Abstract
The integration of artificial intelligence into healthcare has advanced rapidly in recent years, with multimodal approaches emerging as promising tools for improving diagnostic accuracy and clinical decision making. These approaches combine heterogeneous data sources such as medical images, electronic health records, physiological signals, and clinical notes to better capture the complexity of disease processes. Despite this progress, only a limited number of studies offer a unified view of multimodal AI applications in medicine. In this review, we provide a comprehensive and up-to-date analysis of machine learning and deep learning-based multimodal architectures, fusion strategies, and their performance across a range of diagnostic tasks. We begin by summarizing publicly available datasets and examining the preprocessing pipelines required for harmonizing heterogeneous medical data. We then categorize key fusion strategies used to integrate information from multiple modalities and overview representative model architectures, from hybrid designs and transformer-based vision-language models to optimization-driven and EHR-centric frameworks. Finally, we highlight the challenges present in existing works. Our analysis shows that multimodal approaches tend to outperform unimodal systems in diagnostic performance, robustness, and generalization. This review provides a unified view of the field and opens up future research directions aimed at building clinically usable, interpretable, and scalable multimodal diagnostic systems.
1. Introduction
The increasing availability of digitalized medical data has created an opportunity for artificial intelligence (AI) [1] to improve clinical decision making. Medical diagnostics have traditionally relied on unimodal data sources, such as radiological images [2], clinical notes, or physiological signals analyzed independently, while effective in many cases, these unimodal systems do not fully represent the complexity of human diseases, which are often manifested by diverse and interconnected signals. Diseases such as cancer, dementia, cardiovascular disease, and metabolic disorders often require interpretation of data from multiple modalities to ensure accurate diagnosis and treatment.
Multimodal Artificial Intelligence (MAI) addresses these limitations by integrating heterogeneous data sources, including images, signals, structured records, and clinical narratives, into a unified analytical framework. These systems exploit the complementary strengths of each modality to improve diagnostic robustness, prediction accuracy, and clinical interpretability. Recent reviews have focused on MAI’s ability to reduce the fragmentation of healthcare data, capture potential cross-modal interactions, and mimic the general reasoning used by clinicians [3,4,5,6]. Different studies, like those by Simon et al. [7], Demirhan et al. [8], and Adewumi et al. [9] provide complementary perspectives on fusion taxonomies, medical QA systems, fairness and bias, and the transition from large language models to multimodal frameworks. These works underscore the shift toward standardized evaluation, the emergence of general-purpose architectures, and unresolved challenges related to scalability, interpretability, and equitable deployment. Disease-focused reviews such as Isavand et al. [10] further highlight the relevance of multimodal artificial intelligence in oncology, particularly for modeling tumor heterogeneity and therapy response in B-cell non-Hodgkin lymphoma. In addition, MAI has demonstrated high performance gains in medical applications such as tumor classification, dementia subtyping, fetal risk assessment, and critical care prognosis. Beyond traditional hospital-based diagnostics, multimodal artificial intelligence is gaining traction in real-world applications such as portable systems, low-power sensor fusion platforms, and adaptive patient monitoring workflows. Emerging studies have explored the integration of thermal, ultrasound, or SAR sensors with lightweight neural models for real-time diagnostics [11]. Others highlight the potential of hybrid soft-computing approaches for intermediate fusion and benchmarking [12], or adaptive feedback mechanisms in portable patient monitoring systems [13]. These directions signal the expanding scope of MAI toward mobile, efficient, and responsive clinical tools. The fundamental problems in multimodal learning are explained by Baltrušaitis et al. [14] and Liang et al. [5]. The main challenges consist of representation, alignment, inference, generation, transference, and quantification. The clinical domain faces additional challenges because of unbalanced data and modality-specific noise, and the requirement for interpretation. The Barua et al. [4] review further underscores how the research level of MAI faces barriers to clinical use because there are no standardized fusion frameworks, and domain-specific benchmarks do not exist. A recent survey by Huang et al. [15] emphasizes the lack of integration across clinical tasks, recommending future frameworks to better capture temporal dependencies and personalization in MAI systems.
Deep learning based multimodal artificial intelligence systems [16] are increasingly popular because of their ability to extract and combine hierarchical features from unstructured and structured data. As demonstrated in several works, models combining transformers, convolutional networks, and hybrid attention mechanisms show strong performance in disease classification, segmentation, retrieval, and risk prediction tasks [3,17]. However, there are still challenges, especially in dealing with missing modalities, generalizing across institutions, and achieving regulatory approval.
This review explores recent advances in multimodal artificial intelligence for medical diagnostics. It presents a taxonomy of multimodal datasets, examines preprocessing techniques for modality harmonization, categorizes fusion methods, and compares model architectures and their performance. Thereby, it highlights technological advances, interpretability challenges, and real-world implications of MAI in clinical practice.
1.1. Motivation
Multimodal artificial intelligence represents a modern, novel methodology to healthcare. Traditional machine learning pipelines rely heavily on isolated data streams, resulting in an incomplete and potentially misleading understanding of a patient’s health status. Furthermore, data fragmentation increases the risk of delayed intervention, diagnostic errors, and poor outcome prediction.
The growing availability of integrated Electronic Health Record (EHR) systems [18], wearable devices, radiology archives, and biomedical datasets offers a resourceful environment for MAI. However, this opportunity has not been fully exploited due to the lack of a unified framework for managing, matching, and interpreting high-dimensional, heterogeneous inputs. Reviews by Liang et al. [5] and Pei et al. [3] highlight the growing need for architectures that can jointly learn from different modalities while ensuring interpretability and reliability in clinical settings. The clinical value of multimodal artificial intelligence lies in its ability to improve diagnostic confidence, support personalized treatment, and automate time-consuming processes. This shift toward integrated learning matches the real-world diagnostic process, in which doctors typically combine imaging, lab results, and patient history. MAI narrows the gap between algorithmic inference and clinical reasoning.
1.2. Contributions
This review provides a structured and synthetic overview of recent progress in multimodal artificial intelligence for medical diagnosis applications. We review the current literature from different perspectives:
- Availability and characteristics of multimodal datasets used in recent studies.
- Preprocessing techniques to improve data quality and cross-modality coordination, including normalization, resampling, and feature selection.
- Multimodal fusion strategies, including early fusion, intermediate-level feature concatenation, and cross-modal attention mechanisms for representation alignment.
- Deep learning architectures, such as convolutional neural networks (CNNs), transformer-based models, and optimization-based classifiers (e.g., Kernel Extreme Learning Machine (KELM)).
The paper is organized into distinct sections that reflect the pipeline of multimodal learning: from data collection and fusion to model design and application. We conclude with a synthesis of key findings and outline challenges and directions for future research in this rapidly growing field. Our review also summarizes and compares methods across studies, identifies trends in model performance, and emphasizes the importance of clinical applicability and generalization.
2. Methodology
2.1. Selection Criteria
- Time frame: Articles published between late 2023 and 2025 were selected to reflect the most recent developments in MAI for medical diagnostics. This time frame was chosen to capture emerging work on transformer-based models, instruction-tuned LLMs, neural architecture search, and hybrid fusion frameworks.
- Scope: The review focuses on peer-reviewed studies that develop and evaluate machine learning or deep learning models using two or more data modalities for clinical diagnostic purposes. These modalities include imaging (e.g., MRI, CT, and fundus photography), structured EHR data (e.g., lab values and diagnoses), physiological signals (e.g., ECG and CTG), and free-text input (e.g., radiology reports and QA pairs). Papers addressing fusion techniques, preprocessing strategies, model architectures, and real-world evaluation are included.
- Dataset searches: Articles were identified through targeted keyword searches across Google Scholar, IEEE Xplore, and ScienceDirect. Search terms included combinations of the following: “multimodal artificial intelligence”, “multimodal machine learning”, “medical diagnostics”, “fusion techniques”, “deep learning in radiology”, “multimodal EHR”, “vision-language models in healthcare”, and “multimodal health data”.
2.2. Selection Steps
- Titles were initially screened for relevance to multimodal learning and diagnostic applications.
- Abstracts and full texts were reviewed to confirm the use of multiple data modalities and relevance to clinical diagnosis or prediction.
- Articles were excluded if they were outside the medical domain, focused solely on unimodal inputs, or were non-English publications.
- Duplicates were removed, and only the most representative or impactful papers were retained for detailed inclusion.
- Only studies directly cited and analyzed in the present review are included in the tables and synthesis.
To ensure consistency in inclusion, studies were retained only if they presented original, peer-reviewed results, used at least two distinct modalities, and reported quantitative model evaluation for clinical diagnostic tasks. Case reports, editorials, and papers lacking fusion methods or unimodal-only approaches were excluded.
Following this selection process, a final set of articles was obtained for the literature review, ensuring a diverse representation of studies addressing the intersection of multimodal artificial intelligence and medical diagnosis. This review was conducted in accordance with the PRISMA guidelines. The PRISMA flow diagram [19] in Figure 1 summarizes the steps followed during the identification, screening, and inclusion phases that led to the final selection of studies considered in this work.
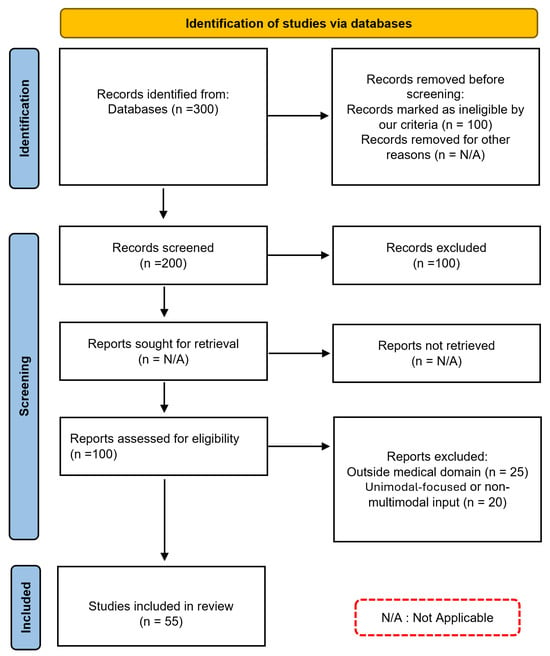
Figure 1.
PRISMA diagram: Systematic Selection Process for our Literature Review.
3. Multimodal Datasets
A variety of multimodal datasets have been developed to support research in medical diagnostics, each combining different types of data such as images, clinical records, and text annotations. These datasets enable the training and evaluation of machine learning models capable of learning from complex, real-world healthcare data. Below, we summarize some of the leading publicly available datasets that have been used to advance multimodal medical AI, as shown in Table 1.
PAD-UFES-20 dataset: Introduced by Pacheco et al. [20], includes 2298 clinical images and associated metadata representing 1373 patients and 1641 skin lesions. Data were collected between 2018 and 2019 through the Dermatological and Surgical Assistance Program (PAD) at the Federal University of Espírito Santo, Brazil. The dataset includes smartphone-captured images in different resolutions and lighting conditions. It covers six diagnostic categories: basal cell carcinoma (BCC), squamous cell carcinoma (SCC), actinic keratosis (ACK), seborrheic keratosis (SEK), Bowen’s disease (BOD), melanoma (MEL), and nevus (NEV). Clinical metadata includes 21 attributes such as age, sex, lesion location, skin type, and lesion size. Approximately 58.4% of lesions are biopsy-proven. Data were quality controlled for image resolution and completeness, and clinical attributes were curated for accuracy and completeness before translation into English.
MedICaT dataset: The MedICaT dataset, published by Subramanian et al. [21], contains 217,060 medical images derived from 131,410 open access biomedical research papers. The dataset contains multiple types of text information which include figure captions together with inline references extracted from full-text papers, sub-figure, and sub-caption annotations. It also contains 75% composite figures, while 74% of images include inline references which enables extensive analysis of image-text relationships. The dataset enables researchers to work on sub-figure-to-sub-caption alignment tasks and figure retrieval. It serves as a challenging evaluation resource for visual medical question answering and document-level image retrieval, and scientific article figure understanding.
FFA-IR dataset: The FFA-IR (Findings-Focus-Alignment with Image Reports) dataset [22] is a large benchmark designed for the generation of descriptive and reliable medical reports. It contains 1330 fundus images paired with Chinese and English diagnostic reports and fine-grained lesion annotations. Each sample contains structured eye disease tags, manually aligned textual descriptions, and bounding box annotations of common lesions (e.g., microaneurysms and hemorrhages). These images and reports were selected by experienced ophthalmologists to promote research in explainable VQA and lesion-based grounding. The dataset enables alignment of report elements with specific image regions, supporting training and evaluation of both generative and retrieval-based multimodal models.
MIMIC-III and MIMIC-IV datasets: The Medical Information Mart for Intensive Care (MIMIC) is a series of publicly available datasets developed by the MIT Lab for Computational Physiology. MIMIC-III [23] includes de-identified health records for over 53,000 ICU patients admitted to Beth Israel Deaconess Medical Center between 2001 and 2012. It contains detailed structured data such as demographics, vital signs, laboratory test results, medications, ICD-9 codes, and time-series physiological signals. MIMIC-IV [24] extends this resource to cover patient data from 2008 to 2019 and introduces a modular format organized into core hospital data, ICU-specific records, and clinical notes. It includes expanded and modernized records of procedures, diagnoses, laboratory events, and medication administrations, with structured linkage between unstructured text (e.g., radiology reports) and tabular observations. Both datasets support multimodal learning tasks involving structured, temporal, and textual EHR components, serving as essential resources for benchmarking and advancing clinical AI research.
