Deep Learning Techniques for Lung Cancer Diagnosis with Computed Tomography Imaging: A Systematic Review for Detection, Segmentation, and Classification
Abstract
1. Introduction
- Granular pipeline analysis: A detailed dissection of the diagnostic pipeline of detection, segmentation, and classification, tailored explicitly for CT imaging;
- Practical challenges: Emphasis on real-world issues, such as data scarcity, data variability, and challenges in clinical integration;
- Forward-looking perspectives: A critical evaluation of performance metrics and future directions was discussed, which includes the potential CNN-transformer hybrid architectures and explainable AI frameworks.
2. Literature Review
2.1. Evolution of CAD and DL
2.2. Deep Convolutional Neural Networks (DCNNs)
2.2.1. Overview of Basic DL Techniques
- Convolutional neural networks (CNNs);
- Fully connected neural networks (FCNNs/DNNs);
- Deep belief networks (DBNs);
- Recurrent neural networks (RNNs);
- Long short-term memory (LSTM) networks;
- Autoencoders (AEs);
- Vision transformers (ViTs).
2.2.2. CNN Model Architecture Figure 2
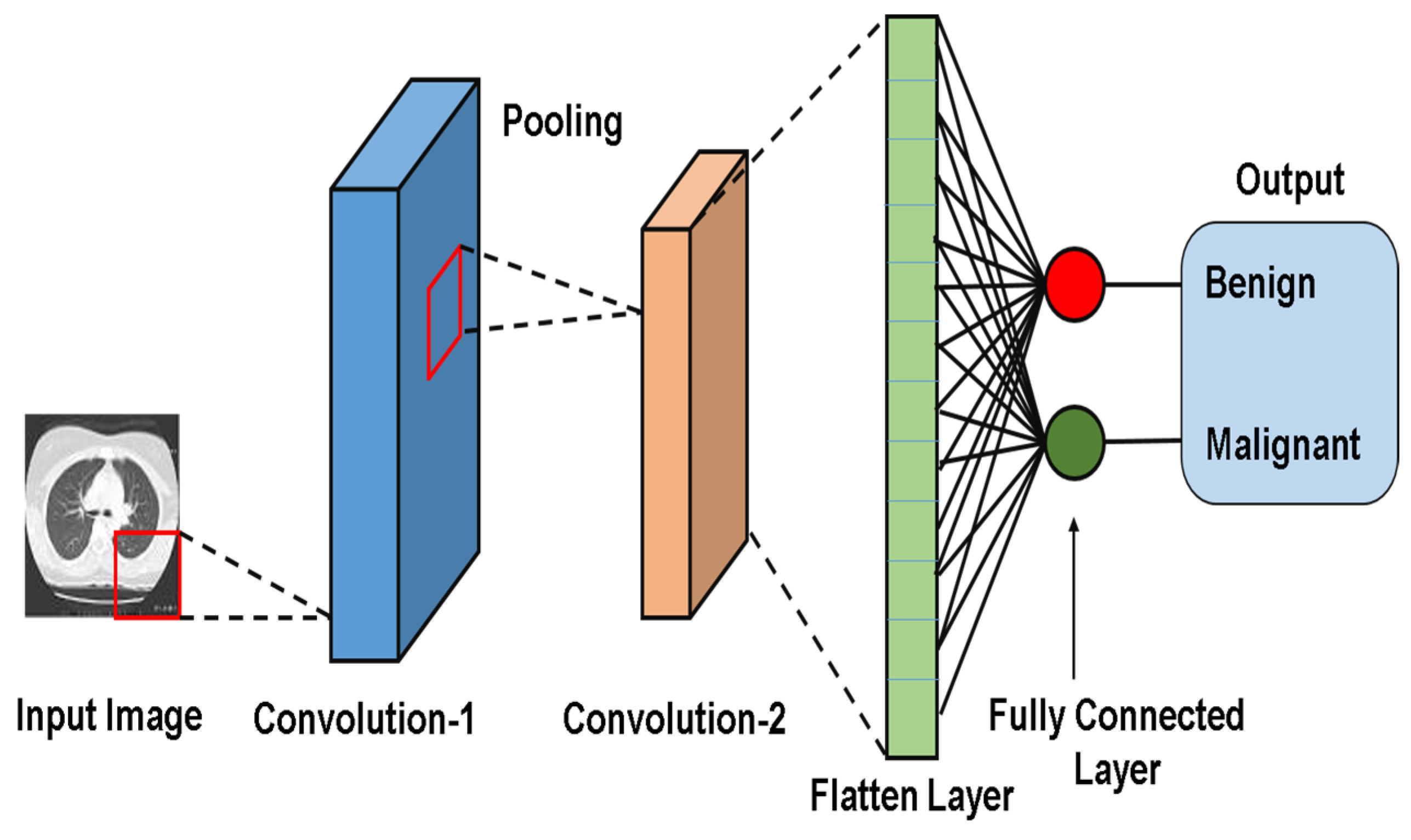
- Convolution (Conv) Layer:Extracts spatial features (e.g., edges, texture) using learnable filters (Kernels);
- Activation Function (Fact):Introduces nonlinearity (e.g., ReLU) to enable the network to model complex patterns.
- Pooling Layer:Reduces spatial dimension while retaining critical features (e.g., max pooling preserves dominant activation);
- Fully Connected (FC) Layer:This layer combines learned features for tasks like classification, regression, and/or feature learning.
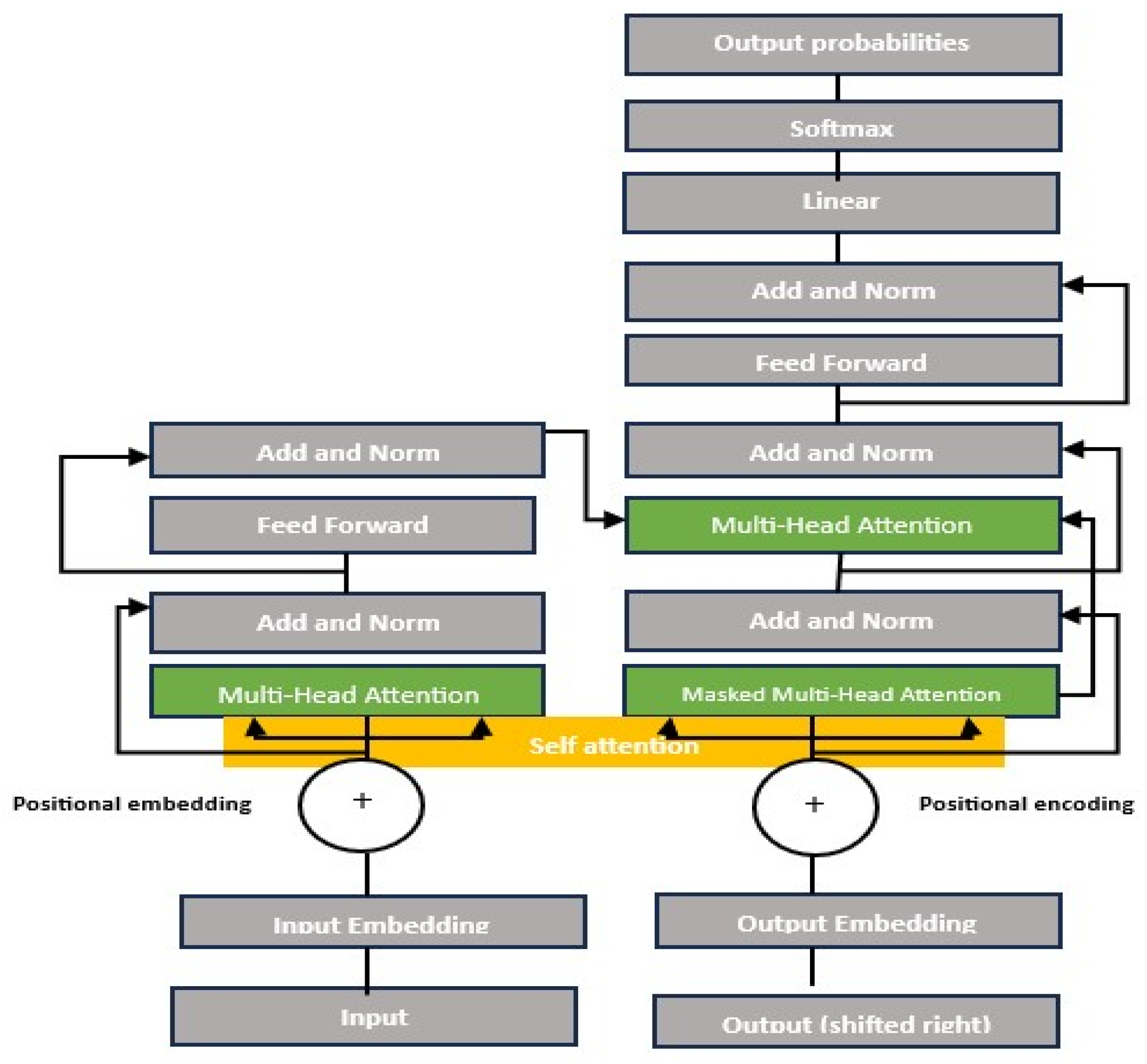
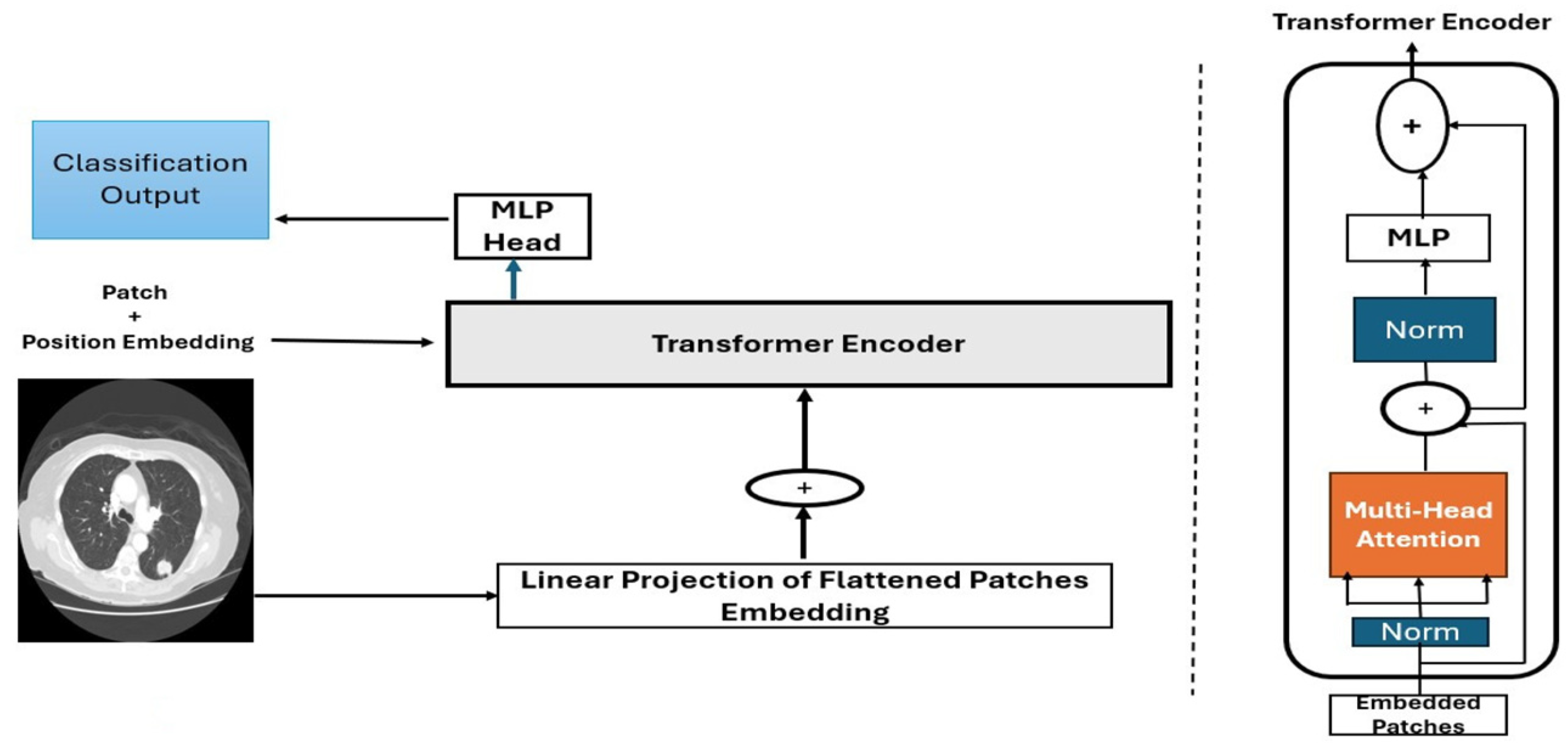
2.2.3. Deep CNNs vs. ViTs
Convolutional Neural Networks
Vision Transformers (ViTs)
- Image patching and embedding
- o
- Patch splitting: An input image (e.g., of size 224 × 224 pixels) is divided into fixed-size, non-overlapping patches. For instance, splitting the image into 16 × 16-pixel patches yield a grid of 224/16 = 14 × 14 = 196 patches.
