
- Wei, M.L.; Tada, M.; So, A.; Torres, R. Artificial intelligence and skin cancer. Front. Med. 2024, 11, 1331895. [Google Scholar] [CrossRef]
- Celebi, M.E.; Codella, N.; Halpern, A. Dermoscopy Image Analysis: Overview and Future Directions. IEEE J. Biomed. Health Inform. 2019, 23, 474–478. [Google Scholar] [CrossRef]
- Stafford, H.; Buell, J.; Chiang, E.; Ramesh, U.; Migden, M.; Nagarajan, P.; Amit, M.; Yaniv, D. Non-Melanoma Skin Cancer Detection in the Age of Advanced Technology: A Review. Cancers 2023, 15, 3094. [Google Scholar] [CrossRef]
- Debelee, T.G. Skin Lesion Classification and Detection Using Machine Learning Techniques: A Systematic Review. Diagnostics 2023, 13, 3147. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.S.; An, H.G.; Oh, B.H.; Yang, S. Artificial Intelligence in Cutaneous Oncology. Front. Med. 2020, 7, 318. [Google Scholar] [CrossRef] [PubMed]
- Choy, S.P.; Kim, B.J.; Paolino, A.; Tan, W.R.; Lim, S.M.L.; Seo, J.; Tan, S.P.; Francis, L.; Tsakok, T.; Simpson, M.; et al. Systematic review of deep learning image analyses for the diagnosis and monitoring of skin disease. NPJ Digit. Med. 2023, 6, 180. [Google Scholar] [CrossRef] [PubMed]
- Hauser, K.; Kurz, A.; Haggenmüller, S.; Maron, R.C.; Von Kalle, C.; Utikal, J.S.; Meier, F.; Hobelsberger, S.; Gellrich, F.F.; Sergon, M.; et al. Explainable artificial intelligence in skin cancer recognition: A systematic review. Eur. J. Cancer 2022, 167, 54–69. [Google Scholar] [CrossRef]
- Takiddin, A.; Schneider, J.; Yang, Y.; Abd-Alrazaq, A.; Househ, M. Artificial Intelligence for Skin Cancer Detection: Scoping Review. J. Med. Internet Res. 2021, 23, e22934. [Google Scholar] [CrossRef]
- Khattar, S.; Kaur, R. Computer assisted diagnosis of skin cancer: A survey and future recommendations. Comput. Electr. Eng. 2022, 104, 108431. [Google Scholar] [CrossRef]
- Painuli, D.; Bhardwaj, S.; köse, U. Recent advancement in cancer diagnosis using machine learning and deep learning techniques: A comprehensive review. Comput. Biol. Med. 2022, 146, 105580. [Google Scholar] [CrossRef]
- Attique Khan, M.; Sharif, M.; Akram, T.; Kadry, S.; Hsu, C.H. A two-stream deep neural network-based intelligent system for complex skin cancer types classification. Int. J. Intell. Syst. 2022, 37, 10621–10649. [Google Scholar] [CrossRef]
- Zahoor, S.; Lali, I.U.; Khan, M.A.; Javed, K.; Mehmood, W. Breast cancer detection and classification using traditional computer vision techniques: A comprehensive review. Curr. Med. Imaging 2020, 16, 1187–1200. [Google Scholar] [CrossRef]
- Malik, S.; Akram, T.; Awais, M.; Khan, M.A.; Hadjouni, M.; Elmannai, H.; Alasiry, A.; Marzougui, M.; Tariq, U. An Improved Skin Lesion Boundary Estimation for Enhanced-Intensity Images Using Hybrid Metaheuristics. Diagnostics 2023, 13, 1285. [Google Scholar] [CrossRef]
- Saba, T.; Khan, M.A.; Rehman, A.; Marie-Sainte, S.L. Region Extraction and Classification of Skin Cancer: A Heterogeneous framework of Deep CNN Features Fusion and Reduction. J. Med. Syst. 2019, 43, 289. [Google Scholar] [CrossRef] [PubMed]