ROCO and ROCOv2 datasets: The Radiology Objects in COntext (ROCO) dataset [25] and its extended version, ROCOv2 [26] were introduced to support multimodal research combining medical imaging with natural language descriptions. ROCO comprises over 81,000 radiology images collected from the Open Access Biomedical Image Search Engine (OpenI) and annotated with corresponding figure captions, MeSH terms, and modality labels. It covers multiple modalities, including X-rays, CT, and MRI, covering diverse anatomical regions and diseases. ROCOv2 enhances this dataset by refining annotations and incorporating an additional 11,000 images from PMC-OAI articles, ensuring a wider domain coverage. Each sample is associated with detailed metadata, such as article titles, captions, and biomedical concepts, enabling supervised and self-supervised learning tasks. These datasets serve as benchmarks for tasks like cross-modal retrieval, vision-language grounding, and medical report generation.
Pediatric Radiology dataset: This dataset comprises 180 radiographic images from pediatric cases paired with clinically oriented multiple-choice diagnostic questions (MCQs). The images and questions were sourced from the public educational platform and textbook: Pediatric Imaging: A Pediatric Radiology Textbook and Digital Library [27]. The dataset includes cases from various anatomical regions such as the chest, abdomen, musculoskeletal system, and head. It is formatted to support research in instruction-following and diagnostic reasoning for pediatric vision-language tasks.
Guangzhou Women and Children’s Medical Center NEC dataset: The data [28] belongs to 2234 infants admitted to the Guangzhou Women and Children’s Medical Center between December 2011 and March 2020. These infants were admitted with symptoms of abdominal distention, bradycardia, or bloody stool, and diagnosed with NEC according to modified Bell’s staging criteria, which is based on radiological signs and clinical parameters. The dataset comprises abdominal radiographs (ARs) (at 6-hour intervals) and 23 structured clinical parameters (demographic, symptomatic, and laboratory-based), all annotated and reviewed by experienced pediatricians. Multiple ARs were obtained per patient, with different sampling strategies depending on the NEC subtype.
Multimodal dataset for Lupus Erythematosus Subtypes: Created by Li et al. [29] from 25 hospitals in China and covers 446 examples. The data contain 800 clinical skin photographs taken by a camera or smartphone, 3786 multicolor immunohistochemistry (multi-IHC) CD4, CD8, CD11b, CD19, DAPI marker images, and clinical data converted to systemic involvement indexed by SII from history and laboratory results. Diagnosis included four types of lupus erythematosus (LE) and eight relevant skin conditions, excluding cases with presentations of various connective tissue pathologies. Validation against the medical facts included cross-checking with medical records, follow-up telephone calls, and review by a dermatology expert.
CTU-UHB Intrapartum CTG dataset: The CTU-UHB Open Access Intrapartum CTG Dataset [30] comprises 552 fetal heart rate (FHR) recordings which were obtained between April 2010 and August 2012 at the obstetrics ward of the University Hospital in Brno, Czech Republic. The OB TraceVue(R) system stores all recordings electronically and starts recording 90 min before delivery. The majority are from vaginal deliveries, and each recording is at most 90 min long. The dataset originated from 9164 available intrapartum CTG recordings after applying clinical and technical selection criteria. It also contains clinical metadata which includes pH values together with Apgar scores and delivery types. The clinical experts performed verification and annotation tasks to guarantee both quality and consistency of the data.
Xinqiao Hospital BPPV dataset: The dataset of Xinqiao Hospital BPPV was developed by Lu et al. [31] in collaboration with the Second Affiliated Hospital of Army Medical University (Xinqiao Hospital). The dataset was collected from 518 BPPV patients who underwent examinations at the hospital from January to March 2021. The data contains eye movement videos together with diagnostic labels that identify the semicircular canal otolith locations into six distinct categories: left posterior canal, right posterior canal, left horizontal canal, right horizontal canal, cupulolithiasis, and cured/asymptomatic. The diagnostic instruments operated with dual rotational axes to enable clinicians to both adjust patient postures and observe eye movements. The hospital ethics committee authorized the anonymized data.
ADNI dataset: The Alzheimer’s Disease Neuroimaging Initiative (ADNI) [32] is a large-scale, longitudinal study launched in 2003 by the National Institute on Aging in collaboration with the NIBIB, FDA, and other agencies. The study has more than 2500 participants in the United States and Canada and is one of the largest and most extensive resources for Alzheimer’s disease research. The dataset contains multimodal data types such as structural MRI, FDG-PET, amyloid PET, diffusion MRI, cognitive assessments (e.g., MMSE, CDR), cerebrospinal fluid (CSF), blood-based biomarkers, and genetic profiles. All imaging data are standardized across sites. ADNI continues to be a vital resource for AD research because of its scope, homogeneity, and availability.
SLAKE-VQA dataset: The SLAKE-VQA dataset, which Liu et al. [33] created, functions as a semantically labeled bilingual (English and Chinese) medical visual question answering (Med-VQA) dataset. It includes 642 radiology images that come from three open datasets which present CT, MRI, and chest X-ray imaging modalities. The dataset covers different body areas, including the brain, neck, chest, abdomen, and pelvic cavity. It contains 14,028 question-answer (QA) pairs distributed between open-ended and closed-ended formats. The questions in this data cover different clinical and visual aspects, such as modality identification and organ recognition, as well as medical reasoning about abnormalities. The knowledge-based QA system of SLAKE uses a structured medical knowledge graph containing more than 5200 curated triplets. SLAKE-VQA is divided into three parts, which contain 9849 training examples and 2109 validation examples, and 2070 test examples.
NACC dataset: The National Alzheimer’s Coordinating Center (NACC) maintains the NACC dataset as its primary resource for data collection [34]. The repository contains standardized multimodal data from more than 40 Alzheimer’s Disease Research Centers (ADRCs) across the United States. The dataset contains standardized multimodal information from more than 19,000 patients through structural brain MRIs and neuropsychological test scores and biospecimens and clinical assessments, and longitudinal follow-ups. The imaging features undergo FreeSurfer processing, while the diagnostic categories include Alzheimer’s disease, frontotemporal dementia, Lewy body dementia, vascular dementia, and related conditions. The extensive nature of this dataset makes it a popular choice for researchers who study dementia classification and progression prediction and develop explainable AI frameworks.
UK Biobank (UKB): The UK Biobank (UKB) [35] is a large biomedical dataset that has recruited more than 500,000 participants aged 40–69 from all over the UK. It contains a wide range of phenotypic, lifestyle, and health-related data, as well as genetic and multimodal imaging data. The available data modalities include structural and functional MRI, DXA scans, retinal imaging, ECGs, and extensive electronic health record (EHR) linkages for long-term disease tracking. The imaging extension of the data is in tens of thousands of participants and is still growing. The UKB is a resource that supports a wide range of population health and disease research, providing harmonized and quality-controlled data across modalities. Its size and richness make it one of the most comprehensive resources for training and validating multimodal machine learning models in clinical research.

Table 1.
Overview of datasets in reviewed studies.
Table 1.
Overview of datasets in reviewed studies.
| Ref. | Dataset | Data Modalities | Data Sources | Dataset Size | Medical Diagnosis | Description |
|---|---|---|---|---|---|---|
| [20] | PAD-UFES-20 Dataset | Clinical images + patient metadata | Federal University of Espírito Santo | 2298 images | Skin Lesions (Various) | Smartphone-acquired lesion images and 21 clinical attributes |
| [28] | Guangzhou NEC Dataset | Radiographs + Clinical | Guangzhou Medical Center | 2234 patients | NEC Diagnosis | Abdominal radiographs with 23 structured clinical parameters |
| [30] | CTU-UHB Intrapartum CTG Dataset | FHR signals + expert features | University Hospital Brno | 552 samples | Fetal Acidosis | Annotated CTG recordings with clinical metadata |
| [31] | Xinqiao Hospital BPPV Dataset | Eye videos + head vectors | Xinqiao Hospital, Army Medical University | 518 patients | BPPV | Eye-tracking video recordings categorized by semicircular canal type |
| [33] | SLAKE-VQA Dataset | X-ray, CT, MRI + QA text | Multiple public sources | 642 images + 14,028 QA pairs | Medical VQA | Bilingual annotated radiology QA pairs with medical knowledge graph |
| [29] | Multimodal Dataset for Lupus Erythematosus Subtypes | Clinical images + multi-IHC + metadata | 25 Hospitals in China | 446 cases | Lupus Erythematosus | Clinical skin photographs, IHC slides, and systemic involvement index |
| [25,26] | ROCO and ROCOv2 | Radiology images + text (captions, MeSH terms) | OpenI and PMC-OAI | 81,000+ (ROCO), +11,000 (ROCOv2) | Radiology-based Disease Identification and Caption Alignment | Annotated radiology image-text pairs across multiple modalities for VQA and retrieval tasks |
| [21] | MedICaT Dataset | Medical figures + captions + inline references | PubMed Central Open Access | 217,060 figures from 131,410 papers | Scientific Radiology Figure Interpretation and Retrieval | Annotated compound figures with captions and inline references for subfigure alignment |
| [22] | FFA-IR Dataset | Fundus images + bilingual reports + lesion annotations | Clinical ophthalmology sources | 1330 samples | Retinal Disease Diagnosis | Multilingual diagnostic reports aligned with fundus images and lesion-level annotations |
| [32] | ADNI Dataset | MRI, PET, CSF biomarkers, cognitive assessments | ADNI Consortium (USA, Canada) | >2500 participants | Alzheimer’s Disease | A comprehensive longitudinal study integrating multimodal neuroimaging and clinical assessments to monitor AD progression. |
| [36] | PMC-VQA Dataset | Biomedical figures + VQA text | PubMed Central | 227,000+ QA pairs | Medical Visual Question Answering | Instruction-tuned large-scale VQA benchmark with domain metadata and UMLS/MeSH alignment |
| [34] | NACC Dataset | MRI + Clinical + Cognitive + Genetic | 40+ US ADRCs | >19,000 patients | Dementia (AD, FTD, DLB, VaD) | Longitudinal multimodal dataset with FreeSurfer imaging features, neuropsychological assessments, and diagnostic labels |
| [23] | MIMIC-III | EHR (structured, time-series) | MIT Lab for Computational Physiology | 53,423 patients | Critical illness, Diabetes, HF, COVID-19 | ICU clinical records with physiological signals, meds, and lab data used for semantic embedding and knowledge-enhanced prediction |
| [37] | Pediatric Radiology Dataset | Radiographic images + diagnostic QA pairs | Pediatric Imaging textbook and digital library | 180 images | Pediatric diagnostic VQA | Pediatric chest, abdominal, and musculoskeletal images with MCQs used in multimodal LLM evaluation |
| [38] | Taiwan Biobank Dataset (TWB) | Genomics + EHR data | Taiwan Biobank | 150,000+ adults | Population health and disease genetics in Taiwan | SNP arrays, physical exams, lifestyle data, family history, longitudinal follow-up |
| [35] | UK Biobank Dataset (UKB) | Genomics + EHR data | UK Biobank | 500,000+ participants | Multimodal disease risk prediction | Genotype arrays, clinical records, health questionnaires, imaging, family history, medication data |
| [24] | MIMIC-IV | Structured EHR, time-series vitals, clinical notes | Beth Israel Deaconess Medical Center | 383,220 admissions (78,275 ICU stays) | ICU risk prediction (e.g., mortality, sepsis) | Publicly available dataset featuring de-identified EHR, vital signs, and notes; spans 2008–2019 with longitudinal hospital data and updated coding standards (ICD-10, LOINC). |
Taiwan Biobank (TWB): The Taiwan Biobank (TWB) [38] represents one of the biggest population-based biomedical datasets in East Asia because it has enrolled more than 150,000 participants between the ages of 20 and 70. The dataset contains detailed phenotypic and genomic information which combines self-reported lifestyle data with physical measurements and laboratory biomarkers, and medical imaging data (such as ultrasound and ECG) that researchers collect during follow-up visits. The TWBv1 and TWBv2 custom SNP arrays provide genotyping data that undergoes imputation and quality control pipelines to achieve genome-wide variant coverage. The Han Chinese population in TWB enables researchers to conduct population-specific association studies while providing access to Taiwan’s National Health Insurance Research Database (NHIRD) for long-term follow-up and comprehensive phenotype analyses of various diseases and traits.
4. Data Preprocessing Techniques
Effective preprocessing is a fundamental step in preparing multimodal data for machine learning models. Preprocessing multimodal medical data requires cleaning and normalization [39], as well as synchronization and feature engineering across image, time-series, and textual data [40]. Since medical data originates from heterogeneous modalities, preprocessing techniques are highly dependent on the structure, resolution, and semantics of each source. In this section, we examine how different studies address the challenges of preparing multimodal medical data. We describe the preprocessing workflows applied to imaging, signal, text, and tabular data, and highlight the techniques used to align, transform, and integrate them before model training, as shown in Table 2. For clarity, the selected studies are grouped according to the dominant modality or task focus within each preprocessing pipeline.