- o
- Patch flattening: Each patch is reshaped into a 1D vector (e.g., 16 × 16 × 3 to give a 196-dimensional vector).
- o
- Patch embedding: Flattened patches were projected into a higher dimensional via a learnable linear projection. This linear transformation enables the model to learn richer feature representations for each patch. The result is a sequence of patch embeddings, each representing a part of the image.
- o
- Positional encoding: Spatial information was retained by adding positional embedding to patch vectors. This embedding enables the model to understand the spatial relationships between the patches.
- Transformer encoder:
- o
- Multi-head self-attention (MSA): The self-attention mechanism allows each patch to attend to others, computing attention scores as follows:where Q represents a query, K is a key, and V represents a value, which are learned linear projections.
- o
- Feedforward network (FFN): After self-attention, features pass through two fully connected layers with a nonlinear activation function (typically GELU activation).
- o
- Residual connection and layer normalization: Stabilize training by preserving information across layers. These techniques ensure that the deeper layers do not lose critical information from the earlier layers.
- Classification head (MLP head): The classification tokens (CLS) are extracted and fed into a multilayer perceptron (MLP) for final classification.
- Practical Limitations and Comparisons
2.3. Lung Nodule Detection and Segmentation
2.4. Lung Nodule Classification
3. Research Methodology
3.1. Overview
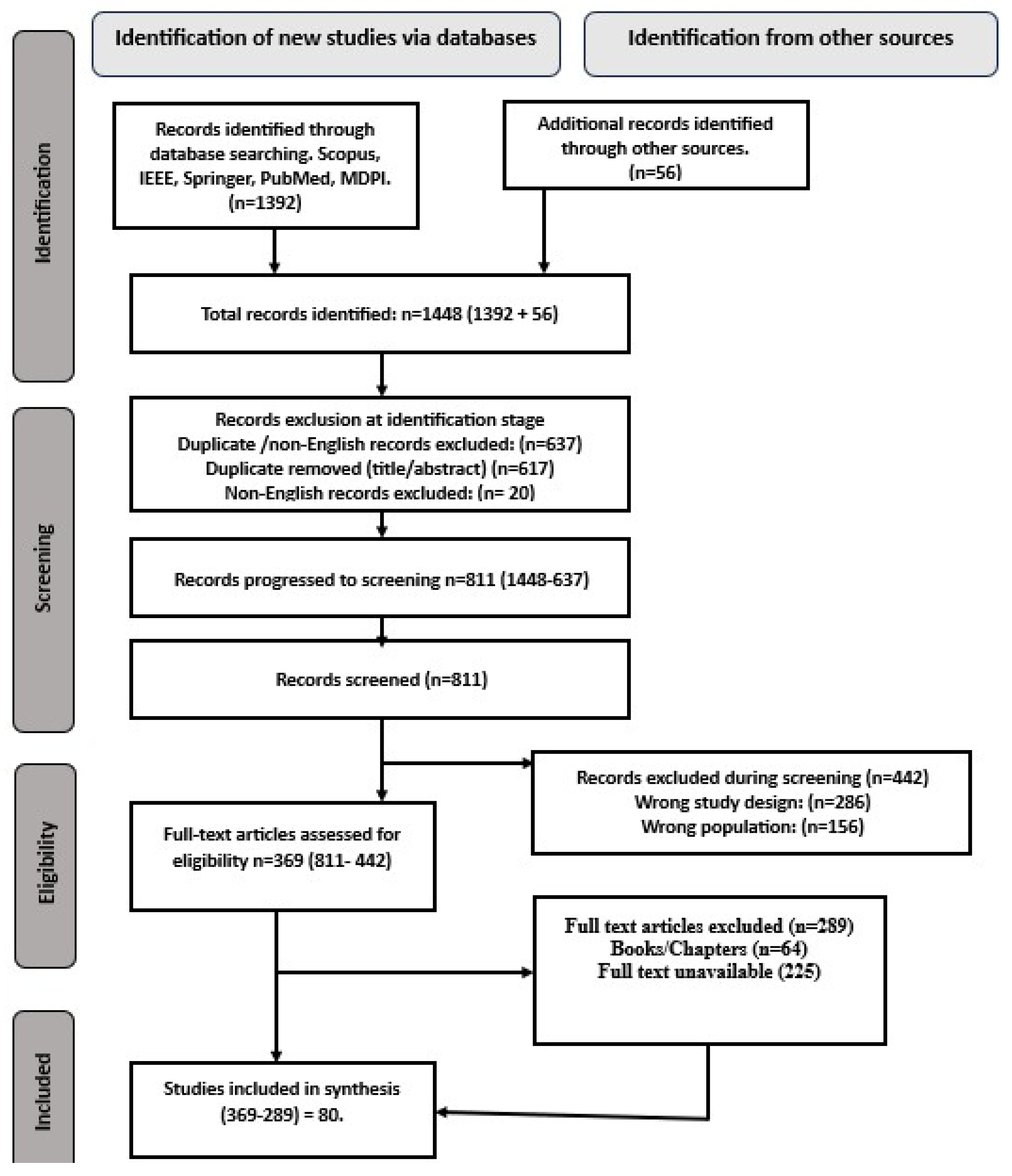
3.2. Literature Search and Selection
3.3. Initial Search Results
3.4. Inclusion/Exclusion Criteria
3.5. Search Strategy
- Screening: Two independent reviewers screened the titles/abstracts and keywords of the English-language articles, and conflicts were resolved through a third reviewer.
- Full-text review: articles meeting the inclusion criteria proceeded to the data extraction stage.
- Timeframe: We prioritized studies published between 2015 and 2024.
- Organization: Full texts were exported to Mendeley, and irrelevant studies (e.g., textbooks, non-peer-reviewed reports, and non-CT modalities) were excluded.
- Data Extraction: We extracted information on global cancer incidence and mortality, model architecture, datasets, performance metrics, and validation methods.
- Tools: we used a custom Excel template and the Mendeley reference manager.
3.6. Quality Assessment
- Performance metrics: accuracy, sensitivity, specificity, F1-score, false positive reduction, Dice similarity coefficient, computational performance (CPM), and AUC-ROC;
- Validation methods: cross-validation and external datasets testing;
- Reproducibility: code availability and transparency in hyperparameter reporting;
- Reporting transparency: adherence to standardized reporting guidelines.