- Bibi, S.; Khan, M.A.; Shah, J.H.; Damaševičius, R.; Alasiry, A.; Marzougui, M.; Alhaisoni, M.; Masood, A. MSRNet: Multiclass Skin Lesion Recognition Using Additional Residual Block Based Fine-Tuned Deep Models Information Fusion and Best Feature Selection. Diagnostics 2023, 13, 63. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Khan, M.A.; Tariq, U.; Armghan, A.; Alenezi, F.; Younus Javed, M.; Aslam, S.M.; Kadry, S. A Computer-Aided Diagnosis System Using Deep Learning for Multiclass Skin Lesion Classification. Comput. Intell. Neurosci. 2021, 2021, 9619079. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Nazir, T.; Khan, M.A.; Alhaisoni, M.; Kim, J.Y.; Nam, Y. MSeg-Net: A Melanoma Mole Segmentation Network Using CornerNet and Fuzzy K -Means Clustering. Comput. Math. Methods Med. 2022, 2022, 7502504. [Google Scholar] [CrossRef]
- Nawaz, M.; Nazir, T.; Masood, M.; Ali, F.; Khan, M.A.; Tariq, U.; Sahar, N.; Damaševičius, R. Melanoma segmentation: A framework of improved DenseNet77 and UNET convolutional neural network. Int. J. Imaging Syst. Technol. 2022, 32, 2137–2153. [Google Scholar] [CrossRef]
- Hussain, M.; Khan, M.A.; Damaševičius, R.; Alasiry, A.; Marzougui, M.; Alhaisoni, M.; Masood, A. SkinNet-INIO: Multiclass Skin Lesion Localization and Classification Using Fusion-Assisted Deep Neural Networks and Improved Nature-Inspired Optimization Algorithm. Diagnostics 2023, 13, 2869. [Google Scholar] [CrossRef]
- Ahmad, N.; Shah, J.H.; Khan, M.A.; Baili, J.; Ansari, G.J.; Tariq, U.; Kim, Y.J.; Cha, J.H. A novel framework of multiclass skin lesion recognition from dermoscopic images using deep learning and explainable AI. Front. Oncol. 2023, 13, 1151257. [Google Scholar] [CrossRef]
- Iqbal, A.; Sharif, M.; Khan, M.A.; Nisar, W.; Alhaisoni, M. FF-UNet: A U-Shaped Deep Convolutional Neural Network for Multimodal Biomedical Image Segmentation. Cogn. Comput. 2022, 14, 1287–1302. [Google Scholar] [CrossRef]
- Khan, M.A.; Muhammad, K.; Sharif, M.; Akram, T.; Albuquerque, V.H.C.D. Multi-Class Skin Lesion Detection and Classification via Teledermatology. IEEE J. Biomed. Health Inform. 2021, 25, 4267–4275. [Google Scholar] [CrossRef]
- Khan, M.A.; Sharif, M.; Akram, T.; Damaševičius, R.; Maskeliūnas, R. Skin lesion segmentation and multiclass classification using deep learning features and improved moth flame optimization. Diagnostics 2021, 11, 811. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Zhang, Y.D.; Sharif, M. Attributes based skin lesion detection and recognition: A mask RCNN and transfer learning-based deep learning framework. Pattern Recognit. Lett. 2021, 143, 58–66. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Sharif, M.; Javed, K.; Rashid, M.; Bukhari, S.A.C. An integrated framework of skin lesion detection and recognition through saliency method and optimal deep neural network features selection. Neural Comput. Appl. 2020, 32, 15929–15948. [Google Scholar] [CrossRef]
- Khan, M.A.; Sharif, M.; Akram, T.; Bukhari, S.A.C.; Nayak, R.S. Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recognit. Lett. 2020, 129, 293–303. [Google Scholar] [CrossRef]
- Afza, F.; Sharif, M.; Khan, M.A.; Tariq, U.; Yong, H.S.; Cha, J. Multiclass Skin Lesion Classification Using Hybrid Deep Features Selection and Extreme Learning Machine. Sensors 2022, 22, 799. [Google Scholar] [CrossRef]
