Abstract
Clinician notes are a rich source of patient information, but often contain inconsistencies due to varied writing styles, abbreviations, medical jargon, grammatical errors, and non-standard formatting. These inconsistencies hinder their direct use in patient care and degrade the performance of downstream computational applications that rely on these notes as input, such as quality improvement, population health analytics, precision medicine, clinical decision support, and research. We present a large-language-model (LLM) approach to the preprocessing of 1618 neurology notes. The LLM corrected spelling and grammatical errors, expanded acronyms, and standardized terminology and formatting, without altering clinical content. Expert review of randomly sampled notes confirmed that no significant information was lost. To evaluate downstream impact, we applied an ontology-based NLP pipeline (Doc2Hpo) to extract biomedical concepts from the notes before and after editing. F1 scores for Human Phenotype Ontology extraction improved from 0.40 to 0.61, confirming our hypothesis that better inputs yielded better outputs. We conclude that LLM-based preprocessing is an effective error correction strategy that improves data quality at the level of free text in clinical notes. This approach may enhance the performance of a broad class of downstream applications that derive their input from unstructured clinical documentation.
1. Introduction
Electronic Health Records (EHRs) have transformed healthcare documentation by improving the legibility and availability of patient data [1]. They have solved the “availability problem”—where paper charts were often inaccessible—and the “legibility problem” of handwritten notes [2,3,4]. Nonetheless, EHRs face major challenges (Table 1) that include physician dissatisfaction with the EHR, poor note quality, and difficulties in converting unstructured free text into a computable format for downstream applications.

Table 1.
Challenges for Medical Record Documentation.
Physician dissatisfaction with EHRs remains high, driven by poor interface design, excessive documentation burden, clerical overload, and limited perceived benefit for patient care [5,6,7,8,9,10,11,12].
Note quality in the EHR remains a persistent problem. Notes may suffer from inconsistent structure, misspellings, excessive abbreviations, slang, and ambiguous terminology [13,14,15,16,17,18]. Up to 20% of note content may be abbreviations or acronyms [19,20,21,22,23,24,25,26]. Additionally, notes are often poorly formatted and unnecessarily verbose. One study of redundancy in EHR notes showed that 78% of the content in sign-out notes and 54% of the content of progress notes was carried over from prior notes [27].
The electronic health record (EHR) serves as the official medical-legal document memorializing the care provided to a patient and the reasoning behind clinical decisions. It also supports billing and reimbursement for services rendered. However, data within the EHR is increasingly being reconfigured and repurposed for a range of secondary uses (Table 2), including clinical decision support, precision medicine, quality improvement, population health management, data exchange, and clinical research [28,29,30,31,32,33,34].

Table 2.
Secondary Uses of EHR Data.
1.1. Secondary Uses of the EHR Require Structured Data as Inputs
Secondary applications of electronic health records (EHRs)—such as precision medicine, population health management, clinical research, decision support, and quality improvement—depend on structured data to function effectively. However, a large portion of clinically meaningful information in the EHR is unstructured narrative text, such as physician notes, discharge summaries, and radiology reports. These free-text data are not readily usable by computational systems. It is estimated that up to 80% of information in the EHR exists in such unstructured text [35,36].
To make this information accessible for secondary use, two main strategies have emerged. The first approach involves transforming textual data into high-dimensional vector representations using language models. These embeddings capture latent semantic relationships and support tasks such as clustering, classification, and semantic similarity search. The second approach extracts biomedical entities from text and maps them to standardized terminologies or ontologies, including SNOMED CT, LOINC, HPO, ICD-10, and RxNorm [35,36,37,38,39,40,41]. While the embedding approach excels at capturing meanings, ontology-based mapping provides superior interpretability. Embeddings and ontology mappings are often used in tandem to leverage their respective strengths.
Nonetheless, the inability to consistently extract and ontologize clinically relevant information from free text remains a significant barrier to realizing the full potential of secondary EHR applications. Without reliable transformation of narrative content into a computable form, downstream systems are constrained by incomplete inputs (Table 2).