3.7. Administrative Information
- ■
- Registration Platform: Adopted the International Prospective Register of Systematic Reviews (PROSPERO)
4. Results
4.1. Study Selection Process
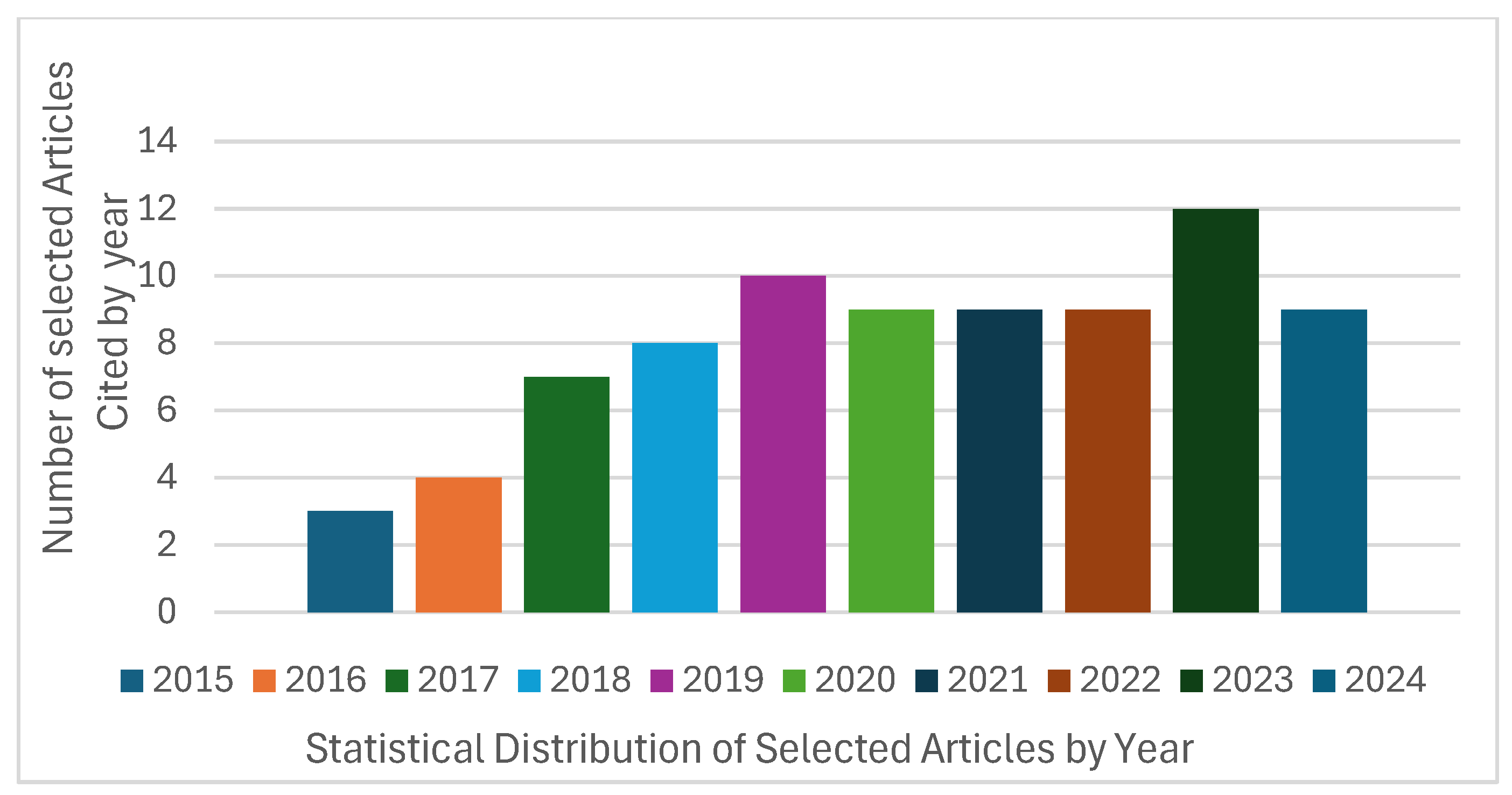
4.2. Study Characteristics
4.3. Data Synthesis and Analysis
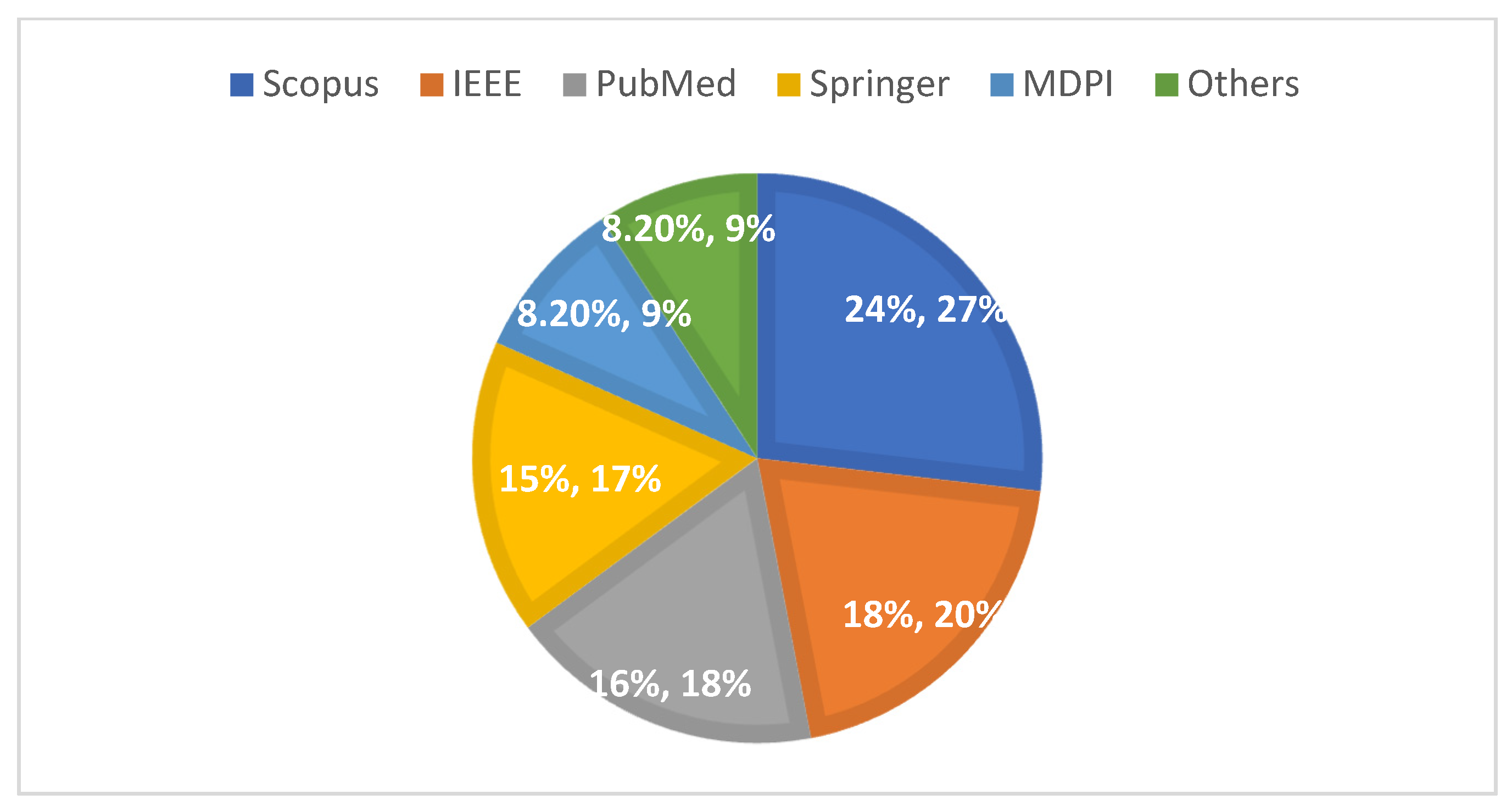
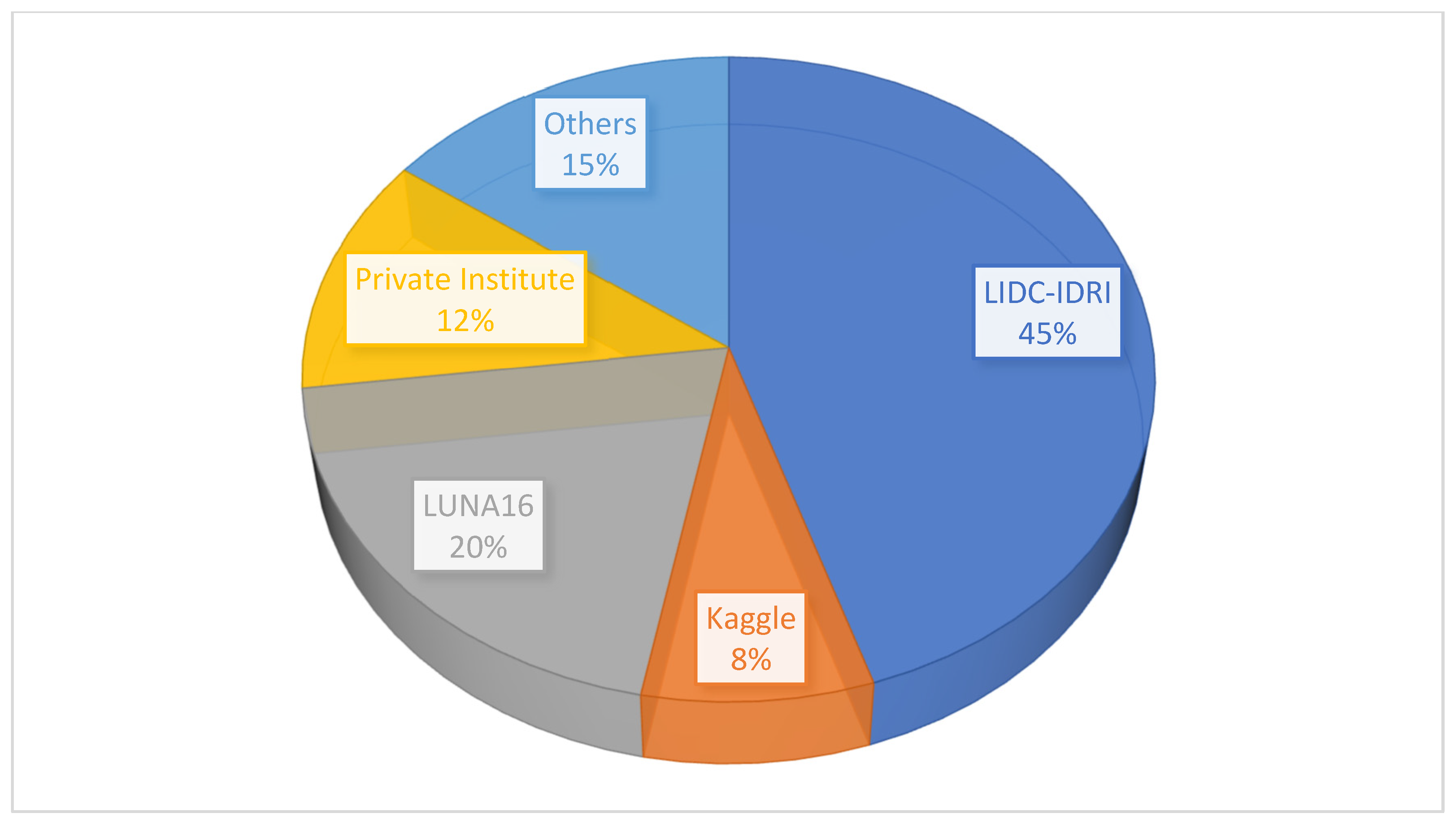
4.4. Databases for Lung Cancer CT Imaging
Datasets Analysis
4.5. Evaluation Metrics
- I.
- Accuracy (Acc):
- II.
- Sensitivity (Recall):
- III.
- Specificity
- IV.
- F1-Score
- V.