- Afza, F.; Sharif, M.; Mittal, M.; Khan, M.A.; Jude Hemanth, D. A hierarchical three-step superpixels and deep learning framework for skin lesion classification. Methods 2022, 202, 88–102. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Zhang, Y.D.; Alhaisoni, M.; Al Hejaili, A.; Shaban, K.A.; Tariq, U.; Zayyan, M.H. SkinNet-ENDO: Multiclass skin lesion recognition using deep neural network and Entropy-Normal distribution optimization algorithm with ELM. Int. J. Imaging Syst. Technol. 2023, 33, 1275–1292. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Sharif, M.; Kadry, S.; Nam, Y. Computer Decision Support System for Skin Cancer Localization and Classification. Comput. Mater. Contin. 2021, 68, 1041–1064. [Google Scholar] [CrossRef]
- Malik, S.; Akram, T.; Ashraf, I.; Rafiullah, M.; Ullah, M.; Tanveer, J. A Hybrid Preprocessor DE-ABC for Efficient Skin-Lesion Segmentation with Improved Contrast. Diagnostics 2022, 12, 2625. [Google Scholar] [CrossRef]
- Malik, S.; Islam, S.M.R.; Akram, T.; Naqvi, S.R.; Alghamdi, N.S.; Baryannis, G. A novel hybrid meta-heuristic contrast stretching technique for improved skin lesion segmentation. Comput. Biol. Med. 2022, 151, 106222. [Google Scholar] [CrossRef]
- Akram, T.; Lodhi, H.M.J.; Naqvi, S.R.; Naeem, S.; Alhaisoni, M.; Ali, M.; Haider, S.A.; Qadri, N.N. A multilevel features selection framework for skin lesion classification. Hum.-Centric Comput. Inf. Sci. 2020, 10, 12. [Google Scholar] [CrossRef]
- Anjum, M.A.; Amin, J.; Sharif, M.; Khan, H.U.; Malik, M.S.A.; Kadry, S. Deep Semantic Segmentation and Multi-Class Skin Lesion Classification Based on Convolutional Neural Network. IEEE Access 2020, 8, 129668–129678. [Google Scholar] [CrossRef]
- Kaur, R.; Gholamhosseini, H.; Sinha, R. Synthetic Images Generation Using Conditional Generative Adversarial Network for Skin Cancer Classification. In Proceedings of the TENCON 2021—2021 IEEE Region 10 Conference (TENCON), Auckland, New Zealand, 7–10 December 2021; pp. 381–386. [Google Scholar] [CrossRef]
- Ali, A.A.; Taha, R.E.; Kaur, R.; Afifi, S.M. Multi-Class Classification of Melanoma on an Edge Device. In Proceedings of the 2023 International Conference on Microelectronics (ICM), Abu Dhabi, United Arab Emirates, 17–20 December 2023; pp. 46–51. [Google Scholar] [CrossRef]
- Dawod, M.I.; Taha, R.; Kaur, R.; Afifi, S.M. Real-time Classification of Skin Cancer on an Edge Device. In Proceedings of the 2023 2nd International Conference on Smart Cities 4.0, Cairo, Egypt, 22–24 October 2023; pp. 184–191. [Google Scholar] [CrossRef]
- Kaur, R.; Gholamhosseini, H.; Sinha, R.; Lindén, M. Melanoma Classification Using a Novel Deep Convolutional Neural Network with Dermoscopic Images. Sensors 2022, 22, 1134. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; GholamHosseini, H.; Sinha, R. Hairlines removal and low contrast enhancement of melanoma skin images using convolutional neural network with aggregation of contextual information. Biomed. Signal Process. Control 2022, 76, 103653. [Google Scholar] [CrossRef]
