Abstract
Artificial Intelligence and Deepfake Technologies have brought a new dimension to the generation of fake data, making it easier and faster than ever before—this fake data could include text, images, sounds, videos, etc. This has brought new challenges that require the faster development of tools and techniques to avoid fraudulent activities at pace and scale. Our focus in this research study is to empirically evaluate the use and effectiveness of deep learning models such as Convolutional Neural Networks (CNNs) and Patch-based Neural Networks in the context of successful identification of real and fake images. We chose the healthcare domain as a potential case study where the fake medical data generation approach could be used to make false insurance claims. For this purpose, we obtained publicly available skin cancer data and used recently introduced stable diffusion approaches—a more effective technique than prior approaches such as Generative Adversarial Network (GAN)—to generate fake skin cancer images. To the best of our knowledge, and based on the literature review, this is one of the few research studies that uses images generated using stable diffusion along with real image data. As part of the exploratory analysis, we analyzed histograms of fake and real images using individual color channels and averaged across training and testing datasets. The histogram analysis demonstrated a clear change by shifting the mean and overall distribution of both real and fake images (more prominent in blue and green) in the training data whereas, in the test data, both means were different from the training data, so it appears to be non-trivial to set a threshold which could give better predictive capability. We also conducted a user study to observe where the naked eye could identify any patterns for classifying real and fake images, and the accuracy of the test data was observed to be 68%. The adoption of deep learning predictive approaches (i.e., patch-based and CNN-based) has demonstrated similar accuracy (~100%) in training and validation subsets of the data, and the same was observed for the test subset with and without StratifiedKFold (k = 3). Our analysis has demonstrated that state-of-the-art exploratory and deep-learning approaches are effective enough to detect images generated from stable diffusion vs. real images.
1. Introduction
The advanced techniques of medical imaging, including computed tomography (CT), X-ray scans, Ultrasound, and magnetic resonance imaging (MRI), play an active role in the effective diagnosis of different diseases and reduce the burden on the healthcare industry. CT scans are more informative than 2D X-ray images due to the different angle views of soft tissues and bones [1]. CT scans, as well as MRI images, are usually stored in a particular format known as Digital Imaging and Communications in Medicine (DICOM) to maintain the high quality of the images [2]. Nowadays, in hospitals, Picture Archiving and Communication System (PACS) servers are being used to store large amounts of medical images [3]. This server serves as the central point to store and retrieve the images as needed, i.e., to transfer data between the scanner, hospital, and workstations. Many medical centers rely on outdated security measures and legacy software which leads to unauthorized access [4]. For example, the researchers from McAfee have used 3D printers to reconstruct the model of a pelvis from unsecured volumetric DICOM data [5]. Deceptive attacks, such as adding tumors to set a benign CT scan image to a malignant scan, can be the cause of unnecessary treatment for patients. Similarly, altering the CT scan image of a malignant to a benign scan can be harmful to the patient and lead to death.
Deepfake was first introduced in 2014 with the training of large amounts of data by Generative Adversarial Networks (GANs) [6]. Deepfake is being used in different tasks, for example, generating images and creating scenes and now it is also being used in the medical field [7]. Different approaches have been proposed to identify Deepfakes using different perspectives, including several methods based on deep learning techniques [8]. In the US, the Defense Advanced Research Projects Agency (DARPA) has launched a forensic project to identify altered media effectively [9]. This project reflects the importance of identifying fake media content. Furthermore, Facebook, with Microsoft’s collaboration, has introduced an AI-based challenge to Deepfake identification. This collaborative effort enhances the need to detect fake content and reduce the risk associated with fake media.
In the US, approximately 80 million scans are performed annually, as per the Harvard Health report [10]. In the past, GAN was primarily used for tampering images and videos but now, with the advancement of artificial intelligence (AI), there are many methods proposed, such as diffusion technology. The identification of such altered images to save human lives as well as to reduce treatment costs and the healthcare industry burden is a time-critical need. Many studies have been proposed in different contexts of fake content identification; however, in the medical field, few Deepfake studies exist. The other concern is the generalizability of the proposed fake media identification model. The existing proposed models are mainly based on GAN and their performance is effective only for the manipulation techniques for which they are trained. More simply, the existing proposed model is not suitable for the latest manipulation techniques, like diffusion.
With the advancement of deep learning, convolutional neural networks (CNNs) have been proposed as effective tools for identifying fake content due to the requirement of less pre-processing, and these models can easily learn filters and features. A solitary pulmonary round tissue mass is one type of lung cancer that is distinct from other types. If the diameter of the mass is greater than 8 mm, then the cancer is malignant, otherwise, it is benign. The two common image tampering methods are copy–move and image splicing. The copy–move method consists of duplicating the non-interesting area. This method can also be used to increase the frequency of the region of interest. Image splicing also works similarly to the copy–move method except that, in the image splicing method, the region of interest originates from an external image. The study [11] employs Mask R-CNN (Mask Region-based Convolutional Neural Network) and the Sobel filter to detect and localize copy–move and picture-splicing forgeries. The Sobel filter allows for the prediction of masks based on the identification of gradients that are similar to those of the real mask.
Deepfake attackers are now capable of learning from the collection of images to create an image that looks like a real image due to the GAN network that is based on two neural networks from which one is a generator and the other is a discriminator [6]. The generator network creates a fake sample and discriminator used to learn the difference between real and fake images. The problem addressed in this study is the identification of fraudulent medical images used to misrepresent medical conditions for insurance claims. The major aim of this study is to evaluate the deep learning models for the identification of fake medical images that are generated with stable diffusion techniques [12]. The key objectives of this study are listed below:
- We have fine-tuned several CNN and transformer-based models to evaluate the performance of the deep learning models for fake medical cancer detection through images.
- We have considered the stable diffusion techniques to generate the fake medical images and to assess the model performance for the identification of these images.
- A detailed histogram analysis was performed to understand distribution changes.
