MedMAE: A Self-Supervised Backbone for Medical Imaging Tasks
Abstract
1. Introduction
- Collecting a large-scale unlabeled medical imaging dataset from various sources that can be used for self-supervised and unsupervised learning techniques;
- Proposing a medical masked autoencoder (MedMAE), a pre-trained backbone that can be used for any medical imaging task;
- Extensive evaluation on multiple medical imaging tasks, demonstrating the superiority of our proposed backbone over existing pre-trained models.
2. Related Work
3. Methodology
3.1. LUMID: Large-Scale Unlabeled Medical Imaging Dataset for Unsupervised and Self-Supervised Learning
3.2. Rationale for Key Architectural Choices
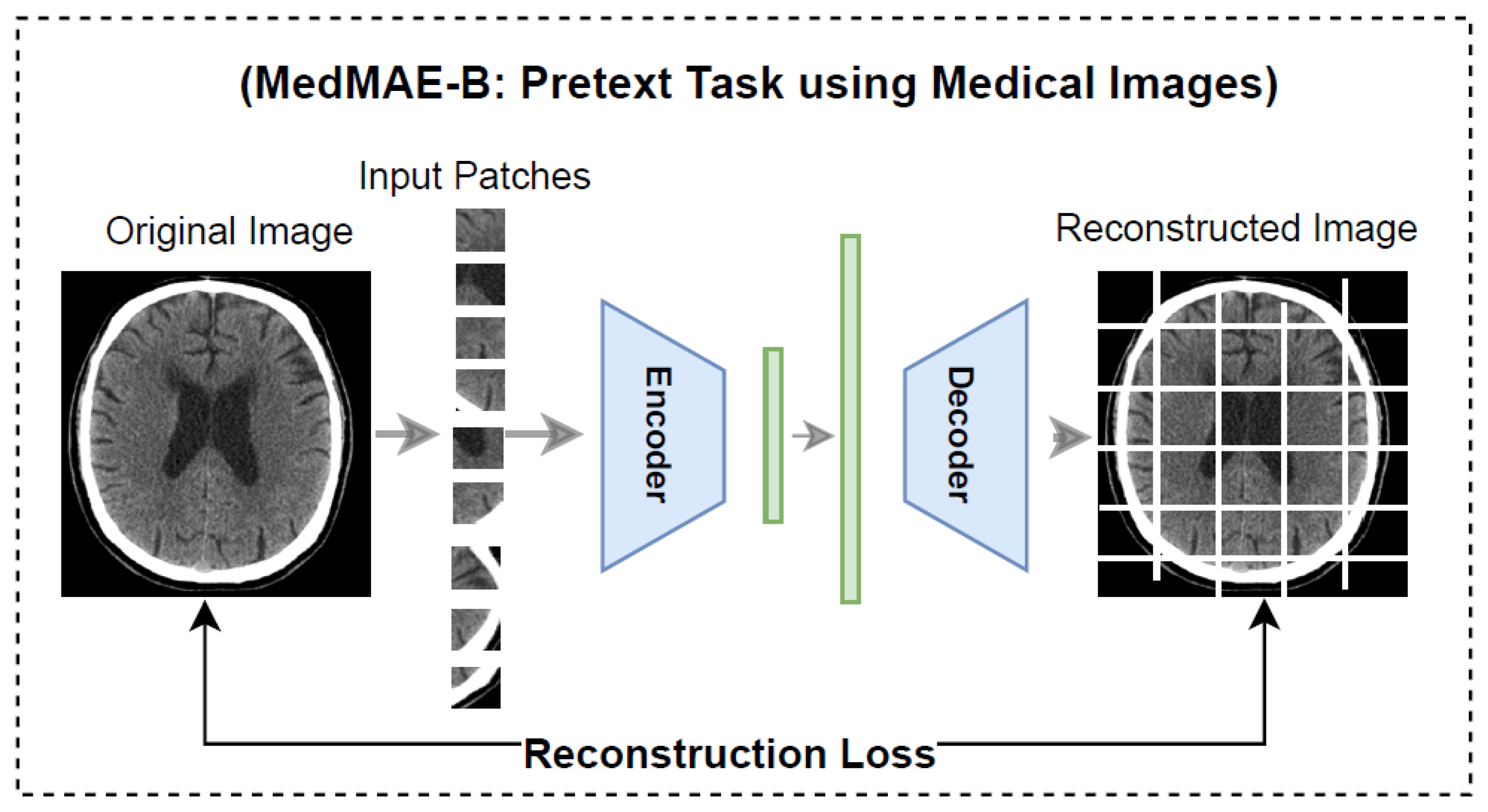
3.3. MedMAE Architecture in Pre-Training
3.4. MedMAE Architecture in Downstream Tasks
4. Experiments and Results
4.1. Implementation Details
4.2. MedMAE Evaluation
4.2.1. Task 1: Automating Quality Control for CT and MRI Scanners
4.2.2. Task 2: Breast Cancer Prediction from CT Images
4.2.3. Task 3: Pneumonia Detection from Chest X-Ray Images
4.2.4. Task 4: Polyp Segmentation in Colonoscopy Sequences
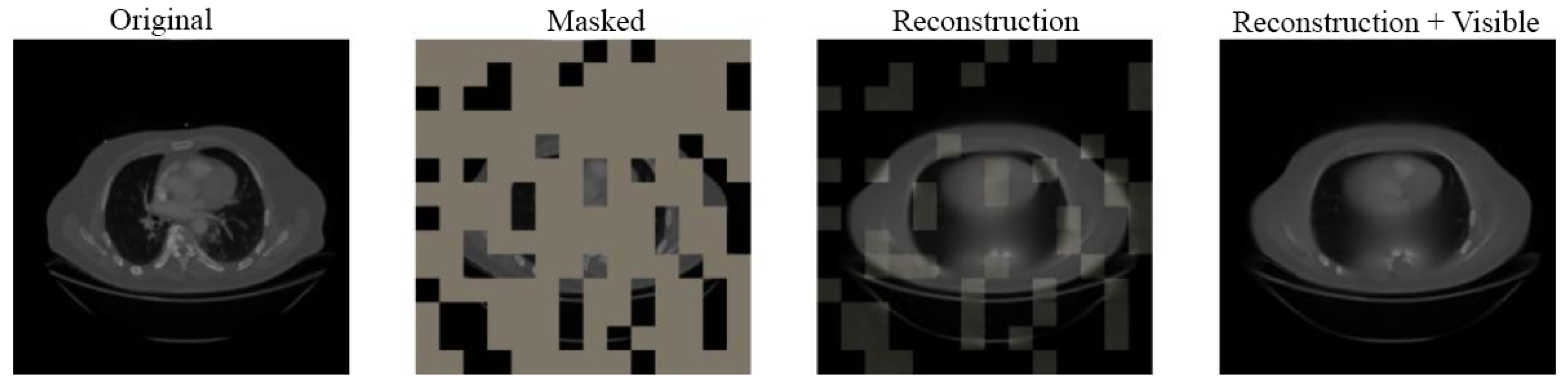
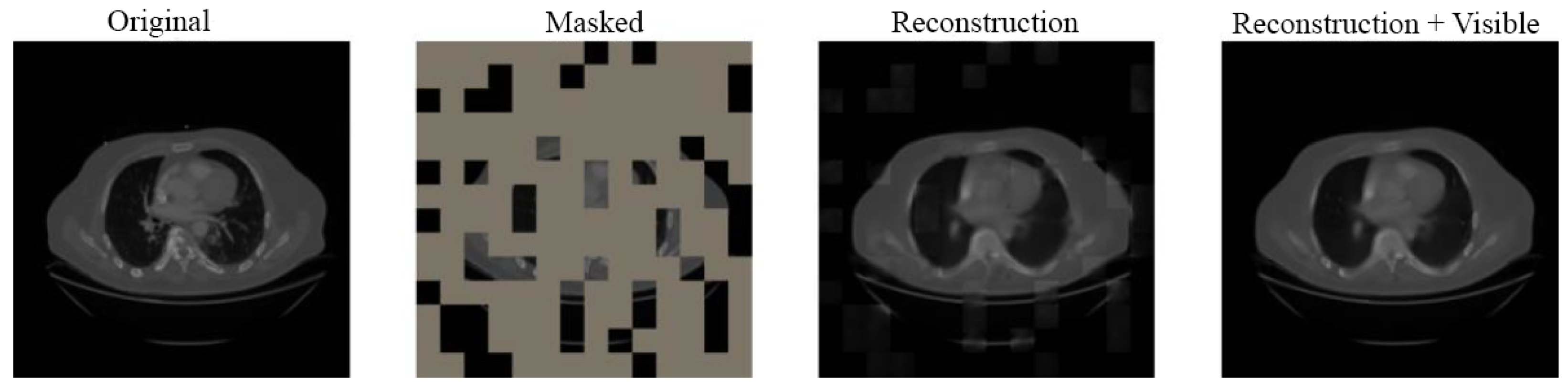
4.3. Visual Results
5. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked autoencoders are scalable vision learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 16000–16009. [Google Scholar]
- Huang, S.C.; Pareek, A.; Jensen, M.; Lungren, M.P.; Yeung, S.; Chaudhari, A.S. Self-supervised learning for medical image classification: A systematic review and implementation guidelines. NPJ Digit. Med. 2023, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Sriram, A.; Muckley, M.; Sinha, K.; Shamout, F.; Pineau, J.; Geras, K.J.; Azour, L.; Aphinyanaphongs, Y.; Yakubova, N.; Moore, W. COVID-19 prognosis via self-supervised representation learning and multi-image prediction. arXiv 2021, arXiv:2101.04909. [Google Scholar]