- Area Under the Curve (AUC):
- VI.
- The Receiver Operating Characteristics (ROC) Curve
- VII.
- Dice Similarity Coefficient (DCS):
5. Discussion
Key Findings, Limitations, and Future Directions
- Limitations:
- Language Exclusion Bias:
- Future Directions:
- Curating multimodal, multicentre datasets: We recommend collaborating globally to build datasets with diverse demographics (e.g., age, ethnicity, sex), imaging protocols (slice thickness: 1–5 mm), and cancer stages (I–IV), for public research utilization. Additionally, we recommend utilizing federated learning (FL) for privacy-preserving data sharing. For example, Liu et al. 2023 [58] demonstrated the efficacy of federated learning with a 3D ResNet18 model, achieving comparable accuracy to centralized training while preserving patient data privacy across institutions, allowing hospitals to collaborate training models without sharing raw data, a vital feature for complying with regulations like the European Union (EU)’s General data Protection Regulation (GDPR), the European Economic Area (EEA)’s regulation for personal data privacy for its citizens regardless of where the data are processed, and the US’s Health Insurance Portability and Accountability (HIPAA) law, which protects healthcare information and patient privacy).
- Develop lightweight, efficient models: To address computational constraints and privacy concerns, developing a lightweight architecture, such as MobileNetV3 or EfficientNet-Lite, for edge deployment is crucial. Lightweight models reduce computational overhead through techniques like pruning (removing redundant neurons), quantization (reducing numerical precision), and knowledge distillation (training compact models to mimic larger ones).
- Expand beyond binary classification: Moving beyond binary classification to multi-class staging (e.g., NSCLC stages I–IV) and histological subtyping (adenocarcinoma vs. squamous cell carcinoma) is crucial for personalized treatment, thereby improving diagnostic granularity. For example, Chang et al. (2024) [66] used a multiview residual network to classify nodule malignancy but did not address staging. We also recommend leveraging federated datasets, such as the Decathlon challenge [90], to pool multi-institutional staging data.
- Ensure real-world validation with explainability: We recommend validating a model on real-world datasets with artefacts (e.g., NSCLC-Radiomics). We also recommend embedding explainable tools, such as Gradient-weighted Class Activation Mapping (GRAD-CAM) and Local Interpretable Model-agnostic Explanations (LIME), into the clinical workflow to build trust.
- Standardize annotation and reporting: We recommend establishing consensus guidelines for nodule labelling (e.g., spatial overlap thresholds), leveraging semi-automated tools to reduce inter-observer variability, and adopting a reporting standard like DECIDE_AI for transparency.
6. Conclusions
- Limited generalizability due to dataset heterogeneity: While public datasets, such as LIDC-IDRI and LUNA16, dominate deep learning research (used in 45% and 20% of the studies, respectively), they lack diversity in terms of demographics, imaging protocols, and scanner specifications. For instance, LUNA16 primarily includes Western populations, with limited representation of Asian or African cohorts, potentially biasing models towards specific ethnic groups. Furthermore, only 155 studies validated their models on multicenter datasets, which can lead to overfitting. For example, models trained on LIDC-IDRI achieved 98% accuracy, but this dropped to 76% when tested on private datasets, such as those from Walter Cantidio Hospital and UFC Brazil, due to differences in slice thickness and contrast protocols. This heterogeneity undermines model robustness in real-world settings, where CT scanners and patient populations exhibit significant variations.
- Lack of standardization in annotation practices: Annotation variability, including inter-radiologist disagreement and differences between manual and automated labeling, introduces systematic bias. For example, in the LIDC-IDRI dataset, nodule boundaries annotated by four radiologists exhibited a 20–30% variance in spatial overlap metrics, with Dice scores ranging from 0.65 to 0.85. Such inconsistencies propagate into model training, as seen in segmentation studies, where U-Net variants achieved a 92% Dice coefficient on the LIDC-IDRI dataset but only 78% on the Decathlon datasets, due to divergent annotation criteria. Additionally, fewer than 10% of studies disclosed annotation guidelines, complicating reproducibility.
- Neglect of cancer staging and subtype classifications: While binary classification (benign vs. malignant) dominates deep learning research (60% of studies), staging and subtype differentiation (e.g., adenocarcinoma vs. squamous cell carcinoma) remain understudied. Only 5% of the reviewed works addressed NSCLC subtyping, despite its clinical relevance for personalized therapy. For instance, Huang et al. (2022) achieved 94.06% AUC for malignancy detection and classification [102] but did not predict stages (I–IV) or metastatic potential. This gap limits clinical utility, as treatment plans rely heavily on stage-specific protocols.
- Lack of transparency and explainability: The black box nature of deep learning models, particularly vision transformers (ViTs), erodes clinician trust. Only 8% of studies incorporated explainable techniques, such as GRAD-CAM [113], LIME or SHAP [72]. For example, Gai et al. (2024) [54] reported that ViTs outperform CNNs in capturing global context, but they offered no visual explanations for nodule localization. In contrast, the CNN-based model provided interpretable feature maps but lacked ViT’s long-range dependency modelling. Bridging this gap is critical for clinical adoption, as radiologists require transparent decision pathways to validate AI outputs.
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Shatnawi, M.Q.; Abuein, Q.; Al-Quraan, R. Deep learning-based approach to diagnose lung cancer using CT-scan images. Intell. Based Med. 2025, 11, 10188. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Wolf, A.M.D.; Oeffinger, K.C.; Shih, T.Y.; Walter, L.C.; Church, T.R.; Fontham, E.T.H.; Elkin, E.B.; Etzioni, R.D.; Guerra, C.E.; Perkins, R.B.; et al. Screening for lung cancer: 2023 guideline update from the American Cancer Society. CA Cancer J. Clin. 2024, 74, 50–81. [Google Scholar] [CrossRef]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584. [Google Scholar] [CrossRef]
- Monkam, P.; Qi, S.; Ma, H.; Gao, W.; Yao, Y.; Qian, W. Detection and Classification of Pulmonary Nodules Using Convolutional Neural Networks: A Survey. IEEE Access 2019, 7, 78075–78091. [Google Scholar] [CrossRef]
- Chen, S.; Cao, Z.; Prettner, K.; Kuhn, M.; Yang, J.; Jiao, L.; Wang, Z.; Li, W.; Geldsetzer, P.; Bärnighausen, T.; et al. Estimates and Projections of the Global Economic Cost of 29 Cancers in 204 Countries and Territories from 2020 to 2050. JAMA Oncol. 2023, 9, 465–472. [Google Scholar] [CrossRef]
- Erefai, O.; Soulaymani, A.; Mokhtari, A.; Obtel, M.; Hami, H. Diagnostic delay in lung cancer in Morocco: A 4-year retrospective study. Clin. Epidemiol. Glob. Health 2022, 16, 101105. [Google Scholar] [CrossRef]
- Ambrosini, V.; Nicolini, S.; Caroli, P.; Nanni, C.; Massaro, A.; Marzola, M.C.; Rubello, D.; Fanti, S. PET/CT imaging in different types of lung cancer: An overview. Eur. J. Radiology. 2012, 81, 988–1001. [Google Scholar] [CrossRef]
- Mahmud, S.H.; Soesanti, I.; Hartanto, R. Deep Learning Techniques for Lung Cancer Detection: A Systematic Literature Review. In Proceedings of the 2023 6th International Conference on Information and Communications Technology, ICOIACT 2023, Yogyakarta, Indonesia, 10 November 2023; pp. 200–205. [Google Scholar] [CrossRef]
- Kvale, P.A.; Johnson, C.C.; Tammemägi, M.; Marcus, P.M.; Zylak, C.J.; Spizarny, D.L.; Hocking, W.; Oken, M.; Commins, J.; Ragard, L.; et al. Interval lung cancers not detected on screening chest X-rays: How are they different? Lung Cancer 2014, 86, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Pereira, T.; Neves, I.; Morgado, J.; Freitas, C.; Malafaia, M.; Sousa, J.; Fonseca, J.; Negrão, E.; de Lima, B.F.; et al. Towards Machine Learning-Aided Lung Cancer Clinical Routines: Approaches and Open Challenges. J. Pers. Med. 2022, 12, 480. [Google Scholar] [CrossRef] [PubMed]
- Brisbane, W.; Bailey, M.R.; Sorensen, M.D. An overview of kidney stone imaging techniques. Nat. Rev. Urol. 2016, 13, 654–662. [Google Scholar] [CrossRef]
- Journy, N.; Rehel, J.-L.; Le Pointe, H.D.; Lee, C.; Brisse, H.; Chateil, J.-F.; Caer-Lorho, S.; Laurier, D.; Bernier, M.-O. Are the studies on cancer risk from CT scans biased by indication? Elements of answer from a large-scale cohort study in France. Br. J. Cancer 2015, 112, 185–193. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, H.; Yang, J.; Wu, J.; Yin, X.; Chen, Y.; Shu, H.; Luo, L.; Coatrieux, G.; Gui, Z.; et al. Improving Low-Dose CT Image Using Residual Convolutional Network. IEEE Access 2017, 5, 24698–24705. [Google Scholar] [CrossRef]
- Li, R.; Xiao, C.; Huang, Y.; Hassan, H.; Huang, B. Deep Learning Applications in Computed Tomography Images for Pulmonary Nodule Detection and Diagnosis: A Review. Diagnostics 2022, 12, 298. [Google Scholar] [CrossRef]
- Forte, G.C.; Altmayer, S.; Silva, R.F.; Stefani, M.T.; Libermann, L.L.; Cavion, C.C.; Youssef, A.; Forghani, R.; King, J.; Mohamed, T.-L.; et al. Deep Learning Algorithms for Diagnosis of Lung Cancer: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 3856. [Google Scholar] [CrossRef]
- Hosseini, S.H.; Monsefi, R.; Shadroo, S. Deep learning applications for lung cancer diagnosis: A systematic review. Multimed. Tools Appl. 2024, 83, 14305–14335. [Google Scholar] [CrossRef]
- Dodia, S.; Annappa, B.; Mahesh, P.A. Recent advancements in deep learning-based lung cancer detection: A systematic review. Eng. Appl. Artificial. Intell. 2022, 116, 105490. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, 71. [Google Scholar] [CrossRef]
- Giger, M.L. Machine learning in medical imaging. J. Am. Coll. Radiol. 2018, 15, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.-C.; Lou, S.-L.; Lin, J.-S.; Freedman, M.; Chien, M.; Mun, S. Artificial Convolution Neural Network Techniques and Applications for Lung Nodule Detection. IEEE Trans. Med. Imaging 1995, 14, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- SPan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Jian, S. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; Available online: http://openaccess.thecvf.com/content_cvpr_2016/html/He_Deep_Residual_Learning_CVPR_2016_paper.html (accessed on 20 September 2024).