- Kaur, R.; GholamHosseini, H.; Sinha, R.; Lindén, M. Automatic lesion segmentation using atrous convolutional deep neural networks in dermoscopic skin cancer images. BMC Med. Imaging 2022, 22, 103. [Google Scholar] [CrossRef]
- Kaur, R.; Hosseini, H.G.; Sinha, R. Lesion Border Detection of Skin Cancer Images Using Deep Fully Convolutional Neural Network with Customized Weights. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Virtual, 1–5 November 2021; pp. 3035–3038. [Google Scholar] [CrossRef]
- Kaur, R.; GholamHosseini, H.; Sinha, R. Skin lesion segmentation using an improved framework of encoder-decoder based convolutional neural network. Int. J. Imaging Syst. Technol. 2022, 32, 1143–1158. [Google Scholar] [CrossRef]
- Kaur, R.; Gholamhosseini, H. Analyzing the Impact of Image Denoising and Segmentation on Melanoma Classification Using Convolutional Neural Networks. In Proceedings of the 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, Australia, 24–27 July 2023. [Google Scholar] [CrossRef]
- Fogelberg, K.; Chamarthi, S.; Maron, R.C.; Niebling, J.; Brinker, T.J. Domain shifts in dermoscopic skin cancer datasets: Evaluation of essential limitations for clinical translation. New Biotechnol. 2023, 76, 106–117. [Google Scholar] [CrossRef]
- Schneider, L.; Wies, C.; Krieghoff-Henning, E.I.; Bucher, T.C.; Utikal, J.S.; Schadendorf, D.; Brinker, T.J. Multimodal integration of image, epigenetic and clinical data to predict BRAF mutation status in melanoma. Eur. J. Cancer 2023, 183, 131–138. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Von Kalle, C. Enhanced classifier training to improve precision of a convolutional neural network to identify images of skin lesions. PLoS ONE 2019, 14, e0218713. [Google Scholar] [CrossRef]
- Maron, R.C.; Hekler, A.; Krieghoff-Henning, E.; Schmitt, M.; Schlager, J.G.; Utikal, J.S.; Brinker, T.J. Reducing the Impact of Confounding Factors on Skin Cancer Classification via Image Segmentation: Technical Model Study. J. Med. Internet Res. 2021, 23, e21695. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Hauschild, A.; Berking, C.; Schilling, B.; Enk, A.H.; Haferkamp, S.; Karoglan, A.; Von Kalle, C.; Weichenthal, M.; et al. Comparing artificial intelligence algorithms to 157 German dermatologists: The melanoma classification benchmark. Eur. J. Cancer 2019, 111, 30–37. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Fröhling, S.; et al. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur. J. Cancer 2019, 111, 148–154. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef]
- Bibi, A.; Khan, M.A.; Javed, M.Y.; Tariq, U.; Kang, B.G.; Nam, Y.; Mostafa, R.R.; Sakr, R.H. Skin lesion segmentation and classification using conventional and deep learning based framework. Comput. Mater. Contin. 2022, 71, 2477–2495. [Google Scholar] [CrossRef]
- Kalyani, K.; Althubiti, S.A.; Ahmed, M.A.; Lydia, E.L.; Kadry, S.; Han, N.; Nam, Y. Arithmetic Optimization with Ensemble Deep Transfer Learning Based Melanoma Classification. Comput. Mater. Contin. 2023, 75, 149–164. [Google Scholar] [CrossRef]