- Lu, M.Y.; Chen, R.J.; Mahmood, F. Semi-supervised breast cancer histology classification using deep multiple instance learning and contrast predictive coding. In Proceedings of the Medical Imaging 2020: Digital Pathology, SPIE, Houston, TX, USA, 15–20 February 2020; Volume 11320, p. 113200J. [Google Scholar]
- Li, X.; Hu, X.; Qi, X.; Yu, L.; Zhao, W.; Heng, P.A.; Xing, L. Rotation-oriented collaborative self-supervised learning for retinal disease diagnosis. IEEE Trans. Med. Imaging 2021, 40, 2284–2294. [Google Scholar] [CrossRef]
- Chen, L.; Bentley, P.; Mori, K.; Misawa, K.; Fujiwara, M.; Rueckert, D. Self-supervised learning for medical image analysis using image context restoration. Med. Image Anal. 2019, 58, 101539. [Google Scholar] [CrossRef]
- Nguyen, X.B.; Lee, G.S.; Kim, S.H.; Yang, H.J. Self-supervised learning based on spatial awareness for medical image analysis. IEEE Access 2020, 8, 162973–162981. [Google Scholar] [CrossRef]
- Sowrirajan, H.; Yang, J.; Ng, A.Y.; Rajpurkar, P. Moco pretraining improves representation and transferability of chest X-ray models. Proc. Med. Imaging Deep Learn. PMLR 2021, 143, 728–744. [Google Scholar]
- Karani, K.; Konukoglu, E. Contrastive learning of global and local features for medical image segmentation with limited annotations. Adv. Neural Inf. Process. Syst. 2020, 33, 12546–12558. [Google Scholar]
- Taleb, A.; Loetzsch, W.; Danz, N.; Severin, J.; Gaertner, T.; Bergner, B.; Lippert, C. 3D self-supervised methods for medical imaging. Adv. Neural Inf. Process. Syst. 2020, 33, 18158–18172. [Google Scholar]
- Xie, Y.; Zhang, J.; Liao, Z.; Xia, Y.; Shen, C. PGL: Prior-guided local self-supervised learning for 3D medical image segmentation. arXiv 2020, arXiv:2011.12640. [Google Scholar]
- Jung, W.; Heo, D.W.; Jeon, E.; Lee, J.; Suk, H.I. Inter-regional high-level relation learning from functional connectivity via self-supervision. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II 24. Springer: Cham, Switzerland, 2021; pp. 284–293. [Google Scholar]
- Jana, A.; Qu, H.; Minacapelli, C.D.; Catalano, C.; Rustgi, V.; Metaxas, D. Liver fibrosis and nas scoring from ct images using self-supervised learning and texture encoding. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1553–1557. [Google Scholar]