Neuroimaging and Cognitive Data: The NACC and ADNI datasets were subjected to neuroimaging preprocessing pipelines which aim to synchronize imaging data with clinical information [41]. Martin et al. [42] performed FreeSurfer-based cortical and subcortical segmentation of NACC MRI scans, followed by volumetric and surface-based normalization and clinical label alignment for dementia staging. The data received stratification based on diagnostic categories, followed by harmonization procedures to minimize inter-site variability. Xue et al. [43] implemented quality control measures through record exclusion and variable sparse removal followed by assessment standardization and domain heuristic value imputation, and dimensionality reduction. Sheng et al. [44] performed spatial resolution normalization on ADNI structural MRI and PET images, followed by z-score normalization. The CSF biochemical markers required standardization before merging, and the Improved Harris Hawks Optimization (ILHHO) performed embedded feature selection to eliminate redundant data. Jahan et al. [45] converted MRI scans into 2D grayscale slices before cropping and resizing them to 224 × 224 pixels and applying min-max normalization to standardize cognitive scores and demographics while performing variable type-based value imputation. The dataset was partitioned into three distinct groups: AD, MCI, and CN.
Cardiology and Radiology Imaging: Feng et al. [46] implemented a structured pipeline to CDW-H by anonymizing structured clinical records, retaining relevant lab variables, and standardizing them using the PCORnet model [47]. A VGG-based classifier selected PLAX and A4C views for echocardiograms, and embeddings were created using CNN layers followed by BiLSTM [48] and attention pooling. Schilcher et al. [49] connected clinical data from the Swedish Fracture Register with radiographs, resizing pelvic X-rays using bounding box detection and applying histogram equalization and rotation during training. Zhao et al. [50] filtered CTG signals shorter than 10,000 samples in CTU-UHB, denoised signals via sparse dictionary learning, and balanced classes using GAN augmentation [51]. The last 30 min of FHR signals were retained. Wang et al. [52] normalized structured EHR features from MIMIC-III and IV, segmented time-series signals into intervals, and converted them to frequency domain using STFT. Clinical notes were tokenized and embedded with pre-trained models. Gemini [53] curated over 600,000 image-report pairs, removing noisy samples through a multi-stage pipeline including exclusion of low-quality images and tokenization of clinical text.
Multimodal Vision-Language Data: Lu et al. [31] divided eye movement videos into six 48-s posture-specific clips while removing segments under 10 s in length and selecting 300 frames from each clip before adding black frames. A self-encoder trained for 3D gaze estimation generated spatial embeddings from head position vectors. Zhang et al. [36] applied templates to parse PMC biomedical figures and captions into structured QA pairs before making them semantically consistent through UMLS [54] and MeSH [55]. Yao et al. [56] performed report tokenization with domain vocabulary and diagnostic finding retention after image resizing for radiology data. The research of Park et al. [57] involved data aggregation from various sources, followed by ResNet-50 [58] filtering for frontal chest X-rays then RadGraph extraction of clinical entities and study quality control.
Clinical and Structured EHR: Cai et al. [59] normalized MIMIC-III features and transformed them into sequences. The temporal encoder [60] received visit sequences for the temporal organization of temporal attributes while static attributes were embedded. The model learned intra-class variance through the sampling of contrastive pairs. Niu et al. [61] standardized EHR data structures through diagnostic categories while they normalized features and used knowledge-based embeddings and UMLS graph alignment through multiple transformation steps. Bampa et al. [62] processed the non-public EHR dataset by removing features with more than 60% missing values and performed bidirectional value imputation and Word2Vec-based categorical variable embedding. The research team of Chung et al. [63] eliminated phenotypic values that were either extreme or missing from TWB before conducting normalization of continuous data and SNP QC filtering and univariate SNP selection. Zeng et al. [64] applied standardization to UKB clinical features and performed imputation for missing entries and genetic data filtering based on call rate and heterozygosity. Lifestyle variables were discretized.
Multi-source and Domain-specific Pipelines: Li et al. [65] performed quality control of hysteroscopic images by removing low-resolution images and standardizing EMR features. The dataset was prepared by assigning each patient a unique identifier and converting categorical variables into one-hot encodings while normalizing the numerical variables. Lee et al. [66] performed quality control on fundus images through resizing and ensured curated EHR features (e.g., HbA1c, eGFR) data quality by selecting relevant features and removing outliers. The researchers tested their model externally using fundus images from the UK Biobank dataset. Zhu et al. [67] performed image resizing of CT and ultrasound data, followed by denoising procedures, before implementing tokenization on clinical summaries. Lin et al. [68] retrieved radiology image-report pairs from PACS before resizing images and normalizing them and using BERT-compatible tokenization on reports while implementing timestamp and ID-based filtering for mismatches. Panagoulias et al. [69] assigned medical images to MCQ mapping while they formatted GPT-4 [70] prompts and tagged image metadata before extracting LLM responses for NER and knowledge graph analysis.
Lupus and Dermatological Imaging: Li et al. [29] applied stain normalization and channel registration techniques to lupus multi-IHC tiles. They performed tile-level classification on extracted patches while they imputed and scaled the structured metadata, which included lab scores and involvement indices. The clinical photographs received standardized treatment for lighting and resolution consistency. The PAD-UFES-20 dermatology dataset received standardized resolution processing, which made all conditions have uniform image quality.

Table 2.
Overview of preprocessing techniques for multimodal data.
Table 2.
Overview of preprocessing techniques for multimodal data.
| Ref. | Dataset | Technique (Summary) |
|---|---|---|
| [28] | Guangzhou NEC Dataset | Radiograph resizing and z-score normalization, clinical feature filtering and LightGBM-based imputation, radiomics extraction and mRMR selection, data augmentation. |
| [50] | CTU-UHB Intrapartum CTG Dataset | FHR denoising with sparse dictionary learning, GAN-based data augmentation, signal truncation to 30 min, morphological feature extraction. |
| [31] | Xinqiao Hospital BPPV Dataset | Video length normalization, uniform frame sampling, head vector transformation, self-encoder-based spatial embedding. |
| [29] | Multimodal Dataset for Lupus Subtypes | Stain normalization, multi-IHC image channel registration, patch tiling, clinical metadata imputation and normalization. |
| [67] | Zhu et al. Urology Dataset | ROI selection from WSIs, resolution standardization, expert verification, triple-sampling for output stability, prompt structuring for VQA. |
| [34] | NACC Dataset | FreeSurfer segmentation, volumetric/surface normalization, inter-site harmonization, domain-based imputation, dimensionality reduction. |
| [23,59,61] | MIMIC-III (EHR-KnowGen) | EHR normalization, semantic embedding using UMLS, EHR encoding with self-attention, contrastive sample generation using supervised contrastive loss, concept alignment via graph embeddings. |
| [69] | Diagnostic VQA Benchmark | Prompt construction for GPT-4V, alignment of medical questions with corresponding images, and later stage analysis using named entity recognition and similarity metrics (RadGraph F1, ROUGE-L, cosine similarity). |
| [45] | ADNI Dataset | MRI resizing and intensity normalization, feature selection on cognitive scores, SHAP-based feature ranking, Grad-CAM applied for CNN interpretability. |
| [49] | Private Hip Fracture Dataset | Radiograph preprocessing with image resizing and augmentation; structured EHR cleaning, normalization, clinical encoding for tabular integration. |
| [68] | Custom Pediatric Appendicitis Dataset | Structured EHR cleaning and feature selection, ultrasound frame sampling, view classifier filtering, clinical-lab alignment. |
| [56] | Internal multimodal dataset (CT + reports) | CT pre-processing, report tokenization, visual-text alignment via ResNet50 and RoBERTa encoders. |
| [64] | UK Biobank | Genetic variants and clinical records were cleaned, encoded, and scaled, lifestyle and outcome features were extracted, and missing values were imputed using statistical methods. |
| [66] | Private dataset + UK Biobank | Fundus images were colored, normalized and resized. Vessel masks were extracted to capture retinal structure. Clinical EHR variables were one-hot encoded and aligned with image features before multimodal integration. |
| [71] | Private multi-institutional dataset | De-identification, low-quality text filtering, standardization into 26 clinical categories, image normalization and resizing. |
| [24] | MIMIC-IV | Time-series vitals were normalized and segmented structured EHRs were encoded using temporal categorical embeddings. Clinical notes were tokenized and embedded via BioClinicalBERT, enabling shared encoder input across modalities. |
| [65] | Private dataset | Temporal frame selection from hysteroscopic videos, image enhancement, manual scoring of injury risk, and structured EMR standardization. |
| [72] | MIMIC-CXR | Preprocessing included filtering uncurated report-image pairs and constructing positive/negative samples for contrastive learning. Free-text reports were tokenized and projected into embeddings. Radiographs were encoded via a vision transformer. A curriculum-based sampling strategy enhanced training robustness. |
The various approaches demonstrate the necessity of developing specialized preprocessing methods that match different data types, disease conditions, and modeling needs.
5. Multimodal Fusion Techniques
Fusion techniques in multimodal learning determine how and when different data sources are integrated into a model. Medical applications primarily use three main fusion strategies, which include early fusion, late fusion, and intermediate fusion. The three fusion frameworks are depicted in Figure 2 and show how feature extraction, integration, and prediction operate at different levels. Attention-based fusion and neural architecture search (NAS) frameworks have become prominent, as they adapt well to complex tasks while delivering good performance. Researchers have developed new hybrid fusion designs (Figure 3) that unite different fusion approaches by combining the raw features of one modality with predictions from another. This section organizes fusion approaches based on their implementation level while presenting notable studies for each category. Research studies that use multiple fusion approaches receive discussion throughout different categories.
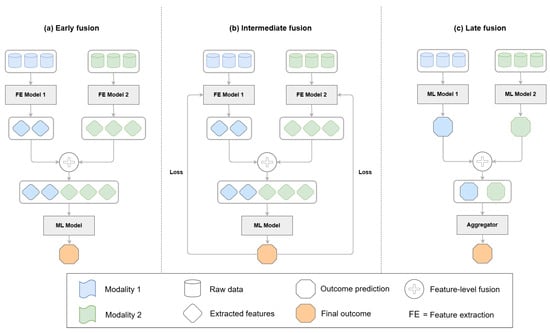
Figure 2.
Data fusion architectures in multimodal learning. (a) Early fusion combines raw features or extracted features before input to the final model. Feature extraction is optional. (b) Intermediate fusion concatenates modality-specific features in an integrated model where the loss is backpropagated through the full network. (c) Late fusion aggregates outputs or features from independently processed modalities at the decision level.
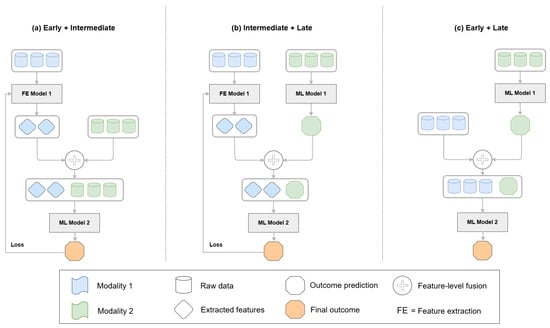
Figure 3.
Examples of mixed fusion architectures. (a) Feature representations from one modality (blue) are used for learning, while others (green) are fused at the feature level without backpropagation. (b) Features from one modality (blue) are extracted but not used in the final prediction, which is based only on the output of the other modality (green). (c) Raw data or features from one modality (blue) are combined with the prediction output of another modality (green) and passed to a downstream model. These configurations illustrate flexible hybrid fusion strategies across different learning stages.
5.1. Early Fusion
Early fusion involves the initial combination of unprocessed or basic features across different modalities before extensive processing takes place. For instance, Park et al. [57] developed a dual-stream architecture to learn multimodal representations through contrastive pretraining which aligned radiographic features with report embeddings. The CNN encoder produced visual features from images, while the transformer-based language model generated textual embeddings. The pretraining process used contrastive loss to align image-report pairs while discriminating them against incorrect samples, avoiding the need for labeled data. This fusion approach allowed the model to develop joint representations, which improved its ability to perform zero-shot report generation and x-ray classification tasks on unprocessed datasets. Similarly, Feng et al. [46] performed early fusion by combining raw echocardiographic views and structured EHR data before feature extraction and model training during their preliminary analysis of fusion strategies.
5.2. Late Fusion
Late fusion refers to integrating outputs or predictions from independently trained modality-specific branches [73]. For example, Gao et al. [28] used a late fusion strategy at the decision level in their NEC prediction framework. After identifying SENet-154 as the optimal DL model for feature extraction from abdominal radiographs, radiomic features were obtained and combined with structured clinical parameters. The fused features were inputs to a LightGBM classifier [74] to support diagnosis and surgical prediction. This model integrated multimodal signals at a later stage, allowing each stream (radiomics and clinical) to independently extract discriminative features before fusion. The radiomic and clinical contributions were evaluated by feature importance analysis, which guided the fusion process and provided interpretability of the joint predictions.