- A user study was performed to compare human perception and deep learning models for detecting fake medical images.
2. Literature Review
The process of creating AI images accelerates due to the advancement of deep learning techniques. Computer Vision has also gained popularity due to the advanced CNN models. However, acquiring the annotated dataset in the domain of medical imaging is a challenging task. To overcome this dataset limitation, Frid-Adar et al. [13] proposed a method to generate synthetic medical images using GANs. The performance of the CNN models, especially for medical image classification, was enhanced with their method. Their strategy was proven using a dataset of 182 liver lesions using computed tomography (CT) images. Training the CNN with both conventional and synthetic data augmentation increases sensitivity from 78.6% to 85.7% and specificity from 88.4% to 92.4%. The approach shows potential for improving medical image classification and could be used more widely in medical diagnostics. With this advancement, the medical field has also been affected due to medical image tampering. From the literature perspective, different studies were proposed based on deep learning and artificial intelligence for the prediction of fake images. In different domains, Deepfake technology is being increasingly used to insert images into videos. On the other hand, a patient’s medical data can be captured by filming a specific area of the body using a machine in a controlled environment. The ease of access raises concerns about the possibility of generating fake medical images. As a result, the demand for an effective model for the prediction of fake medical images is a current need due to the health concerns of patients.
Mirsky et al. [14] proposed a CT-GAN framework in their study that can add or remove tumors in patients’ lungs through CT scan images. To evaluate their proposed model, they selected three radiologists with 2.5 and 7 years of experience, respectively. Also in their study, they selected the trained model that won the 2017 Kaggle Data Science Bowl [15] for the identification of images with and without tampering. Their experiments were based on blind and open trials. In a blind trial, radiologists were unaware of any tampering. The blind trial found high rates of misdiagnosis: 99% of tumor-injected scans were mistaken as malignant, while 94% of tumor-removed scans were misdiagnosed as benign. In an open trial, after learning of the tampering, misdiagnosis rates dropped to 60% and 87%. In the blind study, an AI lung cancer detection program misidentified all manipulated patients. These findings show the increasing susceptibility of today’s medical systems to image-tampering attacks.
Several methods are proposed in response to identifying the tampering medical images. These methods are divided into two main categories, i.e., active and passive detection [16]. In active detection, before transmitting the image through a public channel, a known authentication code, such as a digital signature, is embedded in the image. The image is validated by comparing this embedded code with the original code.
A reign-based hybrid medical image watermarking scheme was introduced by Singh et al. [17]. To enhance security and temper detection accuracy, their proposed technique is based on hybrid domain watermarking techniques. To create a hybrid watermarking technique, Savaridass et al. [18] combined DVT and singular value decomposition in their study. Their proposed approach was tested for different attacks based on Gaussian noise, salt and pepper noise, and filtering. In another study [19], the authors combined DVT and singular value decomposition for a hybrid watermarking technique and tested for different attacks. The active detection techniques are effective, however, for these techniques, special hardware and software are required to add an authentication code in the image before transmitting. Therefore, passive detection techniques are popular as, when using these techniques, no prior information about the image is required. The passive techniques work by identifying local features using statistical and frequency information. Discrete Wavelet Transform (DVT) was used in the study of Thakur et al. [20] to divide the image into sub-images. In their study, speed-up robust features (SURFs) and a support vector machine (SVM) were used to extract features and perform classification tasks, respectively.
The objective of Sharafudeen et al. [21] was to address the detection of altered lung CT scans generated using CT-GANs via a comprehensive case study. They used machine learning algorithms and a unique three-dimensional deep neural architecture to distinguish between tampered and untampered data. On the CT-GAN dataset, the proposed architecture produced an impressive 91.57% accuracy, 91.42% sensitivity, and 97.20% specificity.
Dermoscopic skin lesion images were generated and detected using Conditional Generative Adversarial Networks (CGANs) and Vision Transformers (ViTs) in another study by Sharafudeen et. al. [22]. In their study, Derm-CGAN was created to accurately simulate six different dermoscopic skin lesions. The analysis revealed a strong connection between genuine and synthetic visuals, and they achieved an accuracy of 97.18%.
Albahli et al. [23] proposed a model named MedNet for detecting CT-Scan-based fake lung images. Their proposed model is mainly based on the pre-trained model EfficientNetV2-B4, and they achieved an accuracy of 85.49%.
Amiri et al. [24] proposed a model based on an evolutionary algorithm for the detection of copy–move forgery in medical images. Their proposed model also can detect fake blocks. Their dataset is based on a total of 300 images of which 200 are fake and 100 images are real [25]. They achieved a precision of 90.07% with their proposed model. Furthermore, the literature review summary is available in Table 1.

Table 1.
Literature review summary.
Research Gap
All the previous studies were mainly based on GAN-based fake images, but, as compared to the GAN, stable diffusion is the latest text-to-image manipulation method [12]. It is being used to generate high-quality images based on text descriptions. There are several steps involved, i.e., encoding, synthesis, and refinement in stable diffusion for converting text to image. The encoding part converts the text into the format that is capable of the synthesis part. The word embedding and attention mechanism can also be considered to capture meaningful information from the text. The image synthesis part is used to capture the visual information that is available in the text description. It can capture the controlled information of blurring, smoothing, and denoising the image. Regularization and control mechanisms can be applied to ensure the stability of the image. There are several applications of stable diffusion, including virtual content creation and media production. The use of such technology to generate fake media has increased due to the rapid advancement of stable diffusion. This tends to pose a significant challenge, especially in Deepfake medical images, as such disinformation and manipulation of medical images can lead to death as well as unnecessary treatment. This diffusion model is based on the latent diffusion model, and the equation of LDM can be seen in Equation (1) [12]. In Equation (1), a series of denoising autoencoders are represented by θ(zt, k); k = 1… T. These autoencoders are fine-tuned to predict a refined version of their input zt, where zt is a disturbed reproduction of the original input t, and y is the text prompt.