- Liu, C.; Qiao, M.; Jiang, F.; Guo, Y.; Jin, Z.; Wang, Y. TN-USMA Net: Triple normalization-based gastrointestinal stromal tumors classification on multicenter EUS images with ultrasound-specific pretraining and meta attention. Med. Phys. 2021, 48, 7199–7214. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Ullah, I.; Shabaz, M.; Omer, M.F.; Usman, M.T.; Guellil, M.S.; Koo, J. Visionary vigilance: Optimized YOLOV8 for fallen person detection with large-scale benchmark dataset. Image Vis. Comput. 2024, 149, 105195. [Google Scholar] [CrossRef]
- Arshad, T.; Zhang, J.; Ullah, I. A hybrid convolution transformer for hyperspectral image classification. Eur. J. Remote Sens. 2024, 57, 2330979. [Google Scholar] [CrossRef]
- Yasir, M.; Ullah, I.; Choi, C. Depthwise channel attention network (DWCAN): An efficient and lightweight model for single image super-resolution and metaverse gaming. Expert Syst. 2024, 41, e13516. [Google Scholar] [CrossRef]
- Arshad, T.; Zhang, J.; Ullah, I.; Ghadi, Y.Y.; Alfarraj, O.; Gafar, A. Multiscale feature-learning with a unified model for hyperspectral image classification. Sensors 2023, 23, 7628. [Google Scholar] [CrossRef]
- Li, C.; Wang, M.; Sun, X.; Zhu, M.; Gao, H.; Cao, X.; Ullah, I.; Liu, Q.; Xu, P. A novel dimensionality reduction algorithm for Cholangiocarcinoma hyperspectral images. Opt. Laser Technol. 2023, 167, 109689. [Google Scholar]
- Malik, S.G.; Jamil, S.S.; Aziz, A.; Ullah, S.; Ullah, I.; Abohashrh, M. High-precision skin disease diagnosis through deep learning on dermoscopic images. Bioengineering 2024, 11, 867. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- Rutherford, M.; Mun, S.K.; Levine, B.; Bennett, W.; Smith, K.; Farmer, P.; Jarosz, Q.; Wagner, U.; Freyman, J.; Blake, G.; et al. A DICOM dataset for evaluation of medical image de-identification. Sci. Data 2021, 8, 183. [Google Scholar] [CrossRef]
- Saltz, J.; Saltz, M.; Prasanna, P.; Moffitt, R.; Hajagos, J.; Bremer, E.; Balsamo, J.; Kurc, T. Stony Brook University COVID-19 Positive Cases. 2021. Available online: https://www.cancerimagingarchive.net/collection/covid-19-ny-sbu (accessed on 17 March 2025).