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Rabea, S.; Bhattacharjee, A.; Elkaeed, E.B.; Murugan, R.; Selim, H.M.R.M.; Sahu, R.K.; Shazly, G.A.; Bekhit, M.M.S. A multi-class deep learning model for early lung cancer and chronic kidney disease detection using computed tomography images. Front. Oncol. 2023, 13, 1193746. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 1520170387. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Xia, K.; Wang, J. Recent advances of Transformers in medical image analysis: A comprehensive review. MedComm–Future Med. 2023, 2, e38. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Gu, Y.; Chi, J.; Liu, J.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Lu, X. A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning. Comput. Biol. Med. 2021, 137, 104806. [Google Scholar] [CrossRef] [PubMed]
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Special Section on Soft Computing Techniques for Image Analysis in the Medical Industry Current Trends, Challenges and Solutions Deep Learning Applications in Medical Image Analysis. IEEE Access 2017, 6, 9375–9389. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Xu, W.; Fu, Y.L.; Zhu, D. ResNet and its application to medical image processing: Research progress and challenges. Comput. Methods Programs Biomed. 2023, 240, 107660. [Google Scholar] [CrossRef]
- Durga Bhavani, K.; Ferni Ukrit, M. Design of inception with deep convolutional neural network based fall detection and classification model. Multimed. Tools Appl. 2024, 83, 23799–23817. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Zhao, X.; Wang, L.; Zhang, Y.; Han, X.; Deveci, M.; Parmar, M. A review of convolutional neural networks in computer vision. Artif. Intell. Rev. 2024, 57, 99. [Google Scholar] [CrossRef]
- Naseer, I.; Akram, S.; Masood, T.; Jaffar, A.; Khan, M.A.; Mosavi, A. Performance Analysis of State-of-the-Art CNN Architectures for LUNA16. Sensors 2022, 22, 4426. [Google Scholar] [CrossRef]
- Pang, S.; Meng, F.; Wang, X.; Wang, J.; Song, T.; Wang, X.; Cheng, X. VGG16-T: A novel deep convolutional neural network with boosting to identify pathological type of lung cancer in early stage by CT images. Int. J. Comput. Intell. Syst. 2020, 13, 771–780. [Google Scholar] [CrossRef]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, NI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Lakshmana Prabu, S.K.; Mohanty, S.N.; Shankar, K.; Arunkumar, N.; Ramirez, G. Optimal deep learning model for classification of lung cancer on CT images. Future Gener. Comput. Syst. 2019, 92, 374–382. [Google Scholar] [CrossRef]
- Teramoto, A.; Fujita, H. Fast lung nodule detection in chest CT images using cylindrical nodule-enhancement filter. Int. J. Comput. Assist. Radiol. Surg. 2013, 8, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.E.; Dua, D. Lung Nodule Detection via Optimized Convolutional Neural Network: Impact of Improved Moth Flame Algorithm. Sens. Imaging 2023, 24, 11. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Annual Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Chitty-Venkata, K.T.; Mittal, S.; Emani, M.; Vishwanath, V.; Somani, A.K. A survey of techniques for optimizing transformer inference. Med. Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef]
- Gai, L.; Xing, M.; Chen, W.; Zhang, Y.; Qiao, X. Comparing CNN-based and transformer-based models for identifying lung cancer: Which is more effective? Multimed. Tools Appl. 2024, 83, 59253–59269. [Google Scholar] [CrossRef]
- Zhu, W.; Liu, C.; Fan, W.; Xie, X. Deep Lung: Deep 3D dual path nets for automated pulmonary nodule detection and classification. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision, WACV 2018, Lake Tahoe, NV, USA, 12–15 March 2018; pp. 673–681. [Google Scholar] [CrossRef]
- Gu, Y.; Lu, X.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Gao, L.; Wu, L.; Zhou, T. Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs. Comput. Biol. Med. 2018, 103, 220–231. [Google Scholar] [CrossRef]
- Shelhamer, E.; Long, J.; Darrell, T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef]
- Liu, L.; Fan, K.; Yang, M. Federated learning: A deep learning model based on ResNet18 dual path for lung nodule detection. Multimed. Tools Appl. 2023, 82, 17437–17450. [Google Scholar] [CrossRef]
- Fu, B.; Peng, Y.; He, J.; Tian, C.; Sun, X.; Wang, R. HmsU-Net: A hybrid multi-scale U-net based on a CNN and transformer for medical image segmentation. Comput. Biol. Med. 2024, 170, 108013. [Google Scholar] [CrossRef]
- Jiang, H.; Ma, H.; Qian, W.; Gao, M.; Li, Y. An Automatic Detection System of Lung Nodule Based on Multigroup Patch-Based Deep Learning Network. IEEE J. Biomed. Health Inform. 2018, 22, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Riaz, Z.; Khan, B.; Abdullah, S.; Khan, S.; Islam, S. Lung Tumor Image Segmentation from Computer Tomography Images Using MobileNetV2 and Transfer Learning. Bioengineering 2023, 10, 981. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Fan, X.; Wang, J.; Chen, S.; Meng, J. ParaU-Net: An improved UNet parallel coding network for lung nodule segmentation. J. King Saud. Univ.—Comput. Inf. Sci. 2024, 36, 102203. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Shen, H.; Chen, L.; Liu, K.; Zhao, K.; Li, J.; Yu, L.; Ye, H.; Zhu, W. A subregion-based positron emission tomography/computed tomography (PET/CT) radiomics model for the classification of non-small cell lung cancer histopathological subtypes. Quant. Imaging Med. Surg. 2021, 11, 2918. [Google Scholar] [CrossRef]
- Nasrullah, N.; Sang, J.; Alam, M.S.; Mateen, M.; Cai, B.; Hu, H. Automated lung nodule detection and classification using deep learning combined with multiple strategies. Sensors 2019, 19, 3722. [Google Scholar] [CrossRef]
- Chang, H.-H.; Wu, C.-Z.; Gallogly, A.H. Pulmonary Nodule Classification Using a Multiview Residual Selective Kernel Network. J. Imaging Inform. Med. 2024, 37, 347–362. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, Z.; Gong, L.; Jiang, S.; Wang, L.; Zhang, H. Classification of lung nodules based on CT images using squeeze-and-excitation network and aggregated residual transformations. Radiol. Med. 2020, 125, 374–383. [Google Scholar] [CrossRef]
- Shen, W.; Zhou, M.; Yang, F.; Yu, D.; Dong, D.; Yang, C.; Zang, Y.; Tian, J. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. 2017, 61, 663–673. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, S.; Zhang, B.; Ma, H.; Qian, W.; Yao, Y.; Sun, J. Deep CNN models for pulmonary nodule classification: Model modification, model integration, and transfer learning. J. X-Ray Sci. Technol. 2019, 27, 615–629. [Google Scholar] [CrossRef]
- Huidrom, R.; Chanu, Y.J.; Singh, K.M. Neuro-evolutional based computer aided detection system on computed tomography for the early detection of lung cancer. Multimed. Tools Appl. 2022, 81, 32661–32673. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.; Bogaard, C.D.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wani, N.A.; Kumar, R.; Bedi, J. DeepXplainer: An interpretable deep learning-based approach for lung cancer detection using explainable artificial intelligence. Comput. Methods Programs Biomed. 2024, 243, 107879. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H. LungSeek: 3D Selective Kernel residual network for pulmonary nodule diagnosis. Vis. Comput. 2023, 39, 679–692. [Google Scholar] [CrossRef]
- Chen, Y.; Hou, X.; Yang, Y.; Ge, Q.; Zhou, Y.; Nie, S. A Novel Deep Learning Model Based on Multi-Scale and Multi-View for Detection of Pulmonary Nodules. J. Digit. Imaging 2023, 36, 688–699. [Google Scholar] [CrossRef]
- Cao, H.; Liu, H.; Song, E.; Ma, G.; Xu, X.; Jin, R.; Liu, T.; Hung, C.-C. Multi-Branch Ensemble Learning Architecture Based on 3D CNN for False Positive Reduction in Lung Nodule Detection. IEEE Access 2019, 7, 67380–67391. [Google Scholar] [CrossRef]
- Xie, H.; Yang, D.; Sun, N.; Chen, Z.; Zhang, Y. Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognit. 2019, 85, 109–119. [Google Scholar] [CrossRef]
- Shakeel, P.M.; Burhanuddin, M.; Desa, M.I. Lung cancer detection from CT image using improved profuse clustering and deep learning instantaneously trained neural networks. Measurement 2019, 145, 702–712. [Google Scholar] [CrossRef]
- Ozdemir, O.; Russell, R.L.; Berlin, A.A. A 3D Probabilistic Deep Learning System for Detection and Diagnosis of Lung Cancer Using Low-Dose CT scans. IEEE Trans. Med. Imaging 2020, 39, 1419–1429. [Google Scholar] [CrossRef]
- Masood, A.; Yang, P.; Sheng, B.; Li, H.; Li, P.; Qin, J.; Lanfranchi, V.; Kim, J.; Feng, D.D. Cloud-Based Automated Clinical Decision Support System for Detection and Diagnosis of Lung Cancer in Chest CT. IEEE J. Transl. Eng. Health Med. 2019, 8, 1–13. [Google Scholar] [CrossRef]
- Su, Y.; Li, D.; Chen, X. Lung Nodule Detection based on Faster R-CNN Framework. Comput. Methods Programs Biomed. 2021, 200, 105866. [Google Scholar] [CrossRef] [PubMed]
- Majidpourkhoei, R.; Alilou, M.; Majidzadeh, K.; Babazadehsangar, A. A novel deep learning framework for lung nodule detection in 3d CT images. Multimed. Tools Appl. 2021, 80, 30539–30555. [Google Scholar] [CrossRef]
- Dutande, P.; Baid, U.; Talbar, S. LNCDS: A 2D-3D cascaded CNN approach for lung nodule classification, detection and segmentation. Biomed. Signal Process Control 2021, 67, 102527. [Google Scholar] [CrossRef]