In this study, we have generated medical Deepfake data using cutting-edge diffusion technology, a novel approach in medical Deepfakes. Furthermore, CNNs have shown impressive effectiveness in uncovering Deepfake material, emphasizing their significance in this domain. While adept at extracting features from diminutive elements, CNNs might face difficulties pinpointing crucial areas within an image. Utilizing a ViT model [37] for medical Deepfake detection offers a compelling and promising alternative. ViTs were initially created for image classification tasks and have proven to have excellent performance in several computer vision benchmarks [38]. We have selected the ViT model and CNN-based pre-trained models to identify the robust model for stable-based fake malignant identification. This study’s major significant contributions are listed below.
- Fake Malignant Skin Cancer Dataset Preparation using Stable Diffusion-Based Mode;
- Dataset Preprocessing;
- Feature Analysis using Histogram;
- Training and Evaluation of ViT Model (Global Feature Extraction);
- Training and Evaluating CNN models with and without ImageNet (Weights).
3. Methodology
This section describes the dataset and the proposed methodology. The proposed method is designed to accurately identify the real and fake medical images generated by diffusion technology. The schematic diagram of the proposed study is presented in Figure 1.
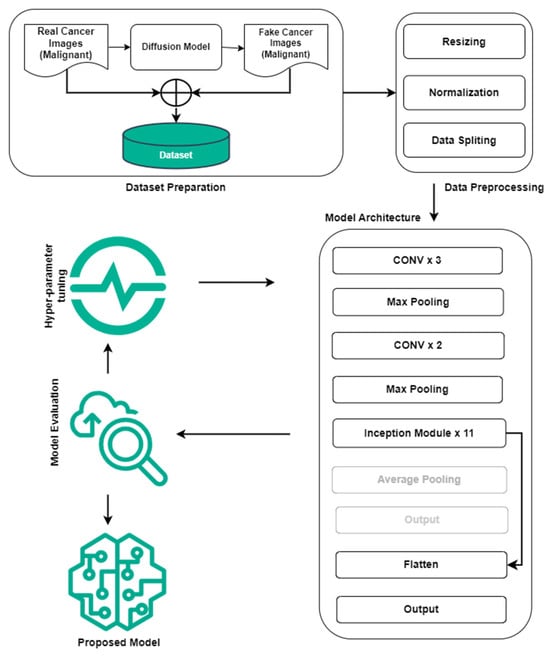
Figure 1.
Abstract diagram of the proposed study for real malignant and fake malignant cancer identification.
3.1. Dataset
We have selected malignant samples of the skin cancer dataset sourced from Kaggle [39]. Furthermore, with these malignant samples fake medical malignant images are generated using stable diffusion [12]. The real malignant class has 1197 samples for training and 300 samples for testing, and the fake malignant class also has 1197 and 300 samples for training and testing, respectively. Due to the balanced classes, and to avoid unnecessary filters, we have not used data augmentation. In the stable diffusion-based model, several parameters are available that need to be fine-tuned for high-quality images. The two important parameters are “num_inference_steps” and “image_guidance_scale”.
- Number of Inference Steps: This parameter determines the number of inference steps or iterations the diffusion model needs to complete to produce the final image. With each inference step, the image is further refined, bringing out more details and better quality. Higher-quality findings are usually obtained by increasing the number of inference stages, which also increases computing time. In our situation, effective images are generated for inference steps of 5.
- Image Guidance Scale: This parameter regulates how strongly the diffusion model receives image guidance. The influence of the starting image (if provided) on the generating process is referred to as image guidance. A greater value for “image_guidance_scale” increases the effect of the original image, which could result in generated images that preserve more aspects of the input image. We have set it to 1 in this study, and the influence of the original image remains at its initial level.
The real malignant and diffusion-based fake malignant skin cancer image samples can be seen in Table 2.

Table 2.
Real malignant and diffusion-based fake malignant skin cancer samples.
For visualization purposes, histograms [40] were generated to illustrate the pixel intensity and frequency distribution within the dataset containing real and synthetic skin cancer images. The average histograms of the training and testing dataset can be seen in Figure 2 and Figure 3, respectively. This graphical representation offers valuable insights into the inherent characteristics of authentic and artificially generated images, aiding in the visual analysis of image fidelity and authenticity.
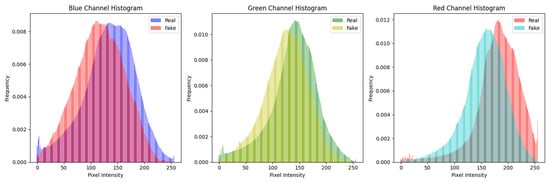
Figure 2.
Histogram of training dataset (real and fake malignant skin cancer images).
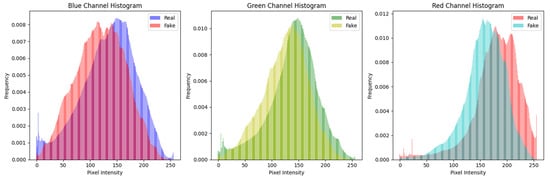
Figure 3.
Histogram of testing dataset (real and fake malignant skin cancer images).
3.2. ViT Network Architecture
In the current image processing domain, the ViT [37] is an effective alternative to CNN for computer vision tasks. Although CNN has been the go-to for image recognition, it has limitations like fixed hierarchical structures, limited receptive fields, and a lack of flexibility in handling images of varying resolutions. ViT overcomes these problems by leveraging self-attention processes to capture both local and global dependencies between picture patches, resulting in potentially higher performance, especially on tasks requiring understanding links between distant areas of the image. The key success of ViT lies in its approach to dealing with the images as patches, its adaptability, and its versatility.
The architecture of ViT is mainly based on tokenization and embedding. The input image is divided into patches, for example, the image will be divided into patches of size 16 × 16. These patches are then normalized after being flattened and converted via a linear operation into a higher-dimensional space. This feature of ViT enables it to learn global and local information of images. The ViT mathematically can be represented as assuming the image size is M × N, and the image is divided into P patches of size k × k. Each patch is represented as a flattened vector of length d. The total number of patches P can be calculated using Equation (2).