Similarly, Li et al. [29] used a decision-level fusion strategy for lupus erythematosus subtype diagnosis by combining representations from two different image encoders (ResNet-50 for multi-IHC tiles and EfficientNet-B1 [75] for clinical photographs) and structured clinical metadata. The outputs from each modality-specific encoder were concatenated and passed to a multilayer perceptron for final classification. This approach allowed the model to independently extract features from each modality while maintaining interpretability, and it was combined with a visualization module to facilitate human-AI collaboration in diagnostic decision-making. Chung et al. [63] implemented a late fusion strategy in which structured phenotypic and genotypic data were processed independently before integration. Phenotypic features were input into a gradient-boosted decision tree (XGBoost), while genotypic data were modeled using a deep neural network. The outputs were merged using a meta-learner, allowing independent optimization of each branch. In another example, Lee et al. [66] employed a two-branch late fusion approach to combine fundus images and structured EHR data for detecting diabetic kidney disease (DKD). They used a ResNet-50 backbone pre-trained on ImageNet to extract image features while a fully connected neural network processed tabular clinical variables. The modality-specific outputs were concatenated and passed through a fusion layer followed by a classification head. The authors selected this approach to maintain the separate contributions of visual and clinical signals and to enable separate optimization of each feature stream before integration. Building on ensemble-based strategies, Zeng et al. [64] used a stacked fusion approach to combine clinical and genetic features to predict hyperuricemia. The clinical features and polygenic risk scores were processed through separate pipelines and then fused at the decision level using ensemble learning. This approach allowed each modality to contribute complementary information, capturing both environmental and genetic risk factors. The stacked architecture allowed for improved risk stratification and was also used to generate an interpretable scalar marker (ISHUA) for individualized outcome prediction. Yildirim et al. [71] evaluated late fusion outcomes within their experiments involving GLoRIA [76] and ConVIRT [77]. Feng et al. [46] also reported results for late fusion by combining separately trained modality-specific models for EHR and imaging in a later stage manner.
5.3. Intermediate Fusion
Intermediate fusion combines modalities between feature extraction and classification stages [78]. This approach is frequently used in clinical tasks that require interaction between feature streams.
Feng et al. [46] introduced a transformer-based intermediate fusion model for detecting cardiac amyloidosis by integrating echocardiographic and EHR data. Features were encoded separately and aligned using attention in a shared latent space. Li et al. [65] used this fusion technique to integrate structured EMR data and hysteroscopic images for reproductive outcome prediction. Features from each modality were first encoded using dedicated ResNet50 branches for image data and fully connected layers for EMR features. These modality-specific representations were then concatenated and passed through a common fusion layer, followed by dense classification layers. Similarly, Schilcher et al. [49] combined radiographic images with structured EHR characteristics to predict mortality in patients with hip fractures. The model first processed clinical variables through a dense feedforward network and extracted visual features from cropped pelvic X-rays using a pre-trained DenseNet-121. Then, it combined modality-specific representations into a shared embedding space before using additional fully connected layers to perform classification. Using this fusion approach, the model combined patient-level clinical risk factors with local visual fracture characteristics. Sheng et al. [44] implemented this strategy in their ILHHO-KELM model by combining MRI features, PET scan features, and CSF biochemical markers into a single feature representation. The radiomic features extracted from MRI and PET images were first normalized and spatially aligned, while the CSF-based features were encoded as numerical vectors. The modality-specific features were then combined into a single multimodal input vector. The model learned joint representations across modalities before classification in this intermediate fusion approach, rather than fusing the raw data at an early stage or relying on separate decision-level outputs. The fused feature space was input to KELM, with feature selection and optimization guided by the ILHHO algorithm to enhance diagnostic robustness and generalization. Martin et al. [42] also employed an intermediate fusion approach to combine structural MRI-derived features with cognitive variables extracted from the NACC dataset. Imaging and clinical features were concatenated before being passed to classification models for dementia staging and subtype discrimination. The fusion process was optimized to balance interpretability and predictive performance, supporting both clinical relevance and model transparency. Jahan et al. [45] implemented feature-level intermediate fusion to combine MRI slices with cognitive scores and demographic features into one diagnostic model. The fusion pipeline performed parallel processing of the image and tabular data streams. The CNN extracted visual features from the pre-processed MRI slices, and fully connected layers processed cognitive and demographic features. The extracted features were concatenated in an intermediate layer before classification. The model gained the ability to discover common representations from diverse sources through this method, which enabled it to map structural information to clinical domains effectively.
Further building on intermediate fusion mechanisms, Cai et al. [59] introduced an approach employing contrastive learning to merge structured EHR segments, using self-attention encoders for shared latent space alignment. Wang et al. [52] merged physiological signals with clinical notes and structured EHR features for risk prediction purposes. The first step involved using modality-specific encoders to derive high-level representations from each data type, where time-series signals went through CNN-based encoders and clinical text went through transformer models, and structured features went through fully connected layers. The shared fusion module received concatenated latent representations to enable cross-modal interactions in a unified feature space. The method allowed extensive multimodal correlation learning between data types while avoiding both early raw data fusion and late decision-level integration. Bampa et al. [62] developed the M-ClustEHR framework using intermediate fusion, where each EHR modality (e.g., diagnosis, medications, lab tests) was passed through a separate modality-specific encoder. These encoders generated latent embeddings that were concatenated into a unified representation before clustering. The fusion was performed after feature extraction but before clustering, allowing for the integration of heterogeneous clinical data types into a common latent space. Along the same direction, Gemini [53] used an intermediate-level fusion strategy to incorporate radiology images and clinical reports. Vision and language features were extracted by modality-specific encoders and projected into a shared semantic space and aligned by attention-based modules. The fusion scheme allowed for bidirectional conditioning between modalities and supported downstream clinical tasks such as classification and vision-language grounding. Niu et al. [61] fused structured EHR data with external UMLS-based knowledge using attention mechanisms. Semantic embeddings from clinical data and knowledge graphs were aligned in a shared space, improving generalization. In addition, Zhang et al. [36] and Yao et al. [56] used intermediate fusion strategies in which modality-specific encoders first processed biomedical images and clinical text, followed by alignment using cross-attention layers. Their architectures supported bidirectional interactions between modalities at the feature level. Although often discussed in terms of vision-language alignment, their design is structurally aligned with intermediate fusion. Similarly, Yildirim et al. [71] employed pre-trained contrastive vision-language models [79] and used joint latent spaces to align representations, while these methods support downstream interpretability and retrieval, the fusion process itself occurs at the intermediate feature level.
5.4. Cross-Modal and Architecture Search Fusion
Recent approaches have introduced fusion models that use attention mechanisms or automated architecture search to move past fixed fusion designs. Models learn dynamic optimal fusion patterns through these methods, often leveraging cross-modal interactions or reinforcement learning-based search strategies.
Among attention-based designs, Lu et al. [31] implemented a multi-layer cross-attention mechanism [80] to combine features from eye-movement videos with head position vectors. In their framework, the TDN-based network encoded eye-movement data, while a self-encoder transformed head position vectors, and cross-attention aligned the resulting features into a shared space. Additionally, they implemented LXMERT-style architecture designs [81] by adding self-attention layers to bidirectional cross-attention modules, enhancing both intra- and inter-modality interactions. The fusion method demonstrated better performance compared to direct concatenation, weighted summation, and self-attention-only models. Furthermore, the removal of the cross-attention module resulted in a performance decrease exceeding 10%, confirming its critical role in detecting joint spatiotemporal patterns for BPPV diagnosis. The hybrid FHR framework [50] included a cross-modal feature fusion (CMFF) mechanism that integrates manually created expert features with deep representations extracted from FHR signals. Each modality was first encoded into a latent representation: expert features via linear projection layers and FHR signals via a custom SE-TCN backbone. These modality-specific embeddings were aligned and fused using a multi-head attention mechanism that computed a cross-modal attention score based on cosine similarity. The final fusion vector was obtained by concatenating pooled (GAP and GMP) representations from both modalities, forming a joint latent space optimized for diagnostic classification.
In the domain of instruction-tuned large models, Reith et al. [37] evaluated multimodal large language models (MLLMs) [82] such as BLIP-2 [83] and MiniGPT-v2 [84] for medical visual question answering. These models fused image and textual modalities through pre-aligned vision encoders and transformer-based language decoders, enabling zero-shot reasoning based on compositional prompts. Although not specifically trained on medical data, their cross-modal fusion capabilities effectively aligned semantic and spatial cues in pediatric radiology tasks. Shifting toward automated architecture design, Cui et al. [85] introduced AutoFM, a neural architecture search (NAS) framework [86] aimed at automatically discovering optimal fusion architectures for structured EHR data. AutoFM used reinforcement learning to dynamically explore hierarchical combinations of input fields such as diagnoses, labs, and procedures. Rather than relying on manually selected fusion strategies, AutoFM constructed task-adaptive networks that generalized across multiple diseases by learning optimal fusion patterns that vary by task and data composition. Yildirim et al. [71] used pre-trained contrastive vision-language models like GLoRIA and ConVIRT to evaluate the alignment of radiographic images with clinical text. These models operate through shared embedding spaces optimized via contrastive objectives, which enables downstream tasks such as disease classification and report generation. Their contrastive design enables both interpretability and task-specific multimodal alignment. Finally, other transformer-based models, including Gemini [53], PMC-VQA [36], and Yao et al. [56], also utilized cross-attention mechanisms for modality alignment. Although discussed primarily under intermediate fusion, their architecture reflects integration occurring after feature extraction, leveraging shared embedding spaces to assess alignment between radiographic images and clinical text. These models operate through shared embedding spaces optimized via contrastive objectives, facilitating downstream tasks such as disease classification and report generation. Their contrastive design enables both interpretability and task-specific multimodal alignment.
6. Multimodal Approaches and Model Architectures
Medical diagnostics employ various model architectures to manage multimodal input complexity through structures that match both input data structures and clinical objectives. In this section, we provide a detailed review of recent model designs, organized according to architectural traits and modeling strategies, as shown in Table 3.

Table 3.
Models and fusion methods across studies.
6.1. Hybrid and Attention-Based Architectures
Feng et al. [46] studied multiple fusion strategies by designing five different model architecture variants for cardiac amyloidosis detection. These included: (a) An EHR only baseline using logistic regression, (b) A CNN+LSTM attention model trained on echocardiographic views, (c) Late fusion of separately trained image models (PLAX and A4C), (d) Late fusion combining image embeddings with EHR features, and (e) A transformer-based intermediate fusion model that integrated features from both modalities using cross-modal attention. The final architecture achieved the highest AUROC of 94.1%, which shows the effectiveness of structured fusion and flexible modality alignment. Figure 4 gives a visual summary of the five architectures and their fusion strategies.
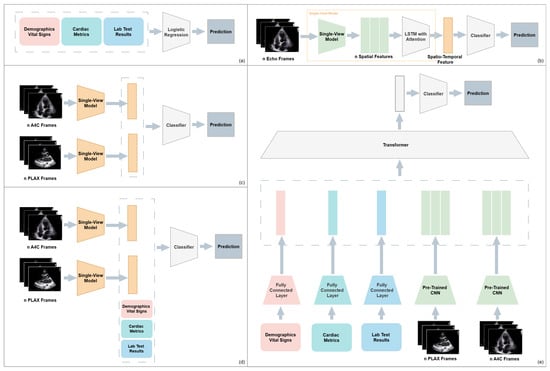
Figure 4.
Overview of five model architectures for multimodal cardiac amyloidosis classification: (a) EHR-only baseline; (b) Single-view CNN-LSTM on echocardiography, (c) Late fusion of PLAX and A4C views, (d) Multimodal late fusion of imaging and EHR features, (e) Multimodal intermediate fusion using a transformer-based architecture.
Gao et al. [28] developed a multi-stage MAI system that used deep learning and machine learning to diagnose NEC rapidly and predict surgical needs. The system consisted of three main components which started with transfer learning to extract radiomic features from abdominal radiographs through pre-trained DL models, followed by feature selection with mRMR and ended with a LightGBM classifier that merged radiographic and clinical features. The model selection process resulted in SENet-154 as the top-performing architecture, which produced the best AUC and other performance metrics results, and its feature embeddings became the input for subsequent prediction tasks. The multimodal model produced diagnostic results with an AUC of 93.37% and an accuracy of 91.57%. The combination of radiographic and clinical data produced an AUC of 94.13% and an accuracy of 88.61% for surgical prediction, which exceeded single-modality models. Lu et al. [31] proposed a hybrid multimodal model called BKTDN, combining a Temporal Difference Network (TDN) with a novel “big kernel” extension and a cross-attention-based fusion module. The BKTDN model extracted motion features from eye-movement videos using short- and long-term convolutional streams with large receptive fields, effectively capturing sparse but discrete signs of oculomotor behavior. Simultaneously, head position vectors were embedded via a pre-trained self-encoder and used in a fusion module to integrate spatial posture context. The final classifier processed the fused representation for multi-class BPPV subtype classification. The BKTDN model achieved superior performance compared to classic time-series baselines such as ResNet, MLSTM, and InceptionTime, with an overall accuracy of 81.7%, precision of 82.1%, sensitivity of 94.1%, and specificity of 96.5%.