- Kadry, S.; Taniar, D.; Damasevicius, R.; Rajinikanth, V.; Lawal, I.A. Extraction of Abnormal Skin Lesion from Dermoscopy Image using VGG-SegNet. In Proceedings of the 2021 Seventh International Conference on Bio Signals, Images, and Instrumentation (ICBSII), Chennai, India, 25–27 March 2021. [Google Scholar] [CrossRef]
- Cheng, X.; Kadry, S.; Meqdad, M.N.; Crespo, R.G. CNN supported framework for automatic extraction and evaluation of dermoscopy images. J. Supercomput. 2022, 78, 17114–17131. [Google Scholar] [CrossRef]
- Jiang, Y.; Dong, J.; Cheng, T.; Zhang, Y.; Lin, X.; Liang, J. iU-Net: A hybrid structured network with a novel feature fusion approach for medical image segmentation. Biodata Min. 2023, 16, 5. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, Y.; Qiao, H.; Wang, M.; Yan, W.; Chen, J. SIL-Net: A Semi-Isotropic L-shaped network for dermoscopic image segmentation. Comput. Biol. Med. 2022, 150, 106146. [Google Scholar] [CrossRef]
- Jiang, Y.; Qiao, H.; Zhang, Z.; Wang, M.; Yan, W.; Chen, J. MDSC-Net: A multi-scale depthwise separable convolutional neural network for skin lesion segmentation. IET Image Process. 2023, 17, 3713–3727. [Google Scholar] [CrossRef]
- Jiang, Y.; Cao, S.; Tao, S.; Zhang, H. Skin Lesion Segmentation Based on Multi-Scale Attention Convolutional Neural Network. IEEE Access 2020, 8, 122811–122825. [Google Scholar] [CrossRef]
- Jiang, Y.; Dong, J.; Zhang, Y.; Cheng, T.; Lin, X.; Liang, J. PCF-Net: Position and context information fusion attention convolutional neural network for skin lesion segmentation. Heliyon 2023, 9, 13942. [Google Scholar] [CrossRef]
- Jiang, Y.; Cheng, T.; Dong, J.; Liang, J.; Zhang, Y.; Lin, X.; Yao, H. Dermoscopic image segmentation based on Pyramid Residual Attention Module. PLoS ONE 2022, 17, e0267380. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, M.; Zhang, Z.; Qiao, H.; Yan, W.; Chen, J. CTDS-Net:CNN-Transformer Fusion Network for Dermoscopic Image Segmentation. In Proceedings of the 2023 5th International Conference on Robotics and Computer Vision (ICRCV), Nanjing, China, 15–17 September 2023; pp. 141–150. [Google Scholar] [CrossRef]
- Maron, R.C.; Haggenmüller, S.; von Kalle, C.; Utikal, J.S.; Meier, F.; Gellrich, F.F.; Hauschild, A.; French, L.E.; Schlaak, M.; Ghoreschi, K.; et al. Robustness of convolutional neural networks in recognition of pigmented skin lesions. Eur. J. Cancer 2021, 145, 81–91. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Xia, Y.; Shen, C. Attention Residual Learning for Skin Lesion Classification. IEEE Trans. Med. Imaging 2019, 38, 2092–2103. [Google Scholar] [CrossRef]
- Gu, R.; Wang, G.; Song, T.; Huang, R.; Aertsen, M.; Deprest, J.; Ourselin, S.; Vercauteren, T.; Zhang, S. CA-Net: Comprehensive Attention Convolutional Neural Networks for Explainable Medical Image Segmentation. IEEE Trans. Med. Imaging 2021, 40, 699–711. [Google Scholar] [CrossRef]
- Li, X.; Yu, L.; Chen, H.; Fu, C.W.; Xing, L.; Heng, P.A. Transformation-Consistent Self-Ensembling Model for Semisupervised Medical Image Segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 523–534. [Google Scholar] [CrossRef]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE 2019, 14, e0217293. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Kim, D.H.; Kim, T.S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed. 2020, 190, 105351. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, J.; Xia, Y.; Shen, C. A Mutual Bootstrapping Model for Automated Skin Lesion Segmentation and Classification. IEEE Trans. Med. Imaging 2020, 39, 2482–2493. [Google Scholar] [CrossRef]