Overall, the ViT architecture consists of the four major components listed below.
Embedding/Patch Embedding: Each patch vector is linearly projected into higher-dimensional space using embedding matrix E (see Equation (3)).
Positional Embedding: To capture the spatial information, positional embeddings () for the N sequences are added to the patch embeddings of N sequences (). Usually, these positional embeddings are sinusoidal functions with different frequencies and phases, see Equation (4).
Transformer Blocks: The patch embeddings, including positional encodings, are transferred across multiple transformer blocks. Position-wise feedforward and multi-head self-attention layers contribute to each block.
Classification Head: The output of the final transformer block is passed to a classification head, which commonly consists of a single linear layer followed by SoftMax activation for classification tasks.
The design of the ViT centers around a multi-head self-attention (MSA). The outputs of the multiple attention heads are then concatenated and linearly processed before being passed into the subsequent layers. This technique enables the model to learn multiple representations from various angles, which enhances its capacity to extract relevant information from the input image. The MSA is based on Q, K, and V, which represent query, key, and value matrices. Transformers rely significantly on the self-attention mechanism, which is the foundation for explicitly modeling interactions and relationships across all sequences in prediction tasks. Unlike CNNs, which depend on local receptive fields, the self-attention layer collects insights and features throughout the input sequence, allowing it to identify local and global information. The different versions of ViT models are available with other parameters, i.e., layers, hidden size, and parameters; see Table 3. The ViT base model was selected for this study due to its low computation and architectural complexity. To avoid computational complexities and limited resources, we have selected the ideal size of the image, which is 224 × 224, and the patch size, which is 16 × 16, and the ViT base model.

Table 3.
ViT model variation specifications [37].
3.3. CNN-Based Pretrained Models
To demonstrate the patch-based technique and local extraction-based traditional CNN model technique, we have considered different well-known models, i.e., MobileNetV2 [41], EfficientNetB0 [42], Xception [43] and InceptionV3 [44], EfficientNetV2B0 [45], ResNet152V2 [46], VGG19 [47], ConvNeXt Base [48], CoAtNetT [49], and ResNeSt [50]. These models are selected because they have a reasonable number of parameters and layers (approximately) (see Table 4).

Table 4.
Pretrained model (CNN-based) variation specifications.
Transfer learning and pre-trained model weights are considered in this study. We have removed the fully connected layers from the network architecture and added one fully connected layer (with sigmoid function [51]) followed by a flattened layer. The fine-tuned InceptionV3 model can be seen in Figure 1. The other CNN-based pretrained models were also fine-tuned, similar to the InceptionV3 model.
4. Results and Discussion
This section consists of evaluation measures, software and hardware requirements, results, discussion, and robustness of the ViT and CNN-based models.
4.1. Evaluation Measures
In machine learning and deep learning, evaluation measures play an active role in measuring the model’s generalizability. This study has considered several measures to confirm the model’s generalizability. In Equations (5)–(7), the TP, TN, FP, and FN represent true positive, true negative, false positive, and false negative, respectively.
Accuracy: The accuracy represents the overall correctness of the model prediction. It is calculated by correctly classifying samples, i.e., True Prediction out of total testing samples (see Equation (5)). In other words, assume we have 100 samples for testing and the model correctly classified 80 samples, i.e., true fake and true real. The model accuracy is 80% in this case.
Precision: Precision represents the model’s ability to identify true positives from predicted ones. It can be calculated by dividing true positive predicted samples by all positive predicted samples, which can be either false or true positive (see Equation (6)).
Recall: Recall is also known as true positive rate or sensitivity. The recall represents the model’s ability to identify true positives from all actual positives.
F1: The harmonic mean of precision and recall is known to be F1. It offers one statistic that balances precision and recall, making it useful when the class distribution is unequal or where both errors are equally important.
4.2. Hardware Specifications
We have used the free version of T4 GPU provided by Google (Google Colab [52]) to conduct the experiments and preparation of the dataset. The hardware specifications related are system RAM: 12.7 GB, GPU RAM: 16 GB, and Disk Space: 78.2 GB.
4.3. Experimental Results and Discussion
The base ViT model was trained with the image size of 224 × 224 and patch size of 16 × 16. The experimental results concerning the training and validation graph, confusion matrix, and classification report are presented in this section with sufficient details. The limitation of the ViT models is that they are eager for datasets. A huge amount of training data is required for effective performance. We have 1197 real images and 1197 fake generated images of malignant cancers in a total of 2394 samples for training, while 300 real images and 300 fake generated images available for testing purposes. We have used 20% of the data as validation from 2394 samples of the training dataset.
The training accuracy and training loss start from 0.9713 and 0.2596, while validation accuracy and validation loss start from 0.9958 and 0.06, respectively. After epoch 5, the model performance is well generalized and at epoch 10 the training accuracy, validation accuracy, training loss, and validation loss were 1.0000, 1.0000, 0.0049, and 0.0049, respectively. The losses and accuracies graph that show the model’s generalizability can be seen in Figure 4.
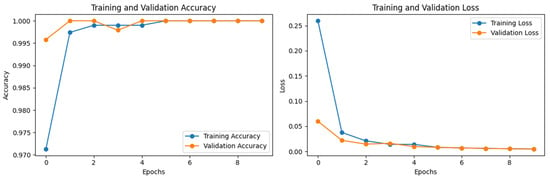
Figure 4.
Training and validation losses and accuracies of ViT base model for real (1) and fake (0) malignant skin cancer.
We have also considered the confusion matrix to evaluate the efficacy of the ViT model. A confusion matrix is an accurate evaluation of how well a classification model performs across classes. It enables practitioners to assess the model’s efficacy beyond basic accuracy measurements (see Figure 5 for the confusion matrix). In Figure 2, 0 represents the real malignant class and 1 represents the stable-based generated fake cancer.