1.2. Current Approaches to Reducing Physician Burden and Improving Note Quality
Efforts to reduce documentation burden and improve note quality have included a range of strategies, each with its own limitations (Table 3). Dictation and use of medical scribes remain costly and logistically complex [42,43]. Templates, smart phrases, and copy-paste functionality—originally intended to streamline documentation—have instead contributed to note bloat and redundant content [44,45]. Spelling and grammar checkers offer piecemeal solutions and may err due to lack of clinical context [18,46,47]. Educational interventions such as documentation training have had limited impact and are often poorly received by physicians [48]. Among emerging innovations, two LLM-based approaches stand out: (1) real-time coaching to guide clinicians toward better documentation [49], and (2) automated note generation via ambient listening [50,51,52]. A recent comparative review by Avendaño et al. [53] emphasizes the growing interest in AI-based documentation tools, including digital scribes and real-time assistants, as scalable solutions. These strategies create new opportunities to reduce clinician documentation burden and to improve the quality and computability of clinical notes (Table 3).

Table 3.
Current Approaches to Reducing Physician Burden and Improving Note Quality.
1.3. Our Proposed Approach to Improving Note Quality
Since the release of ChatGPT in late 2022, interest in large language models (LLMs) has surged across healthcare domains [54,55,56,57,58]. LLMs have been explored for a range of clinical applications, including decision support, diagnostic explanation, text summarization, concept extraction, ontology mapping, and document restructuring [59,60,61,62,63,64]. We propose that LLMs can also play a focused role in improving physician documentation through preprocessing.