- Naseer, I.; Masood, T.; Akram, S.; Jaffar, A.; Rashid, M.; Iqbal, M.A. Lung Cancer Detection Using Modified AlexNet Architecture and Support Vector Machine. Comput. Mater. Contin. 2023, 74, 2039–2054. [Google Scholar] [CrossRef]
- Saha, A.; Ganie, S.M.; Pramanik, P.K.D.; Yadav, R.K.; Mallik, S.; Zhao, Z. VER-Net: A hybrid transfer learning model for lung cancer detection using CT scan images. BMC Med. Imaging 2024, 24, 120. [Google Scholar] [CrossRef]
- Hu, Q.; Souza, L.F.d.F.; Holanda, G.B.; Alves, S.S.; Silva, F.H.d.S.; Han, T.; Filho, P.P.R. An effective approach for CT lung segmentation using mask region-based convolutional neural networks. Artif. Intell. Med. 2020, 103, 101792. [Google Scholar] [CrossRef]
- Song, J.; Yang, C.; Fan, L.; Wang, K.; Yang, F.; Liu, S.; Tian, J. Lung lesion extraction using a toboggan based growing automatic segmentation approach. IEEE Trans. Med. Imaging 2016, 35, 337–353. [Google Scholar] [CrossRef]
- Xu, M.; Qi, S.; Yue, Y.; Teng, Y.; Xu, L.; Yao, Y.; Qian, W. Segmentation of lung parenchyma in CT images using CNN trained with the clustering algorithm generated dataset 08 Information and Computing Sciences 0801 Artificial Intelligence and Image Processing Robert Koprowski. Biomed. Eng. Online 2019, 18, 2. [Google Scholar] [CrossRef]
- Tyagi, S.; Talbar, S.N. CSE-GAN: A 3D conditional generative adversarial network with concurrent squeeze-and-excitation blocks for lung nodule segmentation. Comput. Biol. Med. 2022, 147, 105781. [Google Scholar] [CrossRef]
- Najeeb, S.; Bhuiyan, M.I.H. Spatial feature fusion in 3D convolutional autoencoders for lung tumor segmentation from 3D CT images. Biomed. Signal Process Control 2022, 78, 103996. [Google Scholar] [CrossRef]
- Said, Y.; Alsheikhy, A.A.; Shawly, T.; Lahza, H. Medical Images Segmentation for Lung Cancer Diagnosis Based on Deep Learning Architectures. Diagnostics 2023, 13, 546. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, J.; Xia, Y. Semi-supervised adversarial model for benign–malignant lung nodule classification on chest CT. Med. Image Anal. 2019, 57, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Muzammil, M.; Haq, I.U.; Amir, M.; Abdullah, S.; Khaliq, A.A. Efficient lung nodule classification using transferable texture convolutional neural network. IEEE Access 2020, 8, 175859–175870. [Google Scholar] [CrossRef]
- Musthafa, M.M.; Manimozhi, I.; Mahesh, T.R.; Guluwadi, S. Optimizing double-layered convolutional neural networks for efficient lung cancer classification through hyperparameter optimization and advanced image pre-processing techniques. BMC Med. Inform. Decis. Mak. 2024, 24, 142. [Google Scholar] [CrossRef]
- Song, Q.; Zhao, L.; Luo, X.; Dou, X. Using Deep Learning for Classification of Lung Nodules on Computed Tomography Images. J. Health Eng. 2017, 2017, 8314740. [Google Scholar] [CrossRef]
- Liu, Y.; Hao, P.; Zhang, P.; Xu, X.; Wu, J.; Chen, W. Dense Convolutional Binary-Tree Networks for Lung Nodule Classification. IEEE Access 2018, 6, 49080–49088. [Google Scholar] [CrossRef]
- Liu, L.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.-A. Multi-Task Deep Model with Margin Ranking Loss for Lung Nodule Analysis. IEEE Trans. Med. Imaging 2020, 39, 718–728. [Google Scholar] [CrossRef]
- Asuntha, A.; Srinivasan, A. Deep learning for lung Cancer detection and classification. Multimed. Tools Appl. 2020, 79, 7731–7762. [Google Scholar] [CrossRef]
- Monkam, P.; Qi, S.; Xu, M.; Han, F.; Zhao, X.; Qian, W. CNN models discriminating between pulmonary micro-nodules and non-nodules from CT images. Biomed. Eng. Online 2018, 17, 96. [Google Scholar] [CrossRef]
- Polat, H.; Danaei Mehr, H. Classification of pulmonary CT images by using hybrid 3D-deep convolutional neural network architecture. Appl. Sci. 2019, 9, 940. [Google Scholar] [CrossRef]
- Jena, S.R.; George, S.T.; Ponraj, D.N. Lung cancer detection and classification with DGMM-RBCNN technique. Neural Comput. Appl. 2021, 33, 15601–15617. [Google Scholar] [CrossRef]
- Mastouri, R.; Khlifa, N.; Neji, H.; Hantous-Zannad, S. A bilinear convolutional neural network for lung nodules classification on CT images. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Li, Y.; Wu, R.; Li, Z.; Zhang, J. Benign-malignant classification of pulmonary nodule with deep feature optimization framework. Biomed. Signal Process Control 2022, 76, 103701. [Google Scholar] [CrossRef]
- Sakshiwala; Singh, M.P. A new framework for multi-scale CNN-based malignancy classification of pulmonary lung nodules. J. Ambient Intell. Humaniz. Comput. 2023, 14, 4675–4683. [Google Scholar] [CrossRef]
- Huang, H.; Wu, R.; Li, Y.; Peng, C. Self-Supervised Transfer Learning Based on Domain Adaptation for Benign-Malignant Lung Nodule Classification on Thoracic CT. IEEE J. Biomed. Health Inform. 2022, 26, 3860–3871. [Google Scholar] [CrossRef]
- Mahmood, S.A.; Ahmed, H.A. An improved CNN-based architecture for automatic lung nodule classification. Med. Biol. Eng. Comput. 2022, 60, 1977–1986. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, H.; Ding, L.; Yang, K. A diagnostic classification of lung nodules using multiple-scale residual network. Sci. Rep. 2023, 13, 11322. [Google Scholar] [CrossRef]
- Pandit, B.R.; Alsadoon, A.; Prasad, P.W.C.; Al Aloussi, S.; Rashid, T.A.; Alsadoon, O.H.; Jerew, O.D. Deep learning neural network for lung cancer classification: Enhanced optimization function. Multimed. Tools Appl. 2023, 82, 6605–6624. [Google Scholar] [CrossRef]
- Lima, T.; Luz, D.; Oseas, A.; Veras, R.; Araújo, F. Automatic classification of pulmonary nodules in computed tomography images using pre-trained networks and bag of features. Multimed. Tools Appl. 2023, 82, 42977–42993. [Google Scholar] [CrossRef]
- Bushara, A.R.; Vinod Kumar, R.S.; Kumar, S.S. LCD-Capsule Network for the Detection and Classification of Lung Cancer on Computed Tomography Images. Multimed. Tools Appl. 2023, 82, 37573–37592. [Google Scholar] [CrossRef]
- Naseer, I.; Akram, S.; Masood, T.; Rashid, M.; Jaffar, A. Lung Cancer Classification Using Modified U-Net Based Lobe Segmentation and Nodule Detection. IEEE Access 2023, 11, 60279–60291. [Google Scholar] [CrossRef]
- Alazwari, S.; Alsamri, J.; Asiri, M.M.; Maashi, M.; Asklany, S.A.; Mahmud, A. Computer-aided diagnosis for lung cancer using waterwheel plant algorithm with deep learning. Sci. Rep. 2024, 14, 20647. [Google Scholar] [CrossRef] [PubMed]
- Esha, J.F.; Islam, T.; Pranto, A.M.; Borno, A.S.; Faruqui, N.; Abu Yousuf, M.; Azad, A.; Al-Moisheer, A.S.; Alotaibi, N.; Alyami, S.A.; et al. Multi-View Soft Attention-Based Model for the Classification of Lung Cancer-Associated Disabilities. Diagnostics 2024, 14, 2282. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, S.Y.; Jeya, J.J.; Khan, S.B.; Alzahrani, S.; Alojail, M. Explainable lung cancer classification with ensemble transfer learning of VGG16, Resnet50 and InceptionV3 using grad-cam. BMC Med. Imaging 2024, 24, 176. [Google Scholar] [CrossRef]
- Prasad, U.; Chakravarty, S.; Mahto, G. Lung cancer detection and classification using deep neural network based on hybrid metaheuristic algorithm. Soft Comput. 2024, 28, 8579–8602. [Google Scholar] [CrossRef]

| DL Types | Brief Description | Basic Mode |
|---|---|---|
| Convolutional neural networks (CNNs) | A feedforward network utilizes convolutional and pooling layers to extract spatial features from images. | The architecture consists of input/output layers, hidden layers, and an activation function (e.g., ReLU), which are optimized for classification and segmentation tasks. |
| Fully connected neural networks (FCNNs) | FCNNs, also known as dense neural networks (DNNs), connect every neuron in one layer to every neuron in the next layer. They excel at learning complex patterns from structured data. FCNNs are widely used in classification, regression, and feature learning tasks. | A standard FCNN consists of an input layer, a hidden layer, and an output layer. The input layer receives raw features (e.g., pixel values in an image), the hidden layers compute weighted sums followed by a nonlinear activation function (e.g., ReLU, Sigmoid, or Tanh), and the Output layer produces the final prediction. |
| Deep belief network (DBN) | The probability generation model comprises multiple hidden layers, each constructed using several restricted Boltzmann machines (RBMs). | Multilayer RBM and backpropagation (BP). |
| Recurrent neural network (RNN) | The neural network of short-term memory is designed for sequential data. | It consists of an input layer, a recurrent layer, one or more hidden layers, and an output layer. |
| Autoencoders (AE) | AEs use an encoder–decoder structure to extract and represent features from high-dimensional data through unsupervised learning. | It uses an encoder–decoder. |
| Long Short-Term Memory (LSTM) | It is an RNN that can learn long dependencies, thereby addressing the limitation of short-term memory by incorporating gates that manage long-range dependencies. | Consists of a cell with an input and an output gate. |
| Deep Boltzmann Machine (DBM) | This is a stack of multilayer RBMs, with bidirectional middle layers connected to their adjacent layers, forming a probabilistic generative model. | It consists of multiple layers of Restricted Boltzmann machine (RBMs) stacked together. DBM has an undirected (bi-directional) connection between all adjacent layers. |