- Panayides, A.S.; Amini, A.; Filipovic, N.D.; Sharma, A.; Tsaftaris, S.A.; Young, A.; Foran, D.; Do, N.; Golemati, S.; Kurc, T.; et al. AI in Medical Imaging Informatics: Current Challenges and Future Directions. IEEE J. Biomed. Health Inform. 2020, 24, 1837–1857. [Google Scholar] [CrossRef]
- Wu, H.; Chen, S.; Chen, G.; Wang, W.; Lei, B.; Wen, Z. FAT-Net: Feature adaptive transformers for automated skin lesion segmentation. Med. Image Anal. 2022, 76, 102327. [Google Scholar] [CrossRef]
- Goyal, M.; Oakley, A.; Bansal, P.; Dancey, D.; Yap, M.H. Skin Lesion Segmentation in Dermoscopic Images with Ensemble Deep Learning Methods. IEEE Access 2020, 8, 4171–4181. [Google Scholar] [CrossRef]
- Moturi, D.; Surapaneni, R.K.; Avanigadda, V.S.G. Developing an efficient method for melanoma detection using CNN techniques. J. Egypt. Natl. Cancer Inst. 2024, 36, 6. [Google Scholar] [CrossRef]
- Kaur, R.; GholamHosseini, H. Deep Learning Model with Atrous Convolutions for Improving Skin Cancer Classification. In Image and Video Technology; Series Title: Lecture Notes in Computer Science; Yan, W.Q., Nguyen, M., Nand, P., Li, X., Eds.; Springer Nature: Singapore, 2024; Volume 14403, pp. 422–435. [Google Scholar] [CrossRef]
- Ju, X.; Lin, C.; Lee, S.; Wei, S. Melanoma classification using generative adversarial network and proximal policy optimization. Photochem. Photobiol. 2024. [Google Scholar] [CrossRef]
- Nagadevi, D.; Suman, K.; Lakshmi, P.S. An enhanced skin lesion detection and classification model using hybrid convolution-based ensemble learning model. Res. Biomed. Eng. 2024, 40, 347–372. [Google Scholar] [CrossRef]
- Yuan, W.; Du, Z.; Han, S. Semi-supervised skin cancer diagnosis based on self-feedback threshold focal learning. Discov. Oncol. 2024, 15, 180. [Google Scholar] [CrossRef]
- Wang, W.; Cao, C.; Wu, S.; Liu, X.; Su, H.; Tian, D. SSCD-Net: Semi-supervised Skin Cancer Diagnostical Network Combined with Curriculum Learning, Disease Relation and Clinical Information. In Proceedings of the 2024 International Joint Conference on Neural Networks (IJCNN), Yokohama, Japan, 30 June–5 July 2024; pp. 1–8. [Google Scholar] [CrossRef]
- Munjal, G.; Bhardwaj, P.; Bhargava, V.; Singh, S.; Nagpal, N. SkinSage XAI: An explainable deep learning solution for skin lesion diagnosis. Health Care Sci. 2024, 3, 438–455. [Google Scholar] [CrossRef]
- Vishal; Mehta, S.; Singh, A. Multi-Modal Skin Cancer Diagnosis Using CNN and SVM on Dermoscopic and Clinical Images. In Proceedings of the 2024 3rd International Conference for Advancement in Technology (ICONAT), Goa, India, 6–8 September 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Mehta, S.; Kaur, S. Automated Detection of Skin Lesions Using CNN-SVM: A Comparative Study. In Proceedings of the 2024 15th International Conference on Computing Communication and Networking Technologies (ICCCNT), Kamand, India, 24–28 June 2024; pp. 1–7. [Google Scholar] [CrossRef]