- Chen, L.; Wang, W.; Jin, K.; Yuan, B.; Tan, H.; Sun, J.; Guo, Y.; Luo, Y.; Feng, S.T.; Yu, X.; et al. Special issue “The advance of solid tumor research in China”: Prediction of Sunitinib efficacy using computed tomography in patients with pancreatic neuroendocrine tumors. Int. J. Cancer 2023, 152, 90–99. [Google Scholar]
- Li, M.; Gong, J.; Bao, Y.; Huang, D.; Peng, J.; Tong, T. Special issue “The advance of solid tumor research in China”: Prognosis prediction for stage II colorectal cancer by fusing computed tomography radiomics and deep-learning features of primary lesions and peripheral lymph nodes. Int. J. Cancer 2023, 152, 31–41. [Google Scholar]
- An, P.; Xu, S.; Harmon, S.A.; Turkbey, E.B.; Sanford, T.H.; Amalou, A.; Kassin, M.; Varble, N.; Blain, M.; Anderson, V.; et al. Ct Images in COVID-19 [Data Set]. 2020. Available online: https://www.cancerimagingarchive.net/collection/ct-images-in-covid-19 (accessed on 17 March 2025).
- Tsai, E.; Simpson, S.; Lungren, M.P.; Hershman, M.; Roshkovan, L.; Colak, E.; Erickson, B.J.; Shih, G.; Stein, A.; Kalpathy-Cramer, J.; et al. Data from Medical Imaging Data Resource Center (MIDRC)-RSNA International COVID Radiology Database (RICORD) Release 1C-Chest X-ray, Covid+(MIDRC-RICORD-1C). 2021. Available online: https://www.cancerimagingarchive.net/collection/midrc-ricord-1c (accessed on 17 March 2025).
- Rister, B.; Yi, D.; Shivakumar, K.; Nobashi, T.; Rubin, D.L. CT-ORG, a new dataset for multiple organ segmentation in computed tomography. Sci. Data 2020, 7, 381. [Google Scholar] [CrossRef] [PubMed]
- Yorke, A.A.; McDonald, G.C.; Solis, D.; Guerrero, T. Pelvic Reference Data. 2019. Available online: https://www.cancerimagingarchive.net/collection/pelvic-reference-data (accessed on 17 March 2025).
- Kalpathy-Cramer, J.; Napel, S.; Goldgof, D.; Zhao, B. QIN Multi-Site Collection of Lung CT Data with Nodule Segmentations. 2015. Available online: https://www.cancerimagingarchive.net/collection/qin-lung-ct (accessed on 17 March 2025).