For this submission, we define preprocessing as the enhancement of a clinical note’s structural and linguistic integrity. This includes correcting spelling and grammatical errors [65], expanding abbreviations (e.g., OU →both eyes), replacing colloquial expressions with standardized medical terminology (e.g., upgoing toe→ Babinski sign) [66], and enforcing consistent formatting—all without introducing new content or altering clinical meaning. In contrast to generative documentation systems, preprocessing seeks to improve clarity and consistency while preserving the clinician’s original narrative.
Preprocessing is not intended to reduce physicians’ documentation burden. However, we hypothesize that it offers two main benefits:
- Improved note quality, reflected in fewer spelling, grammar, and formatting errors.
- Greater retrieval of clinical concepts, as measured by Doc2Hpo.
2. Methods
This study evaluates the ability of GPT-4o 2024-11-20 to correct 1618 de-identified clinical notes from a neurology clinic. We hypothesize that preprocessing will find and correct spelling and grammatical errors, correct non-standard terminology, and expand acronyms and abbreviations. We further hypothesize that these corrections will enhance the yield of HPO concepts from the Doc2Hpo pipeline.
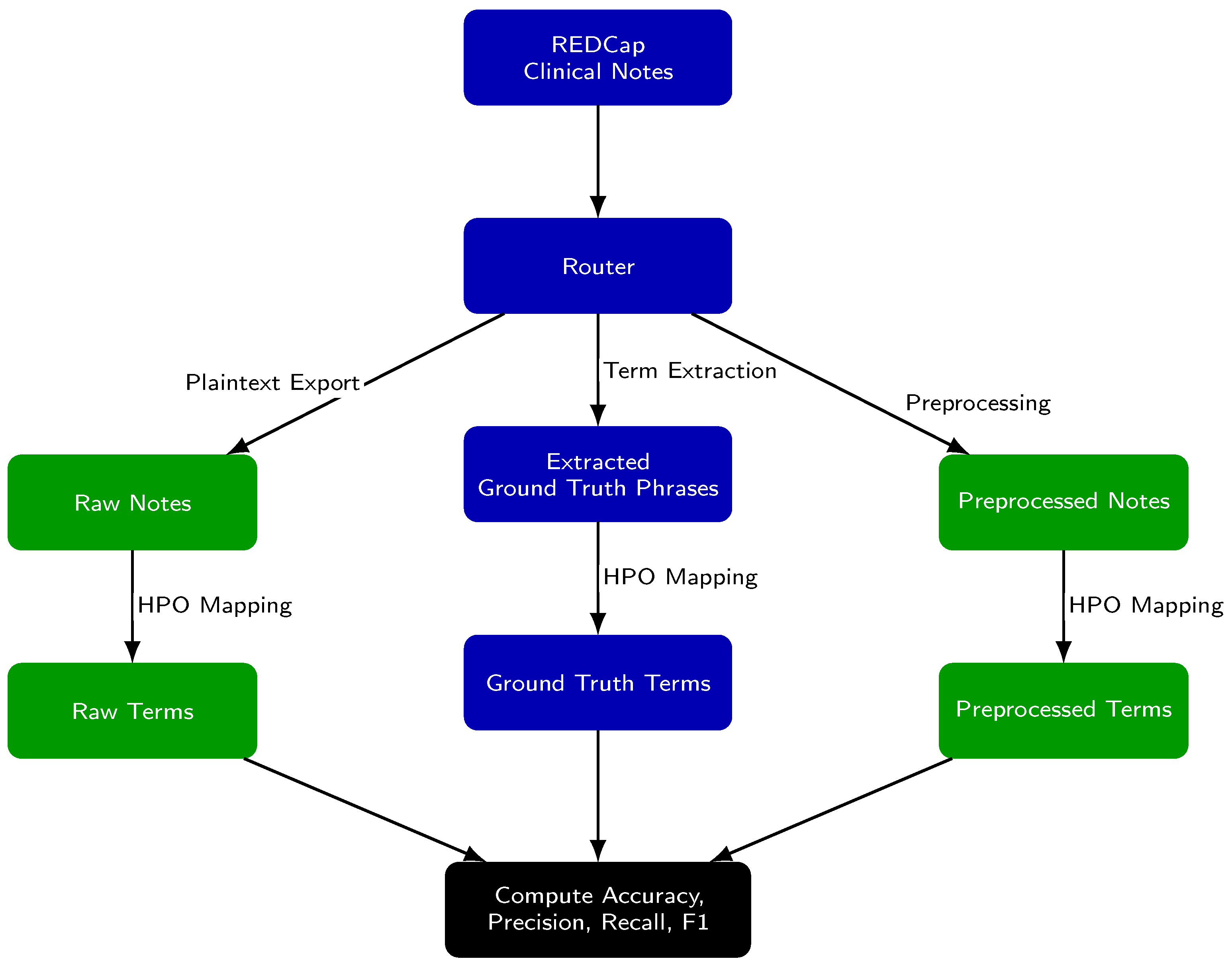
The evaluation framework (Figure 1) assesses the extraction and mapping of Human Phenotype Ontology (HPO) concepts under three conditions: (1) from raw notes, (2) from preprocessed notes, and (3) from GPT-extracted ground truth phrases. We define concept extraction as the identification of relevant clinical terms or phrases within free text. Concept mapping refers to the assignment of each extracted term to a standardized HPO concept and its corresponding identifier.
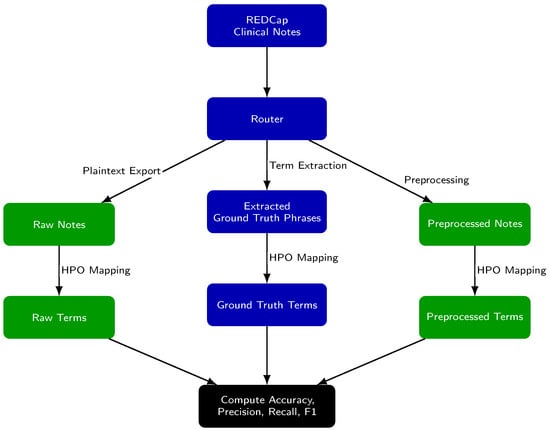
Figure 1.