6.2. Transformer-Based Vision-Language Models
The Gemini model [53] was developed as a scalable, instruction-tuned vision-language model tailored for radiology applications. Its architecture consists of a vision transformer (ViT) [87] encoder for medical images and a transformer-based text encoder for clinical reports. The model was trained using large-scale paired data (radiographs and associated report sentences) with a contrastive loss to align vision and text in a shared embedding space. Instruction tuning further enhanced Gemini’s capacity to perform structured downstream tasks like report generation and visual question answering. On the CheXpert benchmark [88], Gemini achieved a leading zero-shot AUC of 86.7%, outperforming previous vision-language models such as GLoRIA and ConVIRT. Additionally, Gemini demonstrated high image-to-report retrieval performance with a top-1 accuracy of 71.6% and top-5 accuracy of 89.3%. Zhang et al. [36] proposed PMC-VQA, an instruction-tuned vision-language model specifically designed for medical visual question answering (Med-VQA). The architecture uses a frozen vision encoder with a transformer-based language decoder that is fine-tuned using instruction-based learning. The model is trained on synthetic QA pairs from biomedical figures and captions in PMC articles. The QA templates were enriched by aligning image-derived content with MeSH and UMLS knowledge representations. The model achieved 71.2% accuracy in zero-shot generalization on Med-VQA tasks. Yildirim et al. [71] examined the performance of two self-supervised contrastive models, GLoRIA and ConVIRT, for vision-language tasks in radiology on the MIMIC-CXR dataset. GLoRIA applied hierarchical contrastive learning to match local (region-word) and global (image-sentence) embeddings, while ConVIRT applied a transformer-based architecture for global alignment. The models underwent evaluation for multiple downstream tasks which included anomaly classification, phrase grounding, and spatial localization. The pointing game accuracy reached 74% when GLoRIA performed better than ConVIRT in spatial grounding while the abnormality classification mean AUC reached 84%. The retrieval performance of ConVIRT exceeded that of GLoRIA, as it reached 78% top-5 accuracy in phrase matching. Reith et al. [37] evaluated general-purpose instruction-tuned models such as BLIP-2 and MiniGPT-v2 using a curated pediatric radiology dataset comprising 180 radiographs paired with multiple-choice diagnostic questions, while these models were not trained on medical data, their vision-language backbones were tested in a zero-shot multiple-choice setup. BLIP-2 outperformed MiniGPT-v2, achieving an accuracy of 73.3% versus 56.7%, respectively. This evaluation demonstrates that even without domain-specific pretraining, instruction-tuned MLLMs can exhibit emergent diagnostic capabilities, particularly when structured prompts and image-question pairs are well-aligned.
6.3. EHR-Centric and Optimization-Based Models
Cai et al. [59] proposed a contrastive learning framework that enables the learning of robust patient representations from structured EHR data without the need for explicit cross-modal labels. The framework used a transformer-based encoder to process both static and sequential inputs and align them in a shared embedding space. The model was trained using a supervised contrastive loss, where patient visits from the same diagnostic category were pulled together and those from different categories were pushed apart. This method forced the network to learn intra-class similarity and inter-class variability in high-dimensional patient profiles. Wang et al. [52] created a unified multimodal prediction model that used time-series physiological data together with clinical text and structured EHR features from MIMIC-IV to predict patient risk. The model applied CNNs for waveforms and transformer models for clinical notes, and MLPs for structured variables. The shared neural network combined the representations which were optimized through a multitask learning objective. The model reached AUROC values of 91.1% for ICU mortality and 88.5% for sepsis prediction. The research team of Niu et al. [61] presented EHR-KnowGen as a modular knowledge-enhanced multimodal architecture. The model combines EHR data structures with UMLS medical ontology embeddings. The system contains three essential components: The EHR encoder converts tabular data into dense embeddings, while the concept encoder uses graph convolutional networks (GCNs) [89] to transform medical concepts into semantic vectors, and the multimodal fusion network unites these embeddings through attention-based interaction layers. The architecture models patient data relationships with clinical knowledge hierarchically to achieve better generalization performance in disease prediction tasks such as diabetes, heart failure, and COVID-19. The EHR-KnowGen model produced better results than baseline approaches in all experiments, with AUC scores reaching 81.5% for diabetes prediction and 87.8% for heart failure, and 85.1% for COVID-19. Figure 5 illustrates the architecture for multimodal diagnostic generation, incorporating both EHR data and domain knowledge through attention and calibration mechanisms before report decoding. Cui et al. [85] proposed AutoFM, a neural architecture search framework that discovers optimal fusion architectures for multimodal EHR data. The model is composed of a controller that generates candidate architectures and a shared backbone that evaluates their performance using reinforcement learning. This approach allows the architecture to dynamically learn how and where to fuse structured features from different EHR domains, such as diagnoses, lab tests, and procedures. AutoFM is task-agnostic and was evaluated across three disease prediction tasks (heart failure, diabetes, and mortality), outperforming several handcrafted baselines. The resulting architectures varied in connectivity depth and fusion position, demonstrating the benefit of adaptive architecture selection in medical prediction tasks. The model achieved AUROC scores of 84.8% for heart failure, 85.7% for diabetes, and 91.4% for in-hospital mortality. The ILHHO-KELM framework developed by Sheng et al. [44] is a hybrid model that integrates the ILHHO algorithm with the KELM classifier to improve Alzheimer’s disease classification. ILHHO is a metaheuristic optimization algorithm inspired by the cooperative hunting behavior of Harris hawks and, in its improved form, incorporates iterative map-based population initialization and local escaping operators to avoid premature convergence and local optima. This makes it particularly effective for selecting relevant features from high-dimensional multimodal data. After selecting the optimal feature subset through ILHHO, the features move to KELM, which serves as a non-iterative learning algorithm that provides both fast learning speed and good generalization performance. The kernel functions in KELM transform input features into a high-dimensional space to tackle nonlinear classification problems efficiently. The ILHHO-KELM framework resulted in a 99.2% accuracy when diagnosing Alzheimer’s disease from normal controls. Bampa et al. [62] proposed M-ClustEHR, a multimodal deep clustering framework that combines modality-specific autoencoders with joint representation learning to discover disease subtypes from EHR data. Feedforward neural networks encoded each modality separately before the encoded representations were fused into a single joint representation. The system used a dual-branch architecture that combined a reconstruction branch to maintain modality-specific semantics with a clustering branch that used deep clustering goals through pairwise contrastive learning. The approach successfully revealed hidden patterns in diverse data types while avoiding the need for labeled examples to discover phenotypes and model disease trajectories using real-world EHRs.
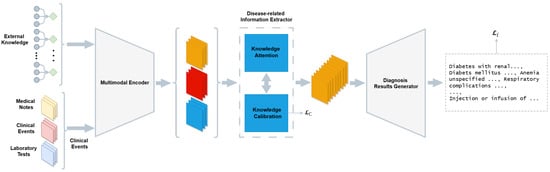
Figure 5.
Overview of the architecture illustrating the diagnostic generation process based on multimodal EHR inputs. The system includes three main components: a multimodal encoder that transforms inputs such as medical notes, clinical events, and lab results into shared representations, a knowledge-driven information extractor, consisting of attention and calibration modules, which refines these representations using external medical knowledge, and a diagnosis generator that produces structured diagnostic outputs.
6.4. Tabular-Image Fusion Architectures
Li et al. [29] developed a human-multimodal deep learning collaborative framework to improve the precise diagnosis of lupus erythematosus subtypes. The model analyzed multiple-scale multi-IHC images together with clinical photographs and structured clinical metadata through a dual-path architecture. The first path analyzed IHC patches through ResNet-50, while the second path analyzed clinical photographs through EfficientNet-B1. The decision fusion layer merged the learned representations with structured clinical features by employing an MLP classifier. The visualization module produced heatmaps for IHC patches and clinical images to support pathologist evaluation through interpretability and human collaboration. The proposed framework reached 82.88% accuracy while achieving an AUROC value of 98.4% for lupus subtype classification. Schilcher et al. [49] developed a multimodal framework that combines hip fracture radiographs with structured EHR data to forecast 30-day mortality. The system uses DenseNet-121 as its backbone to extract features from radiographic images and a feedforward network to process clinical tabular information. The system combines modality-specific embeddings through concatenation before using a shared classification head for processing. The multimodal model produced an AUROC of 84% which exceeded the AUROCs of both the image-only and EHR-only models at 80% and 78% respectively, demonstrating the benefits of combining structured and unstructured data in high-risk orthopedic decision making. Lin et al. [68] proposed a multi-input deep learning model for differential diagnosis of appendicitis in pediatric patients. Their architecture jointly processed two modalities: a ResNet-18-based CNN to extract image features from annotated ultrasound scans, and a multilayer perceptron (MLP) for structured EHR variables. These modality-specific embeddings were concatenated before being passed to fully connected layers for final classification. The joint model achieved an AUROC of 86%, outperforming the EHR-only model (AUROC = 79%) and the image-only model (AUROC = 76%). Martin et al. [42] implemented a dementia diagnosis system with explainable capabilities through a fully connected network architecture that used the NACC dataset. The model processed FreeSurfer-derived neuroimaging features and standardized clinical variables through parallel streams before feature concatenation and dense layers for classification. The system used saliency mapping and attribution analysis to evaluate how individual features and modalities contributed to predictions while maintaining an architecture that prioritized interpretability. The chosen design approach achieved both high diagnostic precision and clinical clarity, which made it appropriate for practical decision support applications. Li et al. [65] proposed a dual-branch architecture to process hysteroscopic images and structured EMR data by using ResNet50 for visual feature extraction and an MLP for EMR encoding. The modality-specific embeddings were concatenated and fed into a dense fusion layer to enable joint inference across inputs. The fused features were passed through fully connected layers for final classification. The multimodal model reached an AUROC of 85.4% for predicting reproductive outcomes which exceeded both single-modality baselines and traditional fusion strategies. Lee et al. [66] developed a multimodal deep learning system that utilized fundus photographs together with structured EHR features to make predictions about diabetic kidney disease. The system consisted of two separate branches where ResNet-50 analyzed fundus images to extract visual features, and a fully connected MLP handled clinical variables. It combined these embeddings before sending them to a fusion classifier. The model reached an AUROC value of 85.8% when tested internally and 81.2% when validated externally using UK Biobank data. Chung et al. [63] developed a dual-branch system that combines structured phenotypic and genotypic features for type 2 diabetes risk prediction. The phenotypic data, consisting of age, BMI, and clinical laboratory measures, underwent XGBoost processing. The SNP-based genotypic data were received by a multilayer perceptron model in parallel processing. The stacking ensemble combined two pipelines using logistic regression as a meta-classifier. The system achieved evaluation results of 83.5% AUROC on Taiwan Biobank data and 79.7% AUROC on UK Biobank data. Yao et al. [56] created a multimodal deep learning architecture that combines CT images with clinical reports for disease classification. The system employed ResNet-50 as its visual feature encoder and RoBERTa-based transformer as its clinical text processor. The model combined modality-specific embeddings through a knowledge-enhanced decoder that used cross-attention and external medical knowledge. It reached 96.42% accuracy together with 98.48% recall and an F1 score of 97% and IoU of 89% which surpassed CNN, Bi-LSTM, and attention-only baseline models. The system showed excellent results in both lesion localization and generating structured reports. Zeng et al. [64] proposed a stacked ensemble model for early detection and risk prediction of hyperuricemia by combining clinical and genetic features from UK Biobank and Nanfang Hospital data. The architecture combined gradient-boosted decision trees (LightGBM) with a neural network component, where outputs from the base models were aggregated using a meta-learner. The model achieved an AUROC of 81.6% on the internal test set and 79.2% on the external test set.
6.5. General-Purpose LLMs and Instruction-Tuned Models
Panagoulias et al. [69] tested GPT-4V for generating diagnostic reports through visual inputs of radiological and dermatological images with patient vignettes. The pipeline converted image understanding into a report generation task while using RadGraph F1 for entity and relation extraction and ROUGE-L for textual overlap and cosine similarity metrics to evaluate embedding alignment with actual reports. The framework demonstrated effective performance across various datasets through its RadGraph F1 score of 77.3% and its cosine similarity score of 93.4% on pediatric radiographs which shows GPT-4V’s evolving ability to generate multimodal diagnoses, although it received no specific medical training. The zero-shot performance trends observed in instruction-tuned vision-language models such as PMC-VQA [36] and Gemini [53] follow a similar pattern. Park et al. [57] also developed a self-supervised multimodal learning framework using large-scale uncurated image-report pairs from the MIMIC-CXR dataset. Their architecture consisted of a CNN-based visual encoder and a transformer-based text encoder, trained jointly using a contrastive learning objective to align radiograph-report embeddings. After pretraining, the shared representation space enabled transfer to multiple tasks, including zero-shot radiograph classification and report generation. On the ChestX-ray14 benchmark, the model achieved a mean AUROC of 78.1% in zero-shot classification.
Zhu et al. [67] evaluated the generalization ability of ChatGPT-4V, a large vision-to-speech model pre-trained on general tasks, in pathology classification tasks. The system operates as a black-box LMM using a common visual-language transformer backbone. It was asked to discriminate between malignant and benign tissue in renal cell carcinoma (RCC) and prostate cancer (PCa) slides. ChatGPT-4V achieved an AUC of 87.1% for RCC classification (sensitivity = 98%, specificity = 70%, F1 score = 86%), but failed to discriminate PCa from benign biopsies (AUC = 51%). Furthermore, Zhu et al. [90] conducted a complementary evaluation of ChatGPT-4V in radiology interpretation tasks. The authors collected 200 diagnostic image-question pairs from open access radiology textbooks and previous clinical studies to assess the model’s reasoning consistency, factual accuracy, and medical relevance. ChatGPT-4V reached 85.0% question-level accuracy on the manually labeled subset and radiologists found it clinically helpful in more than 75% of cases. The study also pointed out the limitations of uncertainty awareness and hallucination control, and the need for domain adaptation and timely tuning. This work further supports the growing diagnostic potential of ChatGPT-4V while pointing out important barriers to clinical implementation.