- Roth, H.R.; Farag, A.; Turkbey, E.; Lu, L.; Liu, J.; Summers, R.M. Data from pancreas-ct. 2016. Available online: https://www.cancerimagingarchive.net/collection/pancreas-ct (accessed on 17 March 2025).
- Grove, O.; Berglund, A.E.; Schabath, M.B.; Aerts, H.J.; Dekker, A.; Wang, H.; Velazquez, E.R.; Lambin, P.; Gu, Y.; Balagurunathan, Y.; et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS ONE 2015, 10, e0118261. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.; Velazquez, E.R.; Leijenaar, R.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Data from NSCLC-Radiomics. Available online: https://www.cancerimagingarchive.net/collection/nsclc-radiomics (accessed on 17 March 2025).
- Teng, X. Improving Radiomic Model Reliability and Generalizability Using Perturbations in Head and Neck Carcinoma. Ph.D. Thesis, Hong Kong Polytechnic University, Hong Kong, 2023. [Google Scholar]
- Smith, K.; Clark, K.; Bennett, W.; Nolan, T.; Kirby, J.; Wolfsberger, M.; Moulton, J.; Vendt, B.; Freymann, J. Data from CT_COLONOGRAPHY. 2015. Available online: https://www.cancerimagingarchive.net/collection/ct-colonography (accessed on 17 March 2025).
- Armato, S.G., III; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Machado, M.A.; Moraes, T.F.; Anjos, B.H.; Alencar, N.R.; Chang, T.M.C.; Santana, B.C.; Menezes, V.O.; Vieira, L.O.; Brandão, S.C.; Salvino, M.A.; et al. Association between increased Subcutaneous Adipose Tissue Radiodensity and cancer mortality: Automated computation, comparison of cancer types, gender, and scanner bias. Appl. Radiat. Isot. 2024, 205, 111181. [Google Scholar] [CrossRef]
- Albertina, B.; Watson, M.; Holback, C.; Jarosz, R.; Kirk, S.; Lee, Y.; Rieger-Christ, K.; Lemmerman, J. The Cancer Genome Atlas Lung Adenocarcinoma Collection (tcga-luad) (Version 4) [Data Set]. Available online: https://www.cancerimagingarchive.net/collection/tcga-luad (accessed on 17 March 2025).
- Desai, S.; Baghal, A.; Wongsurawat, T.; Al-Shukri, S.; Gates, K.; Farmer, P.; Rutherford, M.; Blake, G.; Nolan, T.; Powell, T.; et al. Data from Chest Imaging with Clinical and Genomic Correlates Representing a Rural COVID-19 Positive Population [Data Set]. Available online: https://www.cancerimagingarchive.net/collection/covid-19-ar (accessed on 17 March 2025).
- Biobank, C. Cancer Moonshot Biobank-Lung Cancer Collection (CMB-LCA) (Version 3) [Dataset]. Available online: https://www.cancerimagingarchive.net/collection/cmb-lca (accessed on 17 March 2025).
- Biobank, C. Cancer Moonshot Biobank-Lung Cancer Collection (CMB-CRC) (Version 5) [Dataset]. Available online: https://www.cancerimagingarchive.net/collection/cmb-crc (accessed on 17 March 2025).
- Akin, O.; Elnajjar, P.; Heller, M.; Jarosz, R.; Erickson, B.; Kirk, S.; Lee, Y.; Linehan, M.; Gautam, R.; Vikram, R.; et al. The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma Collection (TCGA-KIRC) (Version 3). Available online: https://www.cancerimagingarchive.net/collection/tcga-kirc (accessed on 17 March 2025).
- Biobank, C. Cancer Moonshot Biobank-Lung Cancer Collection (CMB-PCA) (Version 5) [Dataset]. Available online: https://www.cancerimagingarchive.net/collection/cmb-pca (accessed on 17 March 2025).
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar]
- Roche, C.; Bonaccio, E.; Filippini, J. The Cancer Genome Atlas Sarcoma Collection (TCGA-SARC) (Version 3) [Data Set]. Available online: https://www.cancerimagingarchive.net/collection/tcga-sarc (accessed on 17 March 2025).