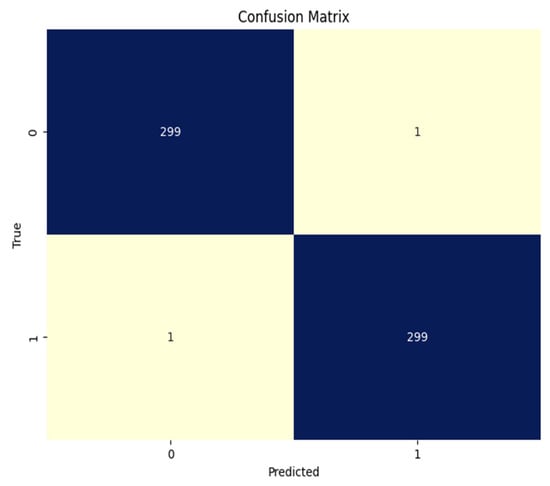
Figure 5.
Confusion matrix base ViT model for real (1) and fake (0) malignant skin cancer.
The classification report can be seen in Table 5, in which the model was evaluated for 600 samples (300 for each class) and performed well. The results are outperformed because accuracy, precision, recall, and f1 are all at 99.66%.

Table 5.
Classification report for real (1) and fake (0) malignant skin cancer.
We have also applied and fine-tuned the CNN-based pre-trained models in this study. In this study, for the case of CNN-based pre-trained models, the best performance was of the InceptionV3 model without weights, see Table 6. The model InceptionV3 without weights achieved training accuracy, train loss, validation accuracy, validation loss, test accuracy, and test loss of 0.995, 0.0017, 0.9979, 0.0062, 0.9966, and 0.0081, respectively; see Figure 6. The weighted precision, recall, and F1 score of InceptionV3 without ImageNet weights all are between 0.9950–0.9966.

Table 6.
Pre-trained (CNN models) scores with and without pretrained weights.
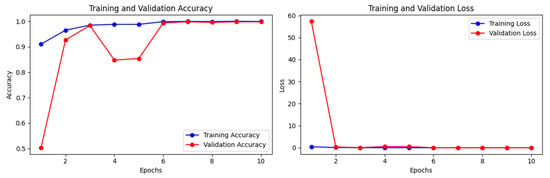
Figure 6.
Training and validation losses and accuracies of the InceptionV3 model without ImageNet for real (1) and fake (0) malignant skin cancer.
The Xception model without weights achieved training accuracy, train loss, validation accuracy, validation loss, test accuracy, and test loss of 1.0000, 0.0017, 0.9979, 0.0023, 0.9950, and 0.0110, respectively (see Figure 7). The weighted precision, recall, and F1 score of InceptionV3 without ImageNet weights are all between 0.9950 and 1.000.
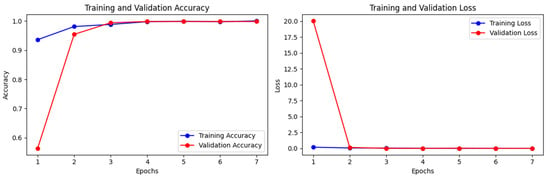
Figure 7.
Training and validation losses and accuracies of the Xception model without ImageNet for real (1) and fake (0) malignant skin cancer.
The ResNet152V2 model without ImageNet weights achieved training accuracy, train loss, validation accuracy, validation loss, test accuracy, and test loss of 1.0000, 0.0014, 0.9958, 0.0272, 0.9933, and 0.0031, respectively; see Figure 8. The weighted precision, recall, and F1 score of InceptionV3 without ImageNet weights are all between 0.9900 and 0.9933.
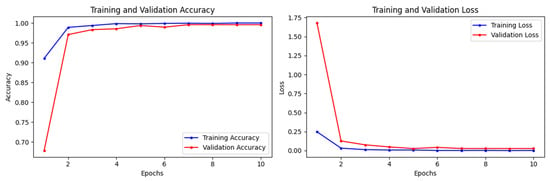
Figure 8.
Training and validation losses and accuracies of ResNet152V2 model without ImageNet for real (1) and fake (0) malignant skin cancer.
From the detailed experiments, we have found that the ViT base model performance and InceptionV3 (without weights) are equal in test accuracy. The training, validation, and test accuracy are 100% for the case of the ViT model; however, the ViT model has 86 M parameters. Furthermore, the ImageNet weights are not used in the InceptionV3 model; it also has 22 M parameters. InceptionV3 has a slightly lower score (point score difference) both in terms of training and validation. The network structure of InceptionV3 is based on 159 layers, and ViT is based on only 12 layers. Figure 9 compares the ViT base model and InceptionV3 (without weights).
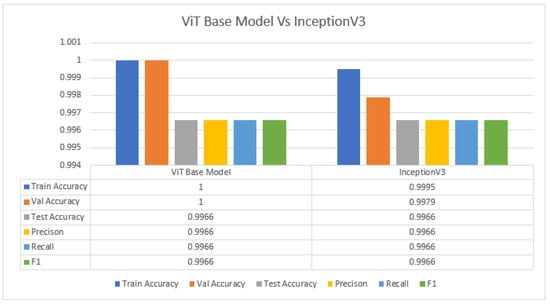
Figure 9.
ViT base model vs. InceptionV3 (without weights) for real (1) and fake (0) malignant skin cancer.
Furthermore, to verify the generalization of the proposed InceptionV3 model, we have also performed experiments with the StratifiedKFold with the k = 3 method. The folds’ results can be seen in Figure 10. Both average train accuracy and average validation accuracy are ~100%, and, after complete training and with the best model-trained weights, the test accuracy is 99.17%, and the test loss is 0.0302.