6.6. Privacy and Security-Oriented Models
Latif et al. [91] presented in their study a fragmented solution to safeguard medical health records that exist within multimodal medical image datasets. This approach resolves an essential problem in multimodal systems by protecting data privacy and security throughout storage and sharing operations. The approach requires EHR encryption followed by block fragmentation, then placement of the blocks into separate medical image channels. Medical images enable secure patient data concealment through their redundancy and tolerance properties while maintaining both image quality and interpretability. The method provides a privacy-preserving solution for sharing multimodal imaging data between systems and institutions in sensitive diagnostic environments.
6.7. Comparative Evaluation
While multimodal architectures in medical AI show significant performance improvements, critical evaluation reveals challenges in terms of interpretability, deployment feasibility, and robustness in real-world clinical settings.
Several studies confirm that multimodal integration significantly improves diagnostic performance. Reported improvements over unimodal baselines typically range from 5% to 15%. For example, Zhao et al. [50] achieved a 6.8% increase in accuracy by combining expert and deep temporal features for fetal risk prediction. Jahan et al. [45] observed a 7.3% improvement through the integration of cognitive scores, demographic information, and MRI slices. Similarly, Gao et al. [28] increased AUC from 88% to 94.13% by fusing radiomics and clinical data. These results highlight the added value of multimodal fusion, particularly when modalities provide complementary insights and are aligned through appropriate fusion strategies.
Interpretability varies widely among models. Some frameworks, such as Martin et al.’s dementia diagnosis system [42], integrate specific interpretability techniques, such as saliency mapping and feature attribution, making them more transparent to clinicians. Similarly, GLoRIA [71] and PMC-VQA [36] offer spatial attention visualizations or concept-linked responses grounded in biomedical ontologies (UMLS, MeSH), which support explanation consistency. Meanwhile, large-scale models like Gemini [53] and ChatGPT-4V [67,90] rely on emergent reasoning and instruction tuning without explicit interpretability mechanisms. This approach can complicate traceability, despite achieving high zero-shot performance.
Regarding the implementation constraints, various approaches prioritize deployability. For example, AutoFM [85] uses neural architecture search to automatically identify lightweight yet performant fusion architectures for structured EHR data, enabling task-specific optimization without the need for manual tuning by humans. Martin et al.’s approach [42] uses simple, dense layers suitable for standard hardware, aligning with clinical workflow constraints. However, instruction-tuned models like Gemini and ChatGPT-4V require significant computational resources and pretraining on large-scale medical or general datasets, which limits immediate clinical integration, especially in low-resource environments. Feng et al.’s model [46] demonstrated that even with only 41 patients, attention-based intermediate fusion could outperform traditional pipelines, indicating the feasibility of effective modeling in data-scarce contexts.
In terms of robustness, Lu et al.’s BKTDN architecture showed a strong dependency on multi-source inputs. Specifically, the removal of the head position vectors caused an accuracy drop of nearly 30%, underlining the importance of modality contribution and the vulnerability of such models to partial input loss. Park et al. [57] demonstrated that self-supervised contrastive learning enables generalization to noisy or unsupervised real-world radiograph-report pairs, offering robustness under weak supervision. Similarly, AutoFM [85] automatically adjusted fusion strategies across disease tasks, confirming adaptability to domain variability. However, ChatGPT-4V [67] showed high variability in its performance, with strong classification results for renal cell carcinoma (AUC = 0.87) but low performance for prostate cancer detection (AUC = 0.51). Underscoring the challenge of task-specific reliability in large generalist models.
This comparative analysis reveals that no single architecture dominates across all axes. Models that are optimized for interpretability or resource constraints may compromise flexibility or scalability, while those designed for generalization may lack domain specificity. Balancing these factors is essential for real-world clinical translation.
7. Discussion
Recent multimodal machine learning frameworks have demonstrated strong diagnostic performance in several clinical domains, including cardiology, neurology, oncology, and maternal-fetal health. Several studies showed that selecting appropriate fusion strategies and modality-specific preprocessing pipelines plays a major role in the success of these systems.
In cardiology, Feng et al. [46] found that transformer-based intermediate fusion of EHR and echocardiography data yielded an AUROC of 94.1%, outperforming simpler fusion schemes. Gao et al. [28] similarly combined SENet-based radiomic features with clinical data and achieved an AUC of 94.13% in predicting surgical eligibility for NEC. These examples reflect how intermediate and decision-level fusion strategies, when coupled with domain-aware preprocessing, can lead to high diagnostic accuracy. Neurological applications have shown comparable gains. Sheng et al. [44] achieved 99.2% accuracy for Alzheimer’s classification by fusing standardized MRI and CSF features using a hybrid optimization framework. Jahan et al. [45] achieved 94.5% by combining cognitive scores, demographic variables, and MRI slices, demonstrating that properly aligned feature-level fusion of structured and unstructured inputs can improve performance while enabling interpretability.
In maternal-fetal and pediatric diagnostics, the Hybrid-FHR model by Zhao et al. [50] surpassed CNN baselines by integrating expert and deep temporal features with a CMFF mechanism, achieving 96.8% accuracy. Lin et al. [68] achieved an AUROC of 86% for appendicitis diagnosis using a dual-branch CNN–MLP model. Li et al. [65] also reported robust performance (AUROC = 85.4%) for reproductive risk prediction by fusing hysteroscopic images with structured EMR data. Models integrating radiographic and tabular data consistently demonstrated better results than unimodal baselines. Schilcher et al. [49] reached 84% AUROC in hip fracture mortality prediction, while Lee et al. [66] showed strong generalization on external datasets for predicting diabetic kidney disease. These studies highlight the importance of modality-specific preprocessing, such as resolution normalization and outlier filtering, and the use of compatible fusion schemes like late or intermediate fusion. Shared encoder designs for EHR-centered tasks are effective for risk stratification. Wang et al. [52] achieved AUROCs of 91.1% and 88.5% for ICU mortality and sepsis prediction, respectively, by merging time-series, clinical notes, and structured features. Cai et al. [59] showed that contrastive learning on EHR segments enhanced classification performance without relying on imaging or text data.
Medical reasoning tasks benefit from the increasing capabilities of Vision-language models (VLMs). The Gemini model [53] achieved 86.7% zero-shot AUROC and high retrieval accuracy, outperforming prior contrastive models such as ConVIRT and GLoRIA. PMC-VQA [36] reached 71.2% accuracy in medical VQA. General-purpose instruction-tuned models like BLIP-2 [37] also performed well (73.3%) even without domain-specific training, though MiniGPT-v2 showed weaker results. GPT-4V, evaluated by Panagoulias et al. [69], demonstrated versatility in multimodal reporting, while ChatGPT-4V [67] showed strong performance in multiple medical tasks but raised concerns about reproducibility and specificity.
Several studies also addressed scalability and robustness. AutoFM [85], a NAS-based system, discovered optimal fusion architectures and achieved AUROC scores above 90% in multiple EHR tasks. Zeng et al. [64] used stacked ensemble learning to combine clinical and genetic features, achieving AUROCs of 81.6% (internal) and 79.2% (external). Bampa et al. [62] introduced M-ClustEHR for unsupervised phenotype discovery using contrastive clustering, without the need for labeled data. Chung et al. [63] showed that combining XGBoost with neural networks improves disease risk prediction in large biobank settings.
In addition to algorithmic performance, several studies have begun addressing the hardware and system-level constraints associated with deploying MAI models in portable or real-time diagnostic settings. For example, Laganà et al. [11] proposed a low-power, lightweight architecture that can integrate sensor-level inputs, such as thermal, ultrasound, or SAR signals, into multimodal diagnostic pipelines. These models aim to operate under latency and power constraints while maintaining robustness to noise or missing data. This approach expands the scope of MAI beyond conventional imaging and EHR data, highlighting new opportunities for early diagnosis using sensor technologies. Integrating MAI into wearable monitors or portable diagnostic kits is a promising approach for real-world deployment, especially when combined with feedback-aware or continuously updating model architectures.
Preprocessing and fusion strategies played a significant role in all of these systems. Studies using structured normalization, quality control, and modality-aligned encoding consistently outperformed those using generic inputs. Intermediate fusion, cross-modal attention, and decision-level ensembles emerged as dominant strategies, while hybrid approaches such as CMFF and AutoFM illustrate the potential of task-adaptive designs.
Despite these advances, several challenges remain. Many models are trained on institution-specific datasets like ADNI, MIMIC-III, and CTU-UHB, limiting generalizability. These datasets exhibit known demographic biases, such as the overrepresentation of certain age groups or ethnicities, which can lead to biased model behavior. For example, MIMIC-III primarily represents ICU patients from a single U.S. hospital system, while ADNI has limited racial diversity and often lacks representation from early disease stages. Few of the reviewed studies explicitly addressed bias mitigation. However, the adoption of balanced sampling, domain adaptation, or subgroup evaluation would be important steps toward improving fairness in real-world applications. Moreover, future research should prioritize the use of geographically and demographically diverse datasets to ensure broader applicability across healthcare systems. Techniques such as domain adaptation, adversarial training, or localized fine-tuning can further improve generalizability by mitigating the effects of distributional shifts between training and deployment environments. In addition to population bias, variation in preprocessing methods can further affect model reliability and comparability. Only a few studies, such as those by Lee et al. [66] and Zeng et al. [64], provide external validation, which is critical for assessing real-world reliability. Preprocessing strategies also vary widely, and their role is often underreported. Studies like Feng et al. [46] and Niu et al. [61] emphasize that task-specific normalization, tokenization, and missing data handling can significantly affect model results.
Fusion strategies also lack standardization, while intermediate fusion and cross-modal attention frequently deliver strong results, few studies perform ablation analysis to measure their precise contributions. Benchmarking remains an open challenge in multimodal learning. Additionally, studies rarely follow a harmonized protocol to evaluate fusion depth, modality contribution, and domain generalizability. To improve reproducibility and comparability, recent work has recommended hybrid soft-computing benchmarking frameworks that explicitly model modality interaction, dropout resilience, and fusion position impact [12]. These approaches provide valuable insights beyond deep learning–based pipelines and underscore the need for interdisciplinary criteria when comparing fusion strategies. An important practical challenge in multimodal diagnostics is the frequent presence of incomplete or missing modalities due to acquisition failure, cost, or patient-specific contraindications, while some studies implement late fusion or decision-level ensembling to handle partial inputs, few perform a systematic evaluation of dropout scenarios. Architectures like AutoFM [85] and Gemini [53] show some resilience through flexible modality alignment or contrastive pretraining. For example, Lu et al.’s BKTDN model [31] reported an accuracy drop of nearly 30% when head position vectors were removed, underscoring both the contribution and sensitivity of certain modalities. Although explicit cases of accuracy degradation due to contradictory modalities are rare, these findings suggest that poor integration or reliance on noisy modalities may negatively impact performance. Integrating robustness to missing or conflicting data as a design criterion in benchmarking and architecture selection will be essential for deployment in heterogeneous clinical environments.
Interpretability methods in multimodal medical AI generally follow modality-specific conventions. In convolutional architectures applied to imaging data, techniques like Grad-CAM or saliency maps are commonly used to visualize spatial attention. For structured inputs such as EHR or tabular features, methods such as SHAP or feature attribution models are typically employed to quantify input contribution, while not all reviewed studies implemented these tools explicitly, this classification reflects prevailing practices in the field. For example, Martin et al. [42] applied saliency mapping and attribution analysis to assess the contribution of neuroimaging and clinical variables in dementia diagnosis, and Li et al. [29] integrated visual heatmaps over IHC and photographic images to support human interpretability. These examples demonstrate how model outputs can be made clinically accessible. However, systematic evaluation of interpretability remains limited in most current MAI systems. Although many reviewed models report strong results, few include a systematic evaluation of their failure modes or domain-specific risks. Overfitting to institution-specific datasets, lack of robustness under real-world noise conditions, or insufficient testing across diverse patient populations remain significant limitations. Moreover, few studies explore how conflicting information across modalities is resolved or flagged by the models, which is a critical area for clinical safety and transparency. The growing use of general-purpose LLMs like ChatGPT-4V introduces additional concerns about reproducibility, hallucination, and domain specificity, especially in high-stakes contexts such as histopathology or surgical trials.
Another emerging area is the development of adaptive MAI systems that incorporate clinician feedback or patient-specific trends over time. These architectures allow for progressive refinement of model predictions or fusion weights based on usage context. For example, Menniti et al. [13] demonstrated a multimodal artificial intelligence framework for portable patient monitoring that continuously adapts to sensor inputs and clinical feedback, offering an early model of human-in-the-loop deployment. Such systems align closely with real-world clinical needs and present opportunities for continuous learning and personalization.
Overall, the reviewed studies demonstrate that multimodal artificial intelligence has the potential to significantly enhance diagnostic performance, provided that models are based on robust preprocessing pipelines, appropriate fusion designs, and validated on diverse patient populations. Future research should prioritize benchmark standardization, cross-institutional validation, model interpretability, and adaptive architecture search. Additionally, future work should examine the ethical implications and regulatory challenges associated with deploying MAI systems in clinical practice, including issues of patient consent, accountability, and model governance. Instruction tuning, contrastive pretraining, and unsupervised learning remain promising directions toward scalable, explainable, and clinically reliable multimodal systems.