| Vision transformers (ViTs) | A model that uses feature extraction based on an attention mechanism to capture long-range dependencies in image data. Consists of a multi-headed attention mechanism. They process images by splitting them into patches, embedding them, and applying transformer encoder layers. | Suitable for long-range dependencies, it often requires position encoding to handle sequence information. Consists of four key stages: Image patching and embedding, positional encoding, transformer encoder, classification head, and multi-layer perceptron (MLP) head. |
| Methods/Approach | Convolutional Neural Networks (CNNs) | Vision Transformers (VITs) |
|---|---|---|
| Core mechanism | Use convolutional filters to extract features. | Utilize a self-attention mechanism to capture global contextual relationships. |
| Scope | CNNs excel at capturing detailed image features using convolutional, pooling, and fully connected layers. Their efficiency makes them well-suited for medical analysis. However, we have a limited understanding of the global context. | ViTs are relatively new in medical imaging but have demonstrated strong potential in capturing global spatial relationships and long-range dependencies. However, they require large datasets and many computational resources. |
| Interpretability | CNNs offer better interpretability by visualizing learned features and activation maps, helping physicians understand model decisions. | ViTs, especially when scaled, can be less interpretable than CNNs because of the complexity of their attention-learned mechanisms. |
| Scalability/computational complexity | CNNs are highly optimized for various computer vision tasks. They benefit from parallel processing and efficient convolutional operation, making them scalable for large datasets and complex image domains. | The self-attention mechanism requires significant computational power and memory due to its complexity. |
| Availability of pre-trained models | Pretrained CNN models (such as ResNet and VGG) trained on large-scale datasets help reduce the need for extensive training data. | ViTs can leverage pretraining on large-scale datasets (e.g., ImageNet) for feature extraction and fine-tuning with relatively less data. |
| Global context | Primarily focus on local features within images. | Capture long-range dependencies and global contextual information. |
| Translational invariance | Possesses translation invariance, insensitive to position. No need for additional encoding. | Lack of translation invariance, sensitive to position. It often requires position encoding to handle sequence information. |
| Database | Articles |
|---|---|
| Scopus | 764 |
| IEEE | 410 |
| Springer | 95 |
| PubMed | 76 |
| MDPI | 47 |
| Others | 56 |
| Criteria | Inclusion | Exclusion | Justification |
|---|---|---|---|
| Language | English | Non-English | We include only English-language articles to ensure global accessibility, as well as due to time constraints in translating non-English publications. |
| Publication year | 2015 to 2024 | Before 2015 | Focus on recent DL advancements. |
| Study focus | DL models (CNNs, ViTs) for lung cancer CT tasks | Non-DL methods or non-lung cancer studies | Aligning with research objectives. |
| Publication type | Peer-reviewed articles or conference papers | Books, technical reports, non-empirical studies | Ensure academic rigour and reproducibility. |
| Data availability | Papers with the full text accessible | Abstracts only | Facilitates detailed analysis of methodologies and results. |
| References | Database | Architecture | Methodology | Validation | Performance Metrics (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Acc. | Sen | Spe | FROC | FP | CPM | |||||
| 2023 [58] | LUNA16 | 3D CNN | Preprocessing, augmentation, federated learning, 3D ResNet18, dual-path Faster R-CNN | N/A | 76.49, 83.41 | 78.44, 83.38 | N/A | N/A | N/A | N/A |
| 2022 [70] | LIDC-IDRI | CNN | Lung segmentation, nodule candidate detection, feature detection and extraction, nodule classification using PSO. | N/A | 95.52 | 95.75 | 95.29 | N/A | N/A | N/A |
| 2017 [71] | LUNA16 | MV-CNN | Preprocessing, data augmentation, fusion | 10-fold | N/A | 95 | N/A | N/A | 1.0 | 0.90 |
| 2024 [72] | Survey lung cancer (SLC) | CNN, | Preprocessing, label encoding, data sampling, CNN with XGBoost, (ConvXGB) deep Xplainer | K-Fold C.V | 97.43 | 98.71 | N/A | N/A | N/A | N/A |
| 2023 [73] | LUNA16 | 3D-CNN | Preprocessing, Lungseek-based 3D selective kernel residual network (SK-ResNet) framework, 3D Res18 and 3D DPN, implement 3D Res 18 with faster R-CNN | N/A | 91.75 | 95.78 | N/A | 89.48 | 25.51 | 89.13 |
| 2023 [74] | LIDC-IDRI | CNN, | Preprocessing, F-Net assisted by 3D, for feature extraction, proposed MSS-Net for detection, MSF, and MVRF for lung nodule fusion | K-Fold C. V | N/A | 98.40 | N/A | N/A | 8.91 | 95.7 |
| 2019 [75] | LUNA16/LIDC-IDRI | 3D-CNN | MBEL-3D-CNN, Multi-branch DenseNet (3DMB-DenseNet), Inception, ResNet, (3DMB-IresNet), 3DMB-VggNet | 10-fold | N/A | N/A | N/A | N/A | 0.5, 0.7 | 87.3 |
| 2019 [76] | LUNA16 | 2D-CNN, | Pre-screening to deal with class imbalance, down-sampling, and adopting augmentation, false positive reduction for the first stage, nodule candidate detection using VGG16, and an improved faster R-CNN | 5-fold | N/A | 86.4 | N/A | 0.954 | 4.67 | 79.0 |
| 2019 [77] | The Cancer Imaging Archive (TCIA) dataset | CNN | Preprocessing, segmentation using improved Profuse clustering (IPCT) feature extraction classification with a deep learning instantaneously trained neural network (DITNN) | N/A | 98.42 | 94 | 97.2 | 96.8 | N/A | N/A |
| 2020 [78] | LUNA16, Kaggle 17 | 3D-CNN | Preprocessing, augmentation, segmentation for candidate extraction, detection, and classification with CADe-CADx. | 10-fold | N/A | 96.5 | N/A | N/A | 19.7 | N/A |
| 2019 [79] | LUNA16, ANODE09, LIDC-IDRI | 3D-Deep CNN | Preprocessing, data augmentation, multi-scale, multi-view, multi-region Proposal network (mRPN) | 10-fold C. V | 98.4 | 98.7 | 94 | 97.43 | 2.1 | N/A |
| 2021 [80] | LIDC-IDRI | CNN (Faster R-CNN) | Data enhancement (flipping, colour transformation), data annotation, network training using Faster R-CNN, feature extraction uses ZF model and Vgg16. | N/A | 91.2% | N/A | N/A | N/A | N/A | N/A |
| 2021 [81] | LIDC-IDRI | CNN | Preprocessing, segmentation, and object detection feature extraction based on a CNN algorithm with a developed model, which is a lightweight model based on LeNet-5 model | 5-fold C. V | 90.1 | 84.1 | 91.7 | N/A | N/A | N/A |
| 2021 [82] | LIDC (LNDb challenges | CNN | In the preprocessing stage, introduce SquExUNet as the segmentation model,3D-NodNet classification, and a 2D-3D cascade CNN, for nodule detection. | K-fold C. V | N/A | 90.0 | N/A | N/A | N/A | N/A |
| 2023 [83] | LUNA16 | CNN + SVM | Preprocessing, modified AlexNet Lung Net-SVM | K-Fold | 97.64 | 96.37 | 99.08 | N/A | N/A | N/A |
| 2024 [84] | Kaggle | CNN | Preprocessing, data augmentation, NasNetlarge, Xception, DenseNet 201, Mobile Net, ResNet101, EfficientNetB0, EfficientNetB4, VGG19, VER-Net | K-fold C. V | 91 | N/A | N/A | N/A | N/A | N/A |
| Reference | Database | Images Type | Main Method | Performance Metrics % |
|---|---|---|---|---|
| 2024 [62] | LIDC | CT | Proposed multi-scale parallel encoding module (MPEM) and original UNet encoder (ParaU-Net), with coordinate feature fusion module (CFFM) to integrate the feature output of the primary auxiliary encoder from different layers, which helps to capture detailed information on global context. | IoU: 87.15 Dice: 92.16 F1-score: 92.24 F2-score: 92.33 F0.5 score: 92.69 |
| 2020 [85] | Walter cantidio hospital university caera (UFC) Brazil | CT | Lung region segmentation with Mask R-CNN combined with SVM, K-means, and Gaussian Mixture Models (GMMs). | Acc: 97.11 Sen: 96.58 Spe 92.18 DC: 97.33 |
| 2016 [86] | LIDC-IDRI | CT | Apply the toboggan-based growing automatic segmentation (TBGA) approach, which consists of three phases: seed election, lesion extraction, and lesion refinement. Conduct 2D seed point location, 3D lesion segmentation, and lesion delineation. | Sen: 96.35 FP: 9.1 Acc: p > 0.05 |
| 2019 [87] | 201 subjects with (4.62 billion patches) of lung cancer | CT | Proposed lung parenchyma segmentation using k-means clustering and cross-shaped verification on two categories to generate the final dataset. CNN architecture to train and optimize the performance. | Acc: 99.08 Sensitivity: 98.8 Spe: 99.5 F1-score: 99.17 AUC: 99.91 DSC: 96.80 |
| 2022 [88] | LUNA16 | CT | Preprocessing, proposed 3D conditional generative adversarial network (CGAN) based on U-Net, with concurrent Squeez excitation (sSCE) and channel excitation module (CSE-GAN). | Dice: 80.74 Sen 85.46 Pre 80.56 Jaccard index 72.52 |
| 2022 [89] | Lung-originated tumor segmentation (LOTUS) dataset | CT | Preprocessing, data augmentation, proposed spatial feature learning at different 2D convolutional autoencoders to create a 3D segmentation network using 3D-Unet, 3D-multi-ResNet, and Recurrent 3D-DenseNet Network. | Dice: 0.866 F1-scorer 0.719 |