Experimental Setup for Evaluating the yield of HPO concepts after preprocessing. Each clinical note was processed along three parallel paths: (1) raw notes are analyzed directly, (2) preprocessed notes are analyzed after editing, and (3) ground truth phrases are medical terms identified in the raw notes that are likely matches to an HPO concept. For all paths, text is mapped to Human Phenotype Ontology (HPO) terms using Doc2Hpo, producing a set of terms from each path. Term sets from the raw notes and preprocessed notes are then compared to the ground truth set to compute accuracy, precision, recall, and F1 score.
Data Acquisition: Institutional Review Board (IRB) approval was obtained from the University of Illinois to use de-identified clinical notes from the neurology clinic. All notes were de-identified using REDCap [67]. Selected notes met the following inclusion criteria:
- A neuroimmunological diagnosis, including multiple sclerosis, myasthenia gravis, neuromyelitis optica, and Guillain–Barré syndrome.
- Patient seen between 2016 and 2022
- Outpatient visit in the neurology clinic
- Examined by a neurology resident or attending physician
- Note length of at least 2000 characters
From an initial pool of 21,028 notes, the final dataset comprised 1618 typed notes exported as ASCII text and stored in a JSON-compatible format (Figure 2).

Figure 2.
Example neurology note converted to JSON format. Note has been truncated for brevity.
Preprocessing: GPT-4o (Open AI) was prompted to preprocess neurology notes (Appendix A and Appendix B). GPT-4o produced a detailed list of all modifications for each note (Appendix C). Character count, word count, sentence count, spelling and grammar corrections, abbreviation expansion, and word substitutions were calculated for each note.
Concept Extraction Using Doc2Hpo: We used the open-source Python implementation of Doc2Hpo [68], executed locally with the Aho-Corasick string-matching algorithm and a custom negation detection module (‘NegationDetector‘) that applies a windowed keyword search within sentence boundaries. We used the HPO release dated 3 March 2025. No modifications were made to the Doc2Hpo source code. Doc2Hpo was applied under three conditions:
- raw notes
- preprocessed notes
- ground truth phrases
To obtain the ground truth terms, GPT-4o was prompted to identify all candidate HPO-eligible phrases in each note (Appendix D). These phrases were combined into a single synthetic sentence for Doc2Hpo input (e.g., “The patient has ataxia, weakness, aphasia, and hyperreflexia.”). Doc2Hpo, which returned a list of matched terms and corresponding HPO IDs. For each of the 1,618 notes, we compiled three term sets based on the output of Doc2Hpo:
- ground truth terms
- raw-note terms
- preprocessed-note terms
Metric Computation: For each extracted term, we classified its match status as follows:
- True Positive (TP): Term present in both the extracted set and the ground truth set.
- False Negative (FN): Ground truth term not captured in extraction set.
- False Positive (FP): Term extracted but not part of ground truth set.
- True Negative (TN): Term present in the note but not successfully mapped to HPO by Doc2Hpo.
Performance metrics (precision, recall, accuracy, and F1 score) were calculated by the method of Powers [69].
3. Results and Discussion
3.1. Preprocessing Finds and Corrects Errors
We used GPT-4o to preprocess 1,618 de-identified clinical notes from a neurology clinic. Each note was converted from plain text to a structured JSON format with high-level section headers (e.g., History, Examination, Impression, Plan). GPT-4o was prompted to preprocess each note by correcting spelling and grammatical errors, expanding acronyms and abbreviations, and replacing non-standard phrasing (Appendix E).