- Albertina, B.; Watson, M.; Holback, C.; Jarosz, R.; Kirk, S.; Lee, Y.; Lemmerman, J. Radiology Data from the Cancer Genome Atlas Lung Adenocarcinoma [tcga-luad] Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-luad (accessed on 17 March 2025).
- Holback, C.; Jarosz, R.; Prior, F.; Mutch, D.G.; Bhosale, P.; Garcia, K.; Lee, Y.; Kirk, S.; Sadow, C.A.; Levine, S.; et al. The Cancer Genome Atlas Ovarian CANCER collection (tcga-ov) (Version 4) [Data Set]. Available online: https://www.cancerimagingarchive.net/collection/tcga-ov (accessed on 17 March 2025).
- McRobbie, D.W.; Semple, S.; Barnes, A.P. Quality Control and Artefacts in Magnetic Resonance Imaging (Update of IPEM Report 80); Institute of Physics and Engineering in Medicine: York, UK, 2017. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. Proc. Int. Conf. Mach. Learn. PMLR 2021, 139, 10096–10106. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Woo, S.; Debnath, S.; Hu, R.; Chen, X.; Liu, Z.; Kweon, I.S.; Xie, S. Convnext v2: Co-designing and scaling convnets with masked autoencoders. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 16133–16142. [Google Scholar]
- Oquab, M.; Darcet, T.; Moutakanni, T.; Vo, H.; Szafraniec, M.; Khalidov, V.; Fernandez, P.; Haziza, D.; Massa, F.; El-Nouby, A.; et al. Dinov2: Learning robust visual features without supervision. arXiv 2023, arXiv:2304.07193. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Dehkordi, A.N.; Koohestani, S. The Influence of Signal to Noise Ratio on the Pharmacokinetic Analysis in DCE-MRI Studies. Front. Biomed. Technol. 2019, 4, 187–196. [Google Scholar]
- Parrish, T.B.; Gitelman, D.R.; LaBar, K.S.; Mesulam, M.M. Impact of signal-to-noise on functional MRI. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 2000, 44, 925–932. [Google Scholar]
- Song, B.; Tan, W.; Xu, Y.; Yu, T.; Li, W.; Chen, Z.; Yang, R.; Hou, J.; Zhou, Y. 3D-MRI combined with signal-to-noise ratio measurement can improve the diagnostic accuracy and sensitivity in evaluating meniscal healing status after meniscal repair. Knee Surg. Sports Traumatol. Arthrosc. 2019, 27, 177–188. [Google Scholar] [CrossRef]
- Rubenstein, J.D.; Li, J.; Majumdar, S.; Henkelman, R. Image resolution and signal-to-noise ratio requirements for MR imaging of degenerative cartilage. AJR Am. J. Roentgenol. 1997, 169, 1089–1096. [Google Scholar] [PubMed]
- Yao, L.; Poblenz, E.; Dagunts, D.; Covington, B.; Bernard, D.; Lyman, K. Learning to diagnose from scratch by exploiting dependencies among labels. arXiv 2017, arXiv:1710.10501. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Blog, G. AutoML for Large Scale Image Classification and Object Detection. Google Research. 2017. Available online: https://research.google/blog/automl-for-large-scale-image-classification-and-object-detection (accessed on 17 March 2025).