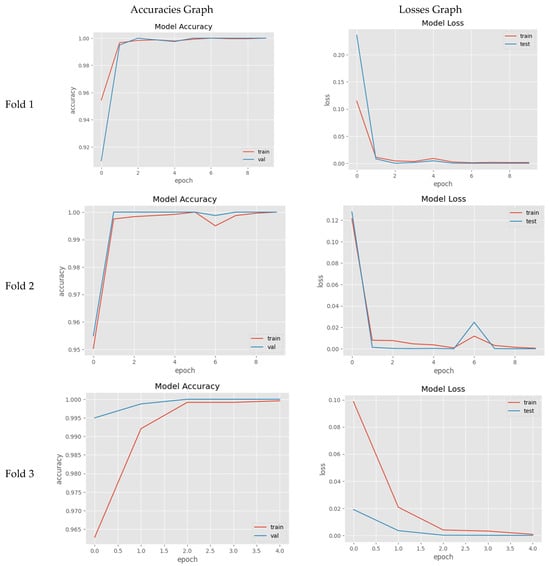
Figure 10.
StratifiedKFold with k = 3 and proposed InceptionV3 (without weights) learning graphs for real (1) and fake (0) malignant skin cancer.
4.4. Practical Implications
Deepfake Technologies serve as crucial ethical problems, especially in the healthcare domain. One of the concerns is that these technologies can be used for medical image manipulation leading to incorrect diagnosis and this situation can be harmful for patients. Furthermore, serious privacy concerns have been raised due to the unauthorized use of personal medical data. To address these concerns, we have proposed a DL-based model for identifying the manipulation in the medical images. The overall implications of our study are significant in the context of combating the proliferation of fake medical images, particularly in the era of AI and Deepfake technology. By demonstrating the effectiveness of our proposed models in identifying fake images generated by diffusion-based technology, we contribute to the advancement of techniques for detecting fraudulent medical data.
Our findings highlight the potential of leveraging deep learning models, such as ViT and InceptionV3, for detecting fake medical images with high accuracy. With the integration of this model in healthcare, fraudulent insurance claims can be reduced. This capability is crucial for safeguarding the integrity of medical data, preventing incorrect diagnoses and unnecessary treatments, and mitigating financial losses associated with medical fraud.
Furthermore, to better explain the robustness of the proposed model, we involved humans in training and testing the identification of real and fake images. Initially, humans were trained (see Table A1) by showing them both real and fake images at the first step, and then they were shown whether the identification they made was correct or not. This activity was repeated over ten image pairs (one real and one fake) so they could learn to identify what features they used to differentiate between them. After the training, they were shown test images to observe how accurately they could classify the test images (see Table A2). A key objective of this activity was to know if humans could utilize any factors to classify this type of task. The average accuracy of 20 users (age between 20–30 years and minimum educational qualification was undergraduates) was observed to be 68%—Figure 11 demonstrates the percentage of accuracy from the total testing images (y-axis) by percentage of users involved (x-axis). Our results indicate that the use of ML approaches, in general, has demonstrated higher accuracy > 88%, except in two cases, in which the demonstrated accuracy was less than the human accuracy in this research study (see Figure 12 for results and comparisons).
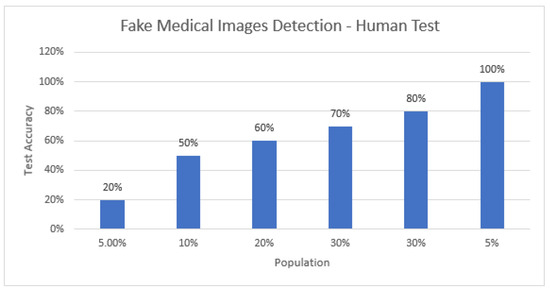
Figure 11.
Percentage of accuracy from the total testing images (y-axis) by percentage of users involved (x-axis).
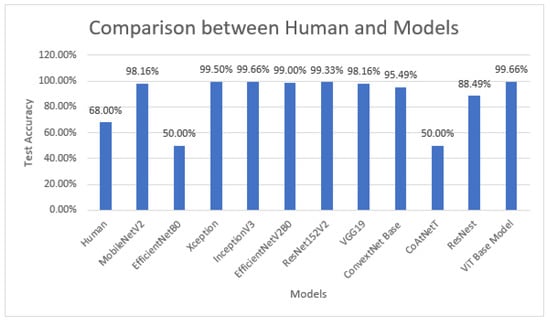
Figure 12.
Human accuracy vs. deep learning models’ accuracy.
5. Conclusions
The process of creating fake images, videos, and audio is easy due to the advancement of AI, neural networks, and Deepfake technology. Manipulating medical images can lead to patient deaths and unnecessary treatments. All the previous studies mainly cover one Deepfake technology, GAN, while diffusion technology is being used nowadays to create fake images and videos. This aspect was unexplored in previous studies, especially for medical Deepfakes, as far as we know. We have collected images of malignant skin cancer from the open-source Kaggle. We have generated the corresponding fake malignant images, i.e., AI-generated images using diffusion-based models. After preparing the dataset, preprocessing is applied to the final dataset. The ViT base model and CNN-based models are trained with and without pre-trained weights (ImageNet weights). The results show that the ViT base model and InceptionV3 model performed best in terms of accuracy, precision, recall, and an F1 of 99.66% for medical Deepfake malignant skin cancer identification. The limitation of this study is binary classification and the focus on skin cancer images generated through stable diffusion. In the future, we aim to expand the dataset with different Deepfake Technologies to consider this problem as multiclass and consider diverse datasets rather than only skin images.