- Faghihi, A.; Fathollahi, M.; Rajabi, R. Diagnosis of skin cancer using VGG16 and VGG19 based transfer learning models. Multimed. Tools Appl. 2023, 83, 57495–57510. [Google Scholar] [CrossRef]
- Ingle, Y.S. Deep Learning for Skin Cancer Classification: A Comparative Study of CNN and Vgg16 on HAM10000 Dataset. Commun. Appl. Nonlinear Anal. 2024, 31, 490–499. [Google Scholar] [CrossRef]
- Vidhyalakshmi, A.M.; Kanchana, M. Skin cancer classification using improved transfer learning model-based random forest classifier and golden search optimization. Int. J. Imaging Syst. Technol. 2024, 34, e22971. [Google Scholar] [CrossRef]
- Bello, A.; Ng, S.C.; Leung, M.F. Skin Cancer Classification Using Fine-Tuned Transfer Learning of DENSENET-121. Appl. Sci. 2024, 14, 7707. [Google Scholar] [CrossRef]
- Mishra, A.K.; Diwan, T.D.; Gupta, I.K.; Agrawal, S. Crow search algorithm with deep transfer learning driven skin lesion detection on dermoscopic images. Intell. Decis. Technol. 2024, 18, 417–426. [Google Scholar] [CrossRef]

| No | Document Type | #Docs |
|---|---|---|
| 1 | Article | 1098 |
| 2 | Conference paper | 556 |
| 3 | Review | 41 |
| 4 | Letter | 1 |
| 5 | Note | 1 |
| No | Country | #Docs |
|---|---|---|
| 1 | India | 316 |
| 2 | China | 275 |
| 3 | United States | 155 |
| 4 | Saudi Arabia | 87 |
| 5 | Pakistan | 84 |
| 6 | United Kingdom | 66 |
| 7 | South Korea | 49 |
| 8 | Egypt | 45 |
| 9 | Germany | 45 |
| 10 | Canada | 41 |
| 11 | Turkey | 40 |
| 12 | Australia | 39 |
| 13 | Bangladesh | 38 |
| 14 | Spain | 34 |
| 15 | Italy | 32 |
| No | Author | Num. of Docs. | Documents | Country |
|---|---|---|---|---|
| 1 | Khan, M.A. | 20 | [28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45] | Pakistan |
| 2 | Akram, T. | 12 | [28,30,39,40,41,42,43,46,47,48,49,50] | Pakistan |
| 3 | Sharif, M. | 9 | [38,39,40,41,42,43,44,47,51] | Pakistan |
| 4 | Kaur, R. | 9 | [52,53,54,55,56,57,58,59,60] | New Zealand |
| 5 | Brinker, T.J. | 9 | [24,61,62,63,64,65,66,67] | Germany |
| 6 | Utikal, J.S. | 7 | [24,62,64,65,66,67] | Germany |
| 7 | Tariq, U. | 7 | [30,33,35,37,44,46,68] | Saudi Arabia |
| 8 | Kadry, S. | 7 | [28,33,47,51,69,70,71] | Norway |
| 9 | Jiang, Y. | 7 | [72,73,74,75,76,77,78] | China |
| 10 | Hekler, A. | 7 | [24,63,64,65,66,67,79] | Germany |
| No | Source | #Docs |
|---|---|---|
| 1 | IEEE Access (IEEE) | 40 |
| 2 | Computers in Biology and Medicine (Elsevier) | 35 |
| 3 | Diagnostics (MDPI) | 34 |
| 4 | Multimedia Tools and Applications (Springer) | 29 |
| 5 | Biomedical Signal Processing and Control (Elsevier) | 25 |
| 6 | Computer Methods and Programs in Biomedicine (Elsevier) | 23 |
| 7 | Sensors (MDPI) | 16 |
| 8 | Cancers (MDPI) | 14 |
| 9 | International Journal of Imaging Systems and Technology (John Wiley and Son Inc.) | 14 |
| 10 | Applied Sciences (MDPI) | 13 |
| 11 | Medical Image Analysis (Elsevier) | 13 |
| 12 | Expert Systems with Applications (Elsevier) | 12 |
| 13 | Computers, Materials and Continua (Tech Science Press) | 12 |
| 14 | IEEE Journal of Biomedical and Health Informatics (IEEE) | 12 |
| 15 | Frontiers in Medicine (Frontiers Media) | 9 |