8. Conclusions
The review analyzed recent developments in multimodal artificial intelligence (MAI) for medical diagnostics by showing how imaging data, together with electronic health records, physiological signals, and free-text reports, enhance disease prediction and clinical decision making. MAI systems across various fields such as cardiology, neurology, oncology, radiology, and pediatrics show better diagnostic results than single-data-method approaches. We discussed the growing adoption of intermediate and attention-based fusion strategies, as well as the use of self-supervised and instruction-tuned vision-language models like BLIP-2 and GPT-4V in answering clinical visual questions. A wide range of datasets, both public and institution-specific, were described, along with preprocessing strategies tailored to each data type. In particular, models integrating structured EHRs with imaging modalities achieved strong predictive accuracy for critical outcomes such as mortality, sepsis, and surgical risk.
Despite these successes, challenges persist related to data harmonization, generalizability, and interpretability. Several studies have addressed these concerns through contrastive learning, knowledge-guided fusion, and architectural innovations such as AutoFM and hybrid ensemble models. Further progress will depend on the integration of scalable training strategies, access to diverse multimodal datasets, and increased emphasis on explainability and clinical relevance. These developments lay the foundation for more robust, generalizable, and transparent AI systems in real-world healthcare environments.
Author Contributions
Conceptualization, B.J. and M.A.A.; methodology, B.J. and M.A.A.; validation, B.J. and M.A.A.; formal analysis, B.J. and M.A.A.; writing—original draft preparation, B.J.; writing—review and editing, M.A.A.; funding acquisition, M.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by the Natural Sciences and Engineering Research Council of Canada (NSERC), under funding reference number RGPIN-2024-05287, and by the AI in Health Research Chair at the Université de Moncton.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| EHR | Electronic Health Record |
| AUROC | Area Under the Receiver Operating Characteristic Curve |
| MRI | Magnetic Resonance Imaging |
| AUC | Area Under the Curve |
| QA | Question Answering |
| CNN | Convolutional Neural Network |
| VQA | Visual Question Answering |
| UK | United Kingdom |
| ADNI | Alzheimer’s Disease Neuroimaging Initiative |
| NEC | Necrotizing Enterocolitis |
| IHC | Immunohistochemistry |
| AI | Artificial Intelligence |
| MAI | Multimodal Artificial Intelligence |
| CTG | Cardiotocography |
| FHR | Fetal Heart Rate |
References
- Albahra, S.; Gorbett, T.; Robertson, S.; D’Aleo, G.; Kumar, S.V.S.; Ockunzzi, S.; Lallo, D.; Hu, B.; Rashidi, H.H. Artificial intelligence and machine learning overview in pathology & laboratory medicine: A general review of data preprocessing and basic supervised concepts. Semin. Diagn. Pathol. 2023, 40, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Najjar, R. Redefining Radiology: A Review of Artificial Intelligence Integration in Medical Imaging. Diagnostics 2023, 13, 2760. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Zuo, K.; Li, Y.; Pang, Z. A review of the application of multi-modal deep learning in medicine: Bibliometrics and future directions. Int. J. Comput. Intell. Syst. 2023, 16, 44. [Google Scholar] [CrossRef]
- Barua, A.; Ahmed, M.U.; Begum, S. A systematic literature review on multimodal machine learning: Applications, challenges, gaps and future directions. IEEE Access 2023, 11, 14804–14831. [Google Scholar] [CrossRef]
- Liang, P.P.; Zadeh, A.; Morency, L.P. Foundations and trends in multimodal machine learning: Principles, challenges, and open questions. ACM Comput. Surv. 2024, 56, 1–42. [Google Scholar] [CrossRef]
- Krones, F.; Marikkar, U.; Parsons, G.; Szmul, A.; Mahdi, A. Review of multimodal machine learning approaches in healthcare. arXiv 2024, arXiv:2402.02460. [Google Scholar] [CrossRef]
- Simon, B.; Ozyoruk, K.; Gelikman, D.; Harmon, S.; Türkbey, B. The future of multimodal artificial intelligence models for integrating imaging and clinical metadata: A narrative review. Diagn. Interv. Radiol. 2024. [Google Scholar] [CrossRef]
- Demirhan, H.; Zadrozny, W. Survey of Multimodal Medical Question Answering. BioMedInformatics 2024, 4, 50–74. [Google Scholar] [CrossRef]
- Adewumi, T.; Alkhaled, L.; Gurung, N.; van Boven, G.; Pagliai, I. Fairness and bias in multimodal ai: A survey. arXiv 2024, arXiv:2406.19097. [Google Scholar]
- Isavand, P.; Aghamiri, S.S.; Amin, R. Applications of Multimodal Artificial Intelligence in Non-Hodgkin Lymphoma B Cells. Biomedicines 2024, 12, 1753. [Google Scholar] [CrossRef]
- Laganà, F.; Bibbò, L.; Calcagno, S.; De Carlo, D.; Pullano, S.A.; Pratticò, D.; Angiulli, G. Smart Electronic Device-Based Monitoring of SAR and Temperature Variations in Indoor Human Tissue Interaction. Appl. Sci. 2025, 15, 2439. [Google Scholar] [CrossRef]
- Mario, V.; Laganá, F.; Manin, L.; Angiulli, G. Soft computing and eddy currents to estimate and classify delaminations in biomedical device CFRP plates. J. Electr. Eng. 2025, 76, 72–79. [Google Scholar] [CrossRef]
- Menniti, M.; Laganà, F.; Oliva, G.; Bianco, M.; Fiorillo, A.S.; Pullano, S.A. Development of Non-Invasive Ventilator for Homecare and Patient Monitoring System. Electronics 2024, 13, 790. [Google Scholar] [CrossRef]
- Baltrušaitis, T.; Ahuja, C.; Morency, L.P. Multimodal machine learning: A survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 41, 423–443. [Google Scholar] [CrossRef]
- Huang, D.; Yan, C.; Li, Q.; Peng, X. From Large Language Models to Large Multimodal Models: A Literature Review. Appl. Sci. 2024, 14, 5068. [Google Scholar] [CrossRef]
- Jabeen, S.; Li, X.; Amin, M.S.; Bourahla, O.; Li, S.; Jabbar, A. A review on methods and applications in multimodal deep learning. ACM Trans. Multimed. Comput. Commun. Appl. 2023, 19, 1–41. [Google Scholar] [CrossRef]
- Li, Y.; Daho, M.E.H.; Conze, P.H.; Zeghlache, R.; Le Boité, H.; Tadayoni, R.; Cochener, B.; Lamard, M.; Quellec, G. A review of deep learning-based information fusion techniques for multimodal medical image classification. Comput. Biol. Med. 2024, 177, 108635. [Google Scholar] [CrossRef]
- Evans, R.S. Electronic health records: Then, now, and in the future. Yearb. Med. Inform. 2016, 25, S48–S61. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Pacheco, A.G.; Lima, G.R.; Salomao, A.S.; Krohling, B.; Biral, I.P.; de Angelo, G.G.; Alves, F.C., Jr.; Esgario, J.G.; Simora, A.C.; Castro, P.B.; et al. PAD-UFES-20: A skin lesion dataset composed of patient data and clinical images collected from smartphones. Data Brief 2020, 32, 106221. [Google Scholar] [CrossRef]
- Subramanian, S.; Wang, L.L.; Mehta, S.; Bogin, B.; van Zuylen, M.; Parasa, S.; Singh, S.; Gardner, M.; Hajishirzi, H. Medicat: A Dataset of Medical Images, Captions, and Textual References; Association for Computational Linguistics: Stroudsburg, PA, USA, 2020; pp. 2112–2120. [Google Scholar] [CrossRef]
- Li, M.; Cai, W.; Liu, R.; Weng, Y.; Zhao, X.; Wang, C.; Chen, X.; Liu, Z.; Pan, C.; Li, M.; et al. Ffa-ir: Towards an explainable and reliable medical report generation benchmark. In Proceedings of the Thirty-Fifth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (Round 2), Online, 6–14 December 2021. [Google Scholar]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Lehman, L.w.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.E.; Bulgarelli, L.; Shen, L.; Gayles, A.; Shammout, A.; Horng, S.; Pollard, T.J.; Hao, S.; Moody, B.; Gow, B.; et al. MIMIC-IV, a freely accessible electronic health record dataset. Sci. Data 2023, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Pelka, O.; Koitka, S.; Rückert, J.; Nensa, F.; Friedrich, C.M. Radiology Objects in COntext (ROCO): A Multimodal Image Dataset. In Proceedings of the Intravascular Imaging and Computer Assisted Stenting and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis, Granada, Spain, 16 September 2018; pp. 180–189. [Google Scholar] [CrossRef]
- Ruckert, J.; Bloch, L.; Brungel, R.; Idrissi-Yaghir, A.; Schäfer, H.; Schmidt, C.S.; Koitka, S.; Pelka, O.; Ben, A.; Abacha, A.G.; et al. ROCOv2: Radiology Objects in COntext Version 2, an Updated Multimodal Image Dataset. Sci. Data 2024, 11, 688. [Google Scholar] [CrossRef] [PubMed]
- Pediatric Imaging: A Pediatric Radiology Textbook and Digital Library. (Source of Pediatric Radiology Dataset). Available online: https://pediatricimaging.org (accessed on 20 May 2025).
- Gao, W.; Pei, Y.; Liang, H.; Lv, J.; Chen, J.; Zhong, W. Multimodal AI System for the Rapid Diagnosis and Surgical Prediction of Necrotizing Enterocolitis. IEEE Access 2021, 9, 51050–51064. [Google Scholar] [CrossRef]
- Li, Q.; Yang, Z.; Chen, K.; Zhao, M.; Long, H.; Deng, Y.; Hu, H.; Jia, C.; Wu, M.; Zhao, Z.; et al. Human-multimodal deep learning collaboration in ‘precise’diagnosis of lupus erythematosus subtypes and similar skin diseases. J. Eur. Acad. Dermatol. Venereol. 2024, 38, 2268–2279. [Google Scholar] [CrossRef]
- Chudáček, V.; Spilka, J.; Burša, M.; Janků, P.; Hruban, L.; Huptych, M.; Lhotská, L. Open access intrapartum CTG database. BMC Pregnancy Childbirth 2014, 14, 16. [Google Scholar] [CrossRef]
- Lu, H.; Mao, Y.; Li, J.; Zhu, L. Multimodal deep learning-based diagnostic model for BPPV. BMC Med. Inform. Decis. Mak. 2024, 24, 82. [Google Scholar] [CrossRef]
- Weiner, M.W.; Aisen, P.S.; Jack, C.R., Jr.; Jagust, W.J.; Trojanowski, J.Q.; Shaw, L.; Saykin, A.J.; Morris, J.C.; Cairns, N.; Beckett, L.A.; et al. The Alzheimer’s disease neuroimaging initiative: Progress report and future plans. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2010, 6, 202–211. [Google Scholar] [CrossRef]
- Liu, B.; Zhan, L.M.; Xu, L.; Ma, L.; Yang, Y.; Wu, X.M. Slake: A semantically-labeled knowledge-enhanced dataset for medical visual question answering. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1650–1654. [Google Scholar] [CrossRef]
- Beekly, D.L.; Ramos, E.M.; Lee, W.W.; Deitrich, W.D.; Jacka, M.E.; Wu, J.; Hubbard, J.L.; Koepsell, T.D.; Morris, J.C.; Kukull, W.A.; et al. The National Alzheimer’s Coordinating Center (NACC) database: The uniform data set. Alzheimer Dis. Assoc. Disord. 2007, 21, 249–258. [Google Scholar] [CrossRef]
- UK Biobank. New Data & Enhancements to UK Biobank. 2024. Available online: https://www.ukbiobank.ac.uk/enable-your-research/about-our-data (accessed on 4 April 2025).