Author Contributions
Conceptualization, M.A.A., S.M., Ș.C.G., N.U., S.A. and C.D.; methodology, M.A.A., S.M., Ș.C.G., N.U., S.A. and C.D.; validation, M.A.A., S.M., Ș.C.G., N.U., S.A. and C.D.; investigation, M.A.A., S.M., Ș.C.G., N.U., S.A. and C.D.; data curation, M.A.A., S.M., Ș.C.G., N.U., S.A. and C.D.; writing—original draft preparation, M.A.A., S.M., Ș.C.G., N.U., S.A. and C.D.; writing—review and editing, M.A.A., S.M., Ș.C.G., N.U., S.A. and C.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The fake malignant images are generated through a stable diffusion technique while the base original malignant images are retrieved from Kaggle (https://www.kaggle.com/datasets/fanconic/skin-cancer-malignant-vs-benign, accessed on 6 May 2024). The complete dataset of this article will be made available by the authors upon request. However, the StratifiedKFold with k = 3 based model weights and test images for verification is available at https://github.com/Muhammad-Asad-Arshed/Detecting-Fake-Medical-Images-Skin-Cancer-, accessed on 21 August 2024).
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Train Samples for Human with Labels.
Table A1.
Train Samples for Human with Labels.
| Image | Label | Image | Label |
|---|---|---|---|
 | Real |  | Fake |
 | Fake |  | Real |
 | Real |  | Fake |
 | Fake |  | Real |
 | Real |  | Fake |
 | Fake |  | Real |
 | Real |  | Fake |
 | Real |  | Fake |
 | Real |  | Fake |
 | Real |  | Fake |

Table A2.
Test Samples for Human (5 Real and 5 Fake Images).
Table A2.
Test Samples for Human (5 Real and 5 Fake Images).
| Image | Label | Image | Label |
|---|---|---|---|
 | Real |  | Fake |
 | Fake |  | Real |
 | Real |  | Fake |
 | Fake |  | Real |
 | Fake |  | Real |
References
- Brenner, D.J.; Hall, E.J. Computed Tomography—An Increasing Source of Radiation Exposure. N. Engl. J. Med. 2007, 357, 2277–2284. [Google Scholar] [CrossRef] [PubMed]
- McLean, I.D.; Martensen, J. Specialized Imaging. In Clinical Imaging: With Skeletal, Chest, & Abdominal Pattern Differentials, 3rd ed.; Mosby: Maryland Heights, MO, USA, 2014; pp. 44–78. [Google Scholar] [CrossRef]
- Strickland, N.H. Current topic: PACS (picture archiving and communication systems): Filmless radiology. Arch. Dis. Child. 2000, 83, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Mejía-Granda, C.M.; Fernández-Alemán, J.L.; Carrillo-De-Gea, J.M.; García-Berná, J.A. Security vulnerabilities in healthcare: An analysis of medical devices and software. Med. Biol. Eng. Comput. 2023, 62, 257–273. [Google Scholar] [CrossRef]
- Christiaan, B. McAfee Researchers Find Poor Security Exposes Medical Data to Cybercriminals|McAfee Blog. Available online: https://www.mcafee.com/blogs/other-blogs/mcafee-labs/mcafee-researchers-find-poor-security-exposes-medical-data-to-cybercriminals/ (accessed on 28 April 2024).
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Sci. Robot 2014, 3, 2672–2680. [Google Scholar] [CrossRef]
- Patel, Y.; Tanwar, S.; Gupta, R.; Bhattacharya, P.; Davidson, I.E.; Nyameko, R.; Aluvala, S.; Vimal, V. Deepfake Generation and Detection: Case Study and Challenges. IEEE Access 2023, 11, 143296–143323. [Google Scholar] [CrossRef]
- Passos, L.A.; Jodas, D.; Costa, K.A.P.; Júnior, L.A.S.; Rodrigues, D.; Del Ser, J.; Camacho, D.; Papa, J.P. A review of deep learning-based approaches for deepfake content detection. Expert Syst. 2024, 41, e13570. [Google Scholar] [CrossRef]
- Media Forensics. Available online: https://www.darpa.mil/program/media-forensics (accessed on 11 July 2023).
- Radiation Risk from Medical Imaging—Harvard Health. Available online: https://www.health.harvard.edu/cancer/radiation-risk-from-medical-imaging (accessed on 28 April 2024).
- Wang, X.; Wang, H.; Niu, S.; Zhang, J. Detection and localization of image forgeries using improved mask regional convolutional neural network. Math. Biosci. Eng. 2019, 16, 4581–4593. [Google Scholar] [CrossRef]
- Rombach, R.; Blattmann, A.; Lorenz, D.; Esser, P.; Ommer, B. High-Resolution Image Synthesis with Latent Diffusion Models. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 10674–10685. [Google Scholar]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Mirsky, Y.; Mahler, T.; Shelef, I.; Elovici, Y. {CT-GAN}: Malicious Tampering of 3D Medical Imagery using Deep Learning. In Proceedings of the 28th USENIX Security Symposium, Santa Clara, CA, USA, 14–16 August 2019. [Google Scholar]
- Liao, F.; Liang, M.; Li, Z.; Hu, X.; Song, S. Evaluate the malignancy of pulmonary nodules using the 3D deep leaky noisy-or network. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3484–3495. [Google Scholar] [CrossRef]
- Birajdar, G.; Mankar, V. Digital image forgery detection using passive techniques: A survey. Digit. Investig. 2013, 10, 226–245. [Google Scholar] [CrossRef]
- Singh, P.; Devi, K.J.; Thakkar, H.K.; Kotecha, K. Region-based hybrid medical image watermarking scheme for robust and secured transmission in IoMT. IEEE Access 2022, 10, 8974–8993. [Google Scholar] [CrossRef]
- Savaridass, M.P.; Deepika, R.; Aarnika, R.; Maniraj, V.; Gokilanandhi, P.; Kowsika, K. Digital watermarking for medical images using DWT and SVD technique. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1084, 012034. [Google Scholar] [CrossRef]
- Mohammed, A.; Jebur, B.A.; Younus, K.M. Hybrid DCT-SVD based digital watermarking scheme with chaotic encryption for medical images. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1152, 012025. [Google Scholar] [CrossRef]
- Thakur, T.; Singh, K.; Yadav, A. Blind approach for digital image forgery detection. Int. J. Comput. Appl. 2018, 179, 34–42. [Google Scholar] [CrossRef]
- Sharafudeen, M.; Chandra, S.S.V. Medical Deepfake Detection using 3-Dimensional Neural Learning. In Proceedings of the Artificial Neural Networks in Pattern Recognition: 10th IAPR TC3 Workshop, ANNPR 2022, Dubai, United Arab Emirates, 24–26 November 2022; Lecture Notes in Computer Science. Volume 13739, pp. 169–180. [Google Scholar] [CrossRef]
- Sharafudeen, M.; Chandra, S.S.V. Leveraging Vision Attention Transformers for Detection of Artificially Synthesized Dermoscopic Lesion Deepfakes Using Derm-CGAN. Diagnostics 2023, 13, 825. [Google Scholar] [CrossRef]
- Albahli, S.; Nawaz, M. MedNet: Medical deepfakes detection using an improved deep learning approach. Multimed. Tools Appl. 2024, 83, 48357–48375. [Google Scholar] [CrossRef]
- Amiri, E.; Mosallanejad, A.; Sheikhahmadi, A. The Optimal Model for Copy-Move Forgery Detection in Medical Images. J. Med Signals Sens. 2024, 14, 5. [Google Scholar] [CrossRef]
- High Resolution Images Create a Pseudo-Pulmonary Embolism (PE) Type Appearance—Chest Case Studies—CTisus CT Scanning. Available online: https://www.ctisus.com/teachingfiles/cases/chest/285194 (accessed on 10 August 2024).