- Lu, Z.; Whalen, I.; Dhebar, Y.; Deb, K.; Goodman, E.D.; Banzhaf, W.; Boddeti, V.N. Multiobjective evolutionary design of deep convolutional neural networks for image classification. IEEE Trans. Evol. Comput. 2020, 25, 277–291. [Google Scholar]
- Lu, Z.; Deb, K.; Boddeti, V.N. MUXConv: Information multiplexing in convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 12044–12053. [Google Scholar]
- Ranjan, E.; Paul, S.; Kapoor, S.; Kar, A.; Sethuraman, R.; Sheet, D. Jointly learning convolutional representations to compress radiological images and classify thoracic diseases in the compressed domain. In Proceedings of the 11th Indian Conference on Computer Vision, Graphics and Image Processing, Hyderabad, India, 18–22 December 2018; pp. 1–8. [Google Scholar]
- Liang, J.; Meyerson, E.; Hodjat, B.; Fink, D.; Mutch, K.; Miikkulainen, R. Evolutionary neural automl for deep learning. In Proceedings of the Genetic and Evolutionary Computation Conference, Prague, Czech Republic, 13–17 July 2019; pp. 401–409. [Google Scholar]
- Zhou, H.Y.; Yu, S.; Bian, C.; Hu, Y.; Ma, K.; Zheng, Y. Comparing to learn: Surpassing imagenet pretraining on radiographs by comparing image representations. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part I 23. Springer: Cham, Switzerland, 2020; pp. 398–407. [Google Scholar]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian error linear units (gelus). arXiv 2016, arXiv:1606.08415. [Google Scholar]

| Collection | Location | Subjects | Data Types |
|---|---|---|---|
| NIH Chest X-Ray [22] | Frontal Chest | 30,805 | X-Ray |
| Pseudo-PHI-DICOM-Data [23] | Various | 21 | CR, CT, DX, MG, MR, PT |
| COVID-19-NY-SBU [24] | Lung | 1384 | CR, CT, DX, MR, PT, NM, OT, SR |
| CTPred-Sunitinib-panNet [25] | Pancreas | 38 | CT |
| Stagell-Colorectal-CT [26] | Abdomen, Pelvis | 230 | CT |
| CT images in COVID19 [27] | Lung | 661 | CT |
| MIDR-RICORD-1a&1b [28] | Lung | 227 | CT |
| CT-ORG [29] | Bladder, Brain, Kidney, Liver | 140 | CT |
| Pelvic-Reference-Data [30] | Pelvis, Prostate, Anus | 58 | CT |
| QIN Lung CT [31] | Lung | 47 | CT |
| Pancreas CT [32] | Pancreas | 82 | CT |
| LungCT-Diagnosis [33] | Lung | 61 | CT |
| NSCLC-Radiomics-Genomics [34] | Lung | 89 | CT |
| RIDER Lung CT [35] | Chest | 32 | CT, CR, DX |
| CT Colon ACRIN 6664 [36] | Colon | 825 | CT |
| LIDC-IDRI [37] | Chest | 1010 | CT, CR, DX |
| TCGA-BLCA [38] | Bladder | 120 | CT, CR, MR, PT, DX, Pathology |
| TCGA-UCEC [39] | Uterus | 65 | CT, CR, MR, PT, Pathology |
| COVID-19-AR [40] | Lung | 105 | CT, DX, CR |
| CMB-LCA [41] | Lung | 10 | CT, DX, MR, NM, US |
| CMB-CRC [42] | Colon | 12 | CT, DX, MR, PT, US |
| TCGA-KIRC [43] | Kidney | 267 | CT, MR, CR, Pathology |
| CMB-PCA [44] | Prostate | 3 | CT, MR, NM |
| CPTAC-CCRCC [45] | Kidney | 222 | CT, MR, Pathology |
| TCGA-SARC [46] | Chest, Abdomen, Pelvis, Leg | 5 | CT, MR, Pathology |
| TCGA-READ [47] | Rectum | 3 | CT, MR, Pathology |
| TCGA-OV [48] | Ovary | 143 | CT. MR, Pathology |
| Pre-Training | Dataset | Backbone | Accuracy (%) | Sensitivity (%) | AUC |
|---|---|---|---|---|---|
| Supervised | IN1k | ResNet | 75.6 | 72.1 | 0.80 |
| IN1k | EfficientNet-S | 71.3 | 68.3 | 0.77 | |
| IN1k | ConvNext-B | 76.8 | 74.1 | 0.82 | |
| IN1k | ConvNextV2-B | 78.1 | 75.9 | 0.83 | |
| IN1k | ViT-B | 78.2 | 76.0 | 0.84 | |
| IN1k | Swin-B | 77.5 | 75.2 | 0.83 | |