- Zhang, X.; Wu, C.; Zhao, Z.; Lin, W.; Zhang, Y.; Wang, Y.; Xie, W. Pmc-vqa: Visual instruction tuning for medical visual question answering. arXiv 2023, arXiv:2305.10415. [Google Scholar]
- Reith, T.P.; D’Alessandro, D.M.; D’Alessandro, M.P. Capability of multimodal large language models to interpret pediatric radiological images. Pediatr. Radiol. 2024, 54, 1729–1737. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.C.A.; Chen, C.Y.; Chen, T.T.; Kuo, P.H.; Hsu, Y.H.; Yang, H.I.; Chen, W.J.; Su, M.W.; Chu, H.W.; Shen, C.Y.; et al. Taiwan Biobank: A rich biomedical research database of the Taiwanese population. Cell Genom. 2022, 2, 100197. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Singh, B. Investigating the impact of data normalization on classification performance. Appl. Soft Comput. 2020, 97, 105524. [Google Scholar] [CrossRef]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Briefings Bioinform. 2018, 19, 1236–1246. [Google Scholar] [CrossRef]
- Li, Y.; Ammari, S.; Balleyguier, C.; Lassau, N.; Chouzenoux, E. Impact of preprocessing and harmonization methods on the removal of scanner effects in brain MRI radiomic features. Cancers 2021, 13, 3000. [Google Scholar] [CrossRef]
- Martin, S.A.; Zhao, A.; Qu, J.; Imms, P.E.; Irimia, A.; Barkhof, F.; Cole, J.H.; Initiative, A.D.N. Explainable artificial intelligence for neuroimaging-based dementia diagnosis and prognosis. medRxiv 2025. [Google Scholar] [CrossRef]
- Xue, C.; Kowshik, S.S.; Lteif, D.; Puducheri, S.; Jasodanand, V.H.; Zhou, O.T.; Walia, A.S.; Guney, O.B.; Zhang, J.D.; Pham, S.T.; et al. AI-based differential diagnosis of dementia etiologies on multimodal data. Nat. Med. 2024, 30, 2977–2989. [Google Scholar] [CrossRef]
- Sheng, J.; Zhang, Q.; Zhang, Q.; Wang, L.; Yang, Z.; Xin, Y.; Wang, B. A hybrid multimodal machine learning model for Detecting Alzheimer’s disease. Comput. Biol. Med. 2024, 170, 108035. [Google Scholar] [CrossRef]
- Jahan, S.; Abu Taher, K.; Kaiser, M.S.; Mahmud, M.; Rahman, M.S.; Hosen, A.S.; Ra, I.H. Explainable AI-based Alzheimer’s prediction and management using multimodal data. PloS ONE 2023, 18, e0294253. [Google Scholar] [CrossRef]
- Feng, Z.; Sivak, J.A.; Krishnamurthy, A.K. Multimodal fusion of echocardiography and electronic health records for the detection of cardiac amyloidosis. In Proceedings of the International Conference on Artificial Intelligence in Medicine, Portorož, Slovenia, 12–15 June 2023; Springer: Berlin/Heidelberg, Germany, 2024; pp. 227–237. [Google Scholar] [CrossRef]
- Fleurence, R.L.; Curtis, L.H.; Califf, R.M.; Platt, R.; Selby, J.V.; Brown, J.S. Launching PCORnet, a national patient-centered clinical research network. J. Am. Med. Inform. Assoc. 2014, 21, 578–582. [Google Scholar] [CrossRef]
- Bin, Y.; Yang, Y.; Shen, F.; Xu, X.; Shen, H.T. Bidirectional long-short term memory for video description. In Proceedings of the 24th ACM International Conference on Multimedia, Amsterdam, The Netherlands, 15–19 October 2016; pp. 436–440. [Google Scholar] [CrossRef]
- Schilcher, J.; Nilsson, A.; Andlid, O.; Eklund, A. Fusion of electronic health records and radiographic images for a multimodal deep learning prediction model of atypical femur fractures. Comput. Biol. Med. 2024, 168, 107704. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhu, J.; Jiao, P.; Wang, J.; Zhang, X.; Lu, X.; Zhang, Y. Hybrid-FHR: A multi-modal AI approach for automated fetal acidosis diagnosis. BMC Med. Inform. Decis. Mak. 2024, 24, 19. [Google Scholar] [CrossRef] [PubMed]
- Bowles, C.; Chen, L.; Guerrero, R.; Bentley, P.; Gunn, R.; Hammers, A.; Dickie, D.A.; Hernández, M.V.; Wardlaw, J.; Rueckert, D. Gan augmentation: Augmenting training data using generative adversarial networks. arXiv 2018, arXiv:1810.10863. [Google Scholar]
- Wang, Y.; Yin, C.; Zhang, P. Multimodal risk prediction with physiological signals, medical images and clinical notes. Heliyon 2024, 10, e26772. [Google Scholar] [CrossRef]
- Yang, L.; Xu, S.; Sellergren, A.; Kohlberger, T.; Zhou, Y.; Ktena, I.; Kiraly, A.; Ahmed, F.; Hormozdiari, F.; Jaroensri, T.; et al. Advancing Multimodal Medical Capabilities of Gemini. arXiv 2024, arXiv:2405.03162. [Google Scholar]
- Bodenreider, O. The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef]
- Lipscomb, C.E. Medical subject headings (MeSH). Bull. Med. Libr. Assoc. 2000, 88, 265. [Google Scholar]
- Yao, Z.; Lin, F.; Chai, S.; He, W.; Dai, L.; Fei, X. Integrating medical imaging and clinical reports using multimodal deep learning for advanced disease analysis. In Proceedings of the 2024 IEEE 2nd International Conference on Sensors, Electronics and Computer Engineering (ICSECE), Jinzhou, China, 29–31 August 2024; pp. 1217–1223. [Google Scholar] [CrossRef]
- Park, S.; Lee, E.S.; Shin, K.S.; Lee, J.E.; Ye, J.C. Self-supervised multi-modal training from uncurated images and reports enables monitoring AI in radiology. Med. Image Anal. 2024, 91, 103021. [Google Scholar] [CrossRef]
- Koonce, B. ResNet 50. In Convolutional Neural Networks with Swift for Tensorflow: Image Recognition and Dataset Categorization; Springer: Berlin/Heidelberg, Germany, 2021; pp. 63–72. [Google Scholar] [CrossRef]
- Cai, Y.; Liu, X.; Fan, M.; Wang, H.; Liu, M.; Yu, Y.; Wu, Y. Contrastive Learning on Multimodal Analysis of Electronic Health Records. Sci. Rep. 2024, 14, 3438. [Google Scholar]
- Suresh, H.; Hunt, N.; Johnson, A.; Celi, L.A.; Szolovits, P.; Ghassemi, M. Clinical intervention prediction and understanding using deep networks. arXiv 2017, arXiv:1705.08498. [Google Scholar]
- Niu, S.; Ma, J.; Bai, L.; Wang, Z.; Guo, L.; Yang, X. EHR-KnowGen: Knowledge-enhanced multimodal learning for disease diagnosis generation. Inf. Fusion 2024, 102, 102069. [Google Scholar] [CrossRef]
- Bampa, M.; Miliou, I.; Jovanovic, B.; Papapetrou, P. M-ClustEHR: A multimodal clustering approach for electronic health records. Artif. Intell. Med. 2024, 154, 102905. [Google Scholar] [CrossRef] [PubMed]
- Chung, R.H.; Onthoni, D.; Lin, H.M.; Li, G.H.; Hsiao, Y.P.; Zhuang, Y.S.; Onthoni, A.; Lai, Y.H.; Chiou, H.Y. Multimodal Deep Learning for Classifying Diabetes: Analyzing Carotid Ultrasound Images from UK and Taiwan Biobanks and Their Cardiovascular Disease Associations. 2024; preprint. [Google Scholar]
- Zeng, L.; Ma, P.; Li, Z.; Liang, S.; Wu, C.; Hong, C.; Li, Y.; Cui, H.; Li, R.; Wang, J.; et al. Multimodal Machine Learning-Based Marker Enables Early Detection and Prognosis Prediction for Hyperuricemia. Adv. Sci. 2024, 11, 2404047. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, H.; Lin, X.; Duan, H. Multimodal Learning system integrating electronic medical records and hysteroscopic images for reproductive outcome prediction and risk stratification of endometrial injury: A multicenter diagnostic study. Int. J. Surg. 2024, 110, 3237–3248. [Google Scholar] [CrossRef]
- Lee, Y.C.; Cha, J.; Shim, I.; Park, W.Y.; Kang, S.W.; Lim, D.H.; Won, H.H. Multimodal deep learning of fundus abnormalities and traditional risk factors for cardiovascular risk prediction. npj Digit. Med. 2023, 6, 14. [Google Scholar] [CrossRef]
- Zhu, L.; Lai, Y.; Ta, N.; Cheng, L.; Chen, R. Multimodal approach in the diagnosis of urologic malignancies: Critical assessment of ChatGPT-4V’s image-reading capabilities. JCO Clin. Cancer Inform. 2024, 8, e2300275. [Google Scholar] [CrossRef]
- Lin, A.C.; Liu, Z.; Lee, J.; Ranvier, G.F.; Taye, A.; Owen, R.; Matteson, D.S.; Lee, D. Generating a multimodal artificial intelligence model to differentiate benign and malignant follicular neoplasms of the thyroid: A proof-of-concept study. Surgery 2024, 175, 121–127. [Google Scholar] [CrossRef]
- Panagoulias, D.P.; Virvou, M.; Tsihrintzis, G.A. Evaluating LLM–Generated Multimodal Diagnosis from Medical Images and Symptom Analysis. arXiv 2024, arXiv:2402.01730. [Google Scholar]
- Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; Anadkat, S.; et al. Gpt-4 technical report. arXiv 2023, arXiv:2303.08774. [Google Scholar]
- Yildirim, N.; Richardson, H.; Wetscherek, M.T.; Bajwa, J.; Jacob, J.; Pinnock, M.A.; Harris, S.; Coelho De Castro, D.; Bannur, S.; Hyland, S.; et al. Multimodal healthcare AI: Identifying and designing clinically relevant vision-language applications for radiology. In Proceedings of the 2024 CHI Conference on Human Factors in Computing Systems, Honolulu, HI, USA, 11–16 May 2024; pp. 1–22. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.y.; Peng, Y.; Lu, Z.; Mark, R.G.; Berkowitz, S.J.; Horng, S. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. PhysioNet 2024. [Google Scholar] [CrossRef]
- Tortora, M.; Cordelli, E.; Sicilia, R.; Nibid, L.; Ippolito, E.; Perrone, G.; Ramella, S.; Soda, P. RadioPathomics: Multimodal learning in non-small cell lung cancer for adaptive radiotherapy. IEEE Access 2023, 11, 47563–47578. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar] [CrossRef]
- Huang, S.C.; Shen, L.; Lungren, M.P.; Yeung, S. Gloria: A multimodal global-local representation learning framework for label-efficient medical image recognition. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021; pp. 3922–3931. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, H.; Miura, Y.; Manning, C.D.; Langlotz, C.P. Contrastive Learning of Medical Visual Representations from Paired Images and Text. Proc. Mach. Learn. Health Care 2022, 182, 1–24. [Google Scholar]
- Guarrasi, V.; Aksu, F.; Caruso, C.M.; Di Feola, F.; Rofena, A.; Ruffini, F.; Soda, P. A systematic review of intermediate fusion in multimodal deep learning for biomedical applications. Image Vis. Comput. 2025, 158, 105509. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, J.; Jin, S.; Lu, S. Vision-language models for vision tasks: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 5625–5644. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Chen, G.; Liu, S.; Yu, Y.; Li, G. Cross-modal self-attention with multi-task pre-training for medical visual question answering. In Proceedings of the 2021 International Conference on Multimedia Retrieval, Taipei, Taiwan, 16–19 November 2021; pp. 456–460. [Google Scholar] [CrossRef]
- Rajabi, N.; Kosecka, J. Towards grounded visual spatial reasoning in multi-modal vision language models. arXiv 2023, arXiv:2308.09778. [Google Scholar]
- Wang, Y.; Chen, W.; Han, X.; Lin, X.; Zhao, H.; Liu, Y.; Zhai, B.; Yuan, J.; You, Q.; Yang, H. Exploring the reasoning abilities of multimodal large language models (mllms): A comprehensive survey on emerging trends in multimodal reasoning. arXiv 2024, arXiv:2401.06805. [Google Scholar]
- Li, J.; Li, D.; Savarese, S.; Hoi, S. Blip-2: Bootstrapping language-image pre-training with frozen image encoders and large language models. In Proceedings of the International Conference on Machine Learning, Zhuhai, China, 17–20 February 2023; pp. 19730–19742. [Google Scholar]
- Chen, J.; Zhu, D.; Shen, X.; Li, X.; Liu, Z.; Zhang, P.; Krishnamoorthi, R.; Chandra, V.; Xiong, Y.; Elhoseiny, M. Minigpt-v2: Large language model as a unified interface for vision-language multi-task learning. arXiv 2023, arXiv:2310.09478. [Google Scholar]
- Cui, S.; Wang, J.; Zhong, Y.; Liu, H.; Wang, T.; Ma, F. Automated fusion of multimodal electronic health records for better medical predictions. In Proceedings of the 2024 SIAM International Conference on Data Mining (SDM), Houston, TX, USA, 18–20 April 2024; pp. 361–369. [Google Scholar] [CrossRef]
- Ren, P.; Xiao, Y.; Chang, X.; Huang, P.Y.; Li, Z.; Chen, X.; Wang, X. A comprehensive survey of neural architecture search: Challenges and solutions. ACM Comput. Surv. (CSUR) 2021, 54, 1–34. [Google Scholar] [CrossRef]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. (CSUR) 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; pp. 590–597. [Google Scholar] [CrossRef]
- Ren, H.; Lu, W.; Xiao, Y.; Chang, X.; Wang, X.; Dong, Z.; Fang, D. Graph convolutional networks in language and vision: A survey. Knowl.-Based Syst. 2022, 251, 109250. [Google Scholar] [CrossRef]
- Zhu, L.; Mou, W.; Lai, Y.; Chen, J.; Lin, S.; Xu, L.; Lin, J.; Guo, Z.; Yang, T.; Lin, A.; et al. Step into the era of large multimodal models: A pilot study on ChatGPT-4V (ision)’s ability to interpret radiological images. Int. J. Surg. 2024, 110, 4096–4102. [Google Scholar] [CrossRef]
- Latif, G.; Alghazo, J.; Mohammad, N.; Abdelhamid, S.E.; Brahim, G.B.; Amjad, K. A Novel Fragmented Approach for Securing Medical Health Records in Multimodal Medical Images. Appl. Sci. 2024, 14, 6293. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