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Proceedings of the Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Lecture Notes in Computer Science. pp. 740–755. [Google Scholar] [CrossRef]
- Wen, B.; Zhu, Y.; Subramanian, R.; Ng, T.-T.; Shen, X.; Winkler, S. COVERAGE—A novel database for copy-move forgery detection. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 161–165. [Google Scholar]
- Hsu, Y.; Chang, S. Detecting image splicing using geometry invariants and camera characteristics consistency. In Proceedings of the 2006 IEEE International Conference on Multimedia and Expo, Toronto, ON, Canada, 9–12 July 2006; Available online: https://ieeexplore.ieee.org/abstract/document/4036658/ (accessed on 4 May 2024).
- Armato, S.G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Med Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef]
- SIPI Image Database. Available online: https://sipi.usc.edu/database/ (accessed on 4 May 2024).
- Kaggle: Your Machine Learning and Data Science Community. Available online: https://www.kaggle.com/ (accessed on 4 May 2024).
- Medical Images Home. Available online: http://www.onlinemedicalimages.com/index.php/en/ (accessed on 4 May 2024).
- The STARE Project. Available online: http://cecas.clemson.edu/~ahoover/stare/ (accessed on 4 May 2024).
- Tagare, H.D.; Jaffe, C.C.; Duncan, J. Medical image databases: A content-based retrieval approach. J. Am. Med. Inform. Assoc. 1997, 4, 184–198. [Google Scholar] [CrossRef]
- Budhiraja, R.; Kumar, M.; Das, M.; Bafila, A.S.; Singh, S. MeDiFakeD: Medical Deepfake Detection using Convolutional Reservoir Networks. In Proceedings of the 2022 IEEE Global Conference on Computing, Power and Communication Technologies (GlobConPT), New Delhi, India, 23–25 September 2022. [Google Scholar]
- The Lung Image Database Consortium image collection (LIDC-IDRI)|IEEE DataPort. Available online: https://ieee-dataport.org/documents/lung-image-database-consortium-image-collection-lidc-idri (accessed on 6 May 2024).
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Y.; Zhang, Y.; Wang, Y.; Hou, F.; Yuan, J.; Tian, J.; Zhang, Y.; Shi, Z.; Fan, J.; He, Z. A survey of visual transformers. IEEE Trans. Neural Netw. Learn. Syst. 2024, 35, 7478–7498. [Google Scholar] [CrossRef]
- Skin Cancer: Malignant vs. Benign. Available online: https://www.kaggle.com/datasets/fanconic/skin-cancer-malignant-vs-benign (accessed on 6 May 2024).
- Javed, R.; Rahim, M.S.M.; Saba, T.; Fati, S.M.; Rehman, A.; Tariq, U. Statistical Histogram Decision Based Contrast Categorization of Skin Lesion Datasets Dermoscopic Images. Comput. Mater. Contin. 2021, 67, 2337–2352. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, ICML 2019, Long Beach, CA, USA, 9–15 June 2019; pp. 10691–10700. Available online: https://arxiv.org/abs/1905.11946v5 (accessed on 10 May 2024).
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNetV2: Smaller Models and Faster Training. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Volume 139, pp. 10096–10106. Available online: https://arxiv.org/abs/2104.00298v3 (accessed on 10 August 2024).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. In Proceedings of the Computer Vision—ECCV 2016—14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; Lecture Notes in Computer Science. Volume 9908, pp. 630–645. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015; Available online: https://arxiv.org/abs/1409.1556v6 (accessed on 12 July 2023).
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11966–11976. [Google Scholar]
- Dai, Z.; Liu, H.; Le, Q.V.; Tan, M. CoAtNet: Marrying Convolution and Attention for All Data Sizes. Adv. Neural Inf. Process. Syst. 2021, 34, 3965–3977. [Google Scholar]
- Zhang, H.; Wu, C.; Zhang, Z.; Zhu, Y.; Lin, H.; Zhang, Z.; Sun, Y.; He, T.; Mueller, J.; Manmatha, R.; et al. ResNeSt: Split-Attention Networks. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), New Orleans, LA, USA, 19–20 June 2022; pp. 2735–2745. [Google Scholar] [CrossRef]
- Nwankpa, C.; Ijomah, W.; Gachagan, A.; Marshall, S. Activation functions: Comparison of trends in practice and research for deep learning. arXiv 2018, arXiv:1811.03378. [Google Scholar]
- Google Colaboratory. Available online: https://colab.google/ (accessed on 9 September 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).