| Self-supervised | IN1k | MAE | 78.5 | 77.2 | 0.85 |
| IN1k | DINOv2 | 81.2 | 79.4 | 0.88 | |
| LUMID | MedMAE (Ours) | 90.2 | 88.0 | 0.95 |
| Pre-Training | Dataset | Backbone | Accuracy (%) | Sensitivity (%) | AUC |
|---|---|---|---|---|---|
| Supervised | IN1k | ResNet | 71.6 | 69.0 | 0.79 |
| IN1k | EfficientNet-S | 66.1 | 63.2 | 0.74 | |
| IN1k | ConvNext-B | 71.9 | 70.4 | 0.80 | |
| IN1k | ConvNextV2-B | 74.2 | 72.8 | 0.82 | |
| IN1k | ViT-B | 72.7 | 71.3 | 0.81 | |
| IN1k | Swin-B | 72.1 | 70.6 | 0.80 | |
| Self-supervised | IN1k | MAE | 74.3 | 73.6 | 0.83 |
| IN1k | DINOv2 | 75.9 | 73.9 | 0.84 | |
| LUMID | MedMAE (Ours) | 85.6 | 84.7 | 0.93 |
| Pre-Training | Dataset | Backbone | Accuracy (%) | Sensitivity (%) | AUC |
|---|---|---|---|---|---|
| Supervised | IN1k | ResNet | 79.9 | 78.8 | 0.86 |
| IN1k | EfficientNet-S | 75.1 | 73.3 | 0.82 | |
| IN1k | ConvNext-B | 83.8 | 82.2 | 0.88 | |
| IN1k | ConvNextV2-B | 84.5 | 83.8 | 0.91 | |
| IN1k | ViT-B | 84.0 | 83.4 | 0.89 | |
| IN1k | Swin-B | 81.3 | 79.1 | 0.87 | |
| Self-supervised | IN1k | MAE | 84.3 | 83.5 | 0.90 |
| IN1k | DINOv2 | 88.1 | 87.8 | 0.93 | |
| LUMID | MedMAE (Ours) | 93.2 | 92.2 | 0.97 |
| Pre-Training | Dataset | Backbone | Accuracy (%) | Sensitivity (%) | AUC |
|---|---|---|---|---|---|
| Supervised | IN1k | ResNet | 66.4 | 64.8 | 0.74 |
| IN1k | EfficientNet-S | 62.9 | 60.0 | 0.71 | |
| IN1k | ConvNext-B | 67.5 | 65.6 | 0.76 | |
| IN1k | ConvNextV2-B | 68.0 | 66.1 | 0.78 | |
| IN1k | ViT-B | 67.8 | 65.9 | 0.77 | |
| IN1k | Swin-B | 67.1 | 65.4 | 0.75 | |
| Self-supervised | IN1k | MAE | 67.9 | 66.7 | 0.78 |
| IN1k | DINOv2 | 69.1 | 68.4 | 0.80 | |
| LUMID | MedMAE (Ours) | 70.1 | 69.1 | 0.81 |
| Training | Method | Accuracy |
|---|---|---|
| Supervised | Wang et al. [22] | 73.8 |
| Yao et al. [61] | 79.8 | |
| CheXNet [62] | 84.4 | |
| Google AutoML [63] | 79.7 | |
| NSGANetV1 [64] | 84.7 | |
| MUXNet-m [65] | 84.1 | |
| AE-CNN [66] | 82.4 | |
| LEAF [67] | 84.3 | |
| SSL-Supervised | ResNet+MoCo [68] | 81.4 |
| DenseNet+C2L [68] | 84.4 | |
| DINOv2 [55] | 83.8 | |
| MedMAE (Ours) | 88.0 |
| Pre-Training | Dataset | Backbone | |
|---|---|---|---|
| Supervised | IN1k | ResNet | 57.9 |
| IN1k | EfficientNet-S | 53.1 | |
| IN1k | ConvNext-B | 61.8 | |
| IN1k | ConvNextV2-B | 64.2 | |
| IN1k | ViT-B | 63.5 | |
| IN1k | Swin-B | 60.2 | |
| Self-supervised | IN1k | MAE | 64.6 |
| IN1k | DINOv2 | 67.3 | |
| LUMID | MedMAE (Ours) | 71.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, A.; Osman, I.; Shehata, M.S.; Braun, W.J.; Feldman, R.E. MedMAE: A Self-Supervised Backbone for Medical Imaging Tasks. Computation 2025, 13, 88. https://doi.org/10.3390/computation13040088
Gupta A, Osman I, Shehata MS, Braun WJ, Feldman RE. MedMAE: A Self-Supervised Backbone for Medical Imaging Tasks. Computation. 2025; 13(4):88. https://doi.org/10.3390/computation13040088
Chicago/Turabian StyleGupta, Anubhav, Islam Osman, Mohamed S. Shehata, W. John Braun, and Rebecca E. Feldman. 2025. "MedMAE: A Self-Supervised Backbone for Medical Imaging Tasks" Computation 13, no. 4: 88. https://doi.org/10.3390/computation13040088
APA StyleGupta, A., Osman, I., Shehata, M. S., Braun, W. J., & Feldman, R. E. (2025). MedMAE: A Self-Supervised Backbone for Medical Imaging Tasks. Computation, 13(4), 88. https://doi.org/10.3390/computation13040088