| 2023 [90] | LIDC-IDRI and decathlon dataset | CT | The proposed cancer segmentation was based on a combination of U-Net and Transformer (UNETR) networks, utilising a 3D network that operates with 3D input CT data. The transformer, acting as an encoder, captures global, multi-scale information, which creates 3D input data; the experiment utilizes a decathlon dataset. | Acc: 97.83 Sen: 96.85 Spe: 97.12 Dice: 96.42 |
| Reference | Database | Modality | Model | Methods/Technique | Classification | Validation | Performance Indicator % |
|---|---|---|---|---|---|---|---|
| 2019 [65] | LUNA 16 and LIDC-IDRI | CT | 3DCNN | Proposed multi-strategy based nodule detection and classification, 3D Faster R-CNN with CmixNet and U-Net-like encoder-decoder for nodule detection, the detected nodules were analyzed through 3D-CMixNet + GBM to classify nodules. | Benign or malignant | 10-fold C. V | Acc 94.17 Sen. 94 Spe 91 |
| 2024 [66] | LIDC-IDRI | CT | 3DCNN | Preprocessing data augmentation, feature computation, Multiview-Residual Selective kernel (MRSKNet), feature computation. | Benign and malignant | 10-fold C. V | ACC 93.6 AUC 97.1 Recall 95.5 Spe 91.7 |
| 2019 [91] | LIDC-IDRI | CT | DCNNs | Semi-supervised adversarial classification (SSAC), adversarial autoencoder (AAE), proposed MV_KBC (MK-SSAC) model. | Benign and malignant | 10-foldC.V | ACC 92.53 AUC 95.81 Sen 84.94 Spe 96.28 |
| 2020 [92] | LIDC-IDRI and LungGx challenge | CT | CNN | Preprocessing, image augmentation, patch generation, batch normalization, and malignancy classification using a CNN model with different patch sizes. | Cancerous or normal | 6-fold C. V | Acc. 96.69, AUC 99.11, Spe 97.37, Recall 96.05 |
| 2024 [93] | Iraq, oncology teaching hospital, national centre for cancer disease (IQ-OTH/NCCD) | CT | CNN | Preprocessing, data augmentation, segmentation, Synthetic Minority Over-sampling Technique (SMOTE), CNN. | Benign, malignant and normal | K-Fold C.V | Acc 99.64, Pre. 96.77 Recall 99.04 F1-score 99.5 |
| 2017 [94] | LIDC-IDRI | CT | CNN | Preprocessing, data augmentation, and construction of three network architectures, CNN, DNN, and SAE | Benign and malignant | K-fold C. V | Acc. 84.15 Sen 83.96 Spe 84.32 |
| 2018 [95] | LIDC-IDRI | CT | CNN | Image preprocessing, augmentation, dense convolutional binary tree network (DenseBTNet). | Benign and malignant | 5-fold C. V | Acc. 88.31 AUC 93.35 |
| 2020 [96] | LIDC-IDRI | CT | CNN | Preprocessing, classification, multi-task deep model with Margin Rankin (MTMR-Net), regression t-SNE for nodule attribute score. | Benign or malignant | 5-fold C. V | Acc 93.5, Sen 93.0, Spe 89.4, AUC 0.97 |
| 2020 [97] | LIDC-IDRI | CT | CNN, | Preprocessing, segmentation using artificial bee colony (ABC), feature selection using fuzzy particle swarm optimization (FPSO), classifier using K-NN, Adaboost, SVM, ELM, and fuzzy particle swarm optimization convolution neural network (FPSOCNN). | Malignant or benign | k-fold C. V | Acc 95.62, Sen 97.93, Spe 96.32, |
| 2018 [98] | LIDC-IDRI | CT | CNN | 13,179, micro-nodules and 21, 315 non nodule patches were extracted with different patch sizes using a CNN. | Nodule and non-nodule | 5-fold C. V | Acc. 88.28, Auc. 0.87 Sen 83.82 F1-score 83.45 |
| 2019 [99] | Bowl and Kaggle dataset | CT | 3D-CNN | Proposed 3D-CNN, AlexNet for Lung CT classification, Hybrid 3D-CNN classifier-based RBF-(SVM). | Benign and malignant | K-fold C.V | Acc. 91.81 Sen 88.53 Pre 91.91 |
| 2021 [100] | LIDC-IDRI | CT | CNN | Preprocessing, segmentation using (MD-PRGS), feature extraction using a deep Gaussian mixture model in region-based CNN classification using (DGMM-RBCNN). | Benign and malignant | K-fold | Acc. 87.79, Pre. 89, Recall 70 F-measure 91 |
| 2021 [101] | LUNA16 | CT | CNN | Preprocessing, bilinear CNN (BCNN) with two streams (VGG16, VGG19) as feature extractors and SVM as a classifier for false positive reduction (FP). | Cancerous/non-cancerous | K-fold C.V | Acc 91.99, AUC 95.9, |
| 2022 [102] | LIDC-IDRI | CT | CNN | Preprocessing, deep feature optimization (DFOF), KNN, SVM, RF. | Benign or malignant | 5-fold C.V | Acc, 90.03, AUC, 94.06, Pre. 96.95, F1-score 93.38 |
| 2023 [103] | LIDC-IDRI | CT | 2D CNN | Data augmentation, multiscale CNN. | Benign or malignant | K-Fold C.V | Acc 93.88, Sen 93.36, Spe 93.26, AUC 93.31 |
| 2022 [104] | LIDC-IDRI | CT | 2D/3D CNN | Data preprocessing using adaptive slice selection (ASS), transfer learning with self-supervised transfer-based domain adaptation (SSTL-DA) 3DCNN. | Benign or malignant | 10-fold C.V | Acc 91.07, Sen 90.93 Spe 91.22 AUC 95.84 F1-score 91.0 |
| 2022 [105] | Kaggle–Bowl dataset | CT | CNN | Preprocessing, segmentation, normalization, and zero centering Proposed model using AlexNet. | Benign or malignant | N/A | Acc 98.77 Sen 98.64 Spe 98.90 |
| 2023 [106] | LIDC-IDRI | CT | CNN | Data preprocessing and splitting multi-scale residual network (MResNet) with pyramid pooling module (PPM) for fused features. | Benign or malignant | K-Fold C. V | Acc 99.12 Sen 98.64 Spe 97.87 AUC99.98 |
| 2023 [107] | Cancer Genome Atlas (CGAD) | CT | CNN | Preprocessing, downsample using maxpooling, feature extraction using autoencoder (AE) model based on CNN, & multiple image reconstruction technique | N/A | N/A | Acc 99.5 |
| 2023 [108] | LIDC-IDRI | CT | CNN | 2D slice,3D, pre-trained VGG16, VGG19,ResNet50, Xception, Inception, PCA for dimesionality reduction, Bag of Features (Bof) for feature extracted from 3D-CNN, classification step use Random Forest (RF). | Benign or malignant | N/A | Acc 95.34 Sen 90.53 Spe 97.26 AUC 99.0 F1-score 91.7 |
| 2023 [109] | LIDC | CT | CNN | Preprocessing, proposed CNN as a feature extractor, image representative is fed into the proposed LCD-CapsNet model to perform classification. | Benign or malignant | k-fold C. V | Acc 94 Spe 99.07, Pre 95 AUC 98.90 Recall 94.5 F1-score 94.5 |
| 2023 [110] | Luna 16 | CT | CNN | Modified U-Net for lobe segmentation, nodule extraction with modified U-Net architecture, classification of candidate nodule using AlexNet-SVM. | Cancerous and non-cancerous | N/A | Acc 97.98 Sen 98.84 Spe 97.47 Pre 97.53 F1-score 97.70 |
| 2024 [111] | LIDC | CT | CNN | Proposed computer-aided diagnosis for lung cancer using waterwheel plant algorithm (CADLC-WWPA) with DL approach, integrated lightweight Mobile Net for feature extraction, and used symmetrical autoencoder (SAE) for classification. | Benign, Malignant and Normal | K-fold C.V | Acc 99.05 Sen 98.55 Spe. 99.35 Pre 98.33 F1-score98.40 |
| 2024 [112] | LIDC-IDRI | CT | CNN | Data preprocessing, data augmentation, feature extraction, and classifier, deep CNN with multiview self-attention mechanism (MVSA-CNN). | Benign primary metastasis | 10. fold C. V | Acc 97.10 Sen 96.31 Spe 97.45 |
| 2024 [113] | IQ-OTH/NCCD | CT | CNN | Data preprocessing, augmentation, feature extraction with PCA, SMOTE and gaussian blur for class imbalance image pretrain network using VGG16, ResNet50, and Inception for classification. | Benign primary metastasis | K. fold C. V | Acc 98.18 Pre. 1.00 F1-score 99.6 Recall 99.2 |
| 2024 [114] | LIDC-IDRI | CT | CNN | preprocessing, segmentation, feature selection hybrid algorithm approach combine Spotted Hyena Optimization and Seagul Algorithm (SH-SOA), classification CNN-LSTM | Benign primary metastasis | K. fold C. V | Acc 99.6 Sen 99.8 Spe 99.3 Pre. 99.14 Recall 99.2 |
| Datasets | Year of Dataset Released | Image Modality | Number of Images | Number of Samples | Annotation | Image Format | Dataset Size (GB) |
|---|---|---|---|---|---|---|---|
| Lung TIME | 1998 | CT | N/A | 157 | Pixel-based | DICOM | 18.9 |
| I-ELCAP | 2003 | CT | N/A | 50 | Pixel-based | DICOM | 4.76 |
| NELSON | 2003 | CT | 7557 | N/A | Pixel-based | DICOM | N/A |
| ANODE09 | 2009 | CT | N/A | 55CT | N/A | Meta | 5.61 |
| Rider Lung CT | 2009 | CT | 15,419 | 32 | N/A | DICOM | 7.55 |
| NLST | 2009 | CT, Pathology | 26,254 | 451 | N/A | DICOM | 11.3 TB |
| LIDC-IDRI | 2011 | CT, DX, CR | 244,527 | 1018 | Pixel-based | DICOM | 124 |
| Lung CT Diagnosis | 2014 | CT | 4682 | 61 | N/A | DICOM | 2.5 |
| Qin Lung CT | 2015 | CT | 3954 | 47 | N/A | DICOM | 2.08 |
| LUNA 16 | 2016 | CT, DX, CR | 36,378 | 888 | Pixel-based | DICOM | 116 |
| LNDb 2020 | 2019 | CT | 294 | N/A | Available | DICOM | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullahi, K.; Ramakrishnan, K.; Ali, A.B. Deep Learning Techniques for Lung Cancer Diagnosis with Computed Tomography Imaging: A Systematic Review for Detection, Segmentation, and Classification. Information 2025, 16, 451. https://doi.org/10.3390/info16060451
Abdullahi K, Ramakrishnan K, Ali AB. Deep Learning Techniques for Lung Cancer Diagnosis with Computed Tomography Imaging: A Systematic Review for Detection, Segmentation, and Classification. Information. 2025; 16(6):451. https://doi.org/10.3390/info16060451
Chicago/Turabian StyleAbdullahi, Kabiru, Kannan Ramakrishnan, and Aziah Binti Ali. 2025. "Deep Learning Techniques for Lung Cancer Diagnosis with Computed Tomography Imaging: A Systematic Review for Detection, Segmentation, and Classification" Information 16, no. 6: 451. https://doi.org/10.3390/info16060451
APA StyleAbdullahi, K., Ramakrishnan, K., & Ali, A. B. (2025). Deep Learning Techniques for Lung Cancer Diagnosis with Computed Tomography Imaging: A Systematic Review for Detection, Segmentation, and Classification. Information, 16(6), 451. https://doi.org/10.3390/info16060451