| No | Organization | #Docs |
|---|---|---|
| 1 | Skin Cancer Unit, German Cancer Research Center (DKFZ), Heidelberg, Germany | 7 |
| 2 | Dept. of Dermatology, Heidelberg University, Mannheim, Germany | 6 |
| 3 | Dept. of Dermatology, University Hospital Essen, Essen, Germany | 6 |
| 4 | Chitkara University Institute of Engineering and Technology, Chitkara University, Punjab, India | 5 |
| 5 | Dept. of Dermatology, University Hospital Regensburg, Regensburg, Germany | 5 |
| No | Title | Year | Source | #Cit |
|---|---|---|---|---|
| 1 | Attention Residual Learning for Skin Lesion Classification [80] | 2019 | IEEE Transactions on Medical Imaging | 370 |
| 2 | Ca-Net: Comprehensive Attention Convolutional Neural Networks for Explainable Medical Image Segmentation [81] | 2021 | IEEE Transactions on Medical Imaging | 343 |
| 3 | Deep Learning Outperformed 136 Of 157 Dermatologists in A Head-To-Head Dermoscopic Melanoma Image Classification Task [67] | 2019 | European Journal of Cancer | 287 |
| 4 | Transformation-Consistent Self-Ensembling Model for Semisupervised Medical Image Segmentation [82] | 2021 | IEEE Transactions on Neural Networks and Learning Systems | 247 |
| 5 | Classification of Skin Lesions using Transfer Learning and Augmentation with Alex-Net [83] | 2019 | PLoS ONE | 223 |
| 6 | Multiple Skin Lesions Diagnostics via Integrated Deep Convolutional Networks for Segmentation and Classification [84] | 2020 | Computer Methods and Programs in Biomedicine | 221 |
| 7 | A Mutual Bootstrapping Model for Automated Skin Lesion Segmentation and Classification [85] | 2020 | IEEE Transactions on Medical Imaging | 218 |
| 8 | AI in Medical Imaging Informatics: Current Challenges and Future Directions [86] | 2020 | IEEE Journal of Biomedical and Health Informatics | 205 |
| 9 | Fat-Net: Feature Adaptive Transformers for Automated Skin Lesion Segmentation [87] | 2022 | Medical Image Analysis | 198 |
| 10 | Skin Lesion Segmentation in Dermoscopic Images with Ensemble Deep Learning Methods [88] | 2020 | IEEE Access | 189 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Supriyanto, C.; Salam, A.; Zeniarja, J.; Utomo, D.W.; Dewi, I.N.; Paramita, C.; Wijaya, A.; Safar, N.Z.M. A Bibliometric Review of Deep Learning Approaches in Skin Cancer Research. Computation 2025, 13, 78. https://doi.org/10.3390/computation13030078
Supriyanto C, Salam A, Zeniarja J, Utomo DW, Dewi IN, Paramita C, Wijaya A, Safar NZM. A Bibliometric Review of Deep Learning Approaches in Skin Cancer Research. Computation. 2025; 13(3):78. https://doi.org/10.3390/computation13030078
Chicago/Turabian StyleSupriyanto, Catur, Abu Salam, Junta Zeniarja, Danang Wahyu Utomo, Ika Novita Dewi, Cinantya Paramita, Adi Wijaya, and Noor Zuraidin Mohd Safar. 2025. "A Bibliometric Review of Deep Learning Approaches in Skin Cancer Research" Computation 13, no. 3: 78. https://doi.org/10.3390/computation13030078
APA StyleSupriyanto, C., Salam, A., Zeniarja, J., Utomo, D. W., Dewi, I. N., Paramita, C., Wijaya, A., & Safar, N. Z. M. (2025). A Bibliometric Review of Deep Learning Approaches in Skin Cancer Research. Computation, 13(3), 78. https://doi.org/10.3390/computation13030078