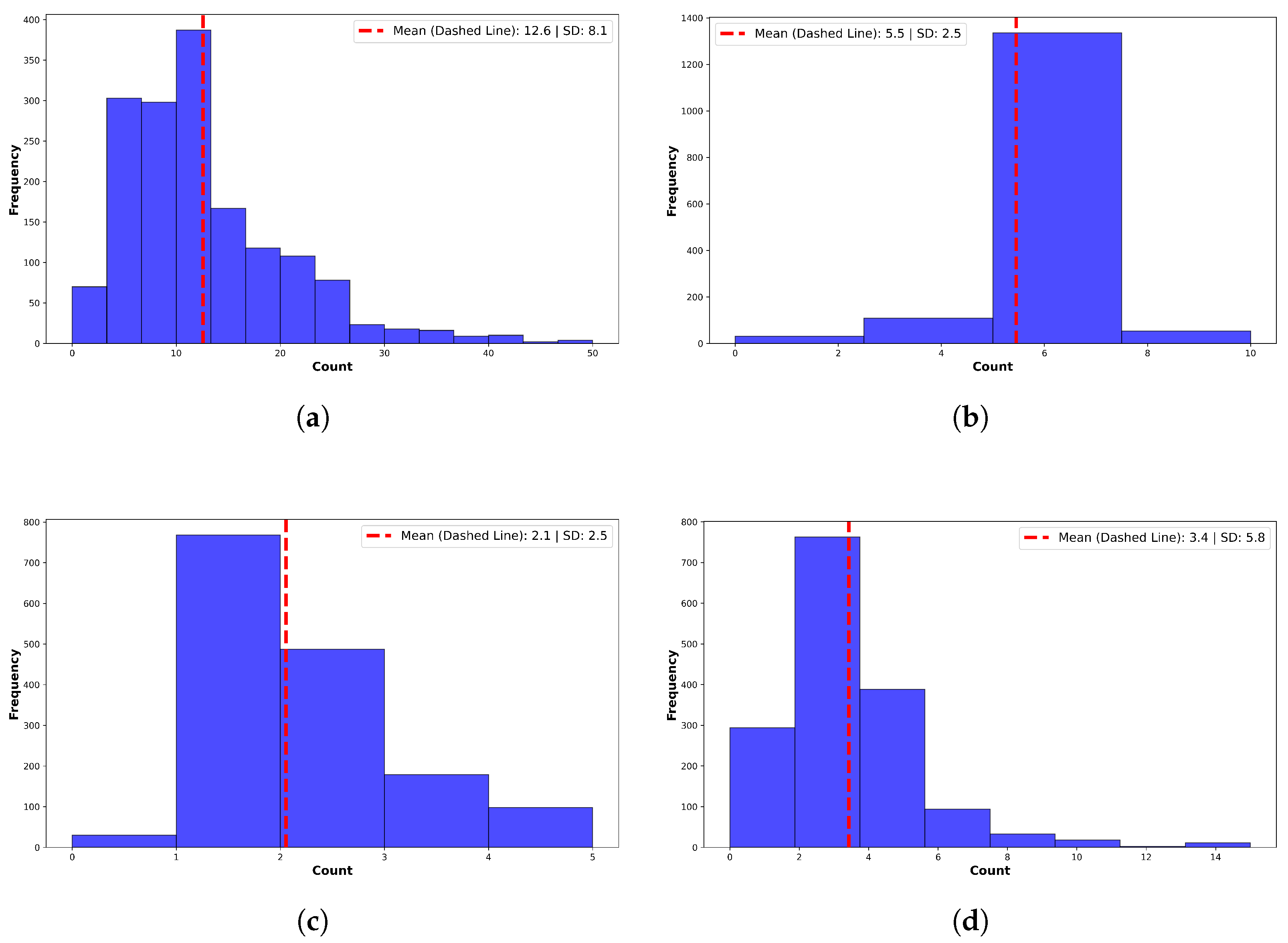
On average, GPT-4o corrected 4.9 ± 1.8 grammatical errors, 3.3 ± 5.2 spelling errors, and expanded 12.6 ± 8.1 abbreviations or acronyms per note (Figure 3). Additionally, it replaced 3.1 ± 3.0 instances of non-standard language or jargon with standardized clinical terminology. A manual review of 100 randomly selected notes confirmed preservation of clinical meaning, along with improvements in clarity, structure, and readability. No hallucinated or spurious content was identified by expert reviewers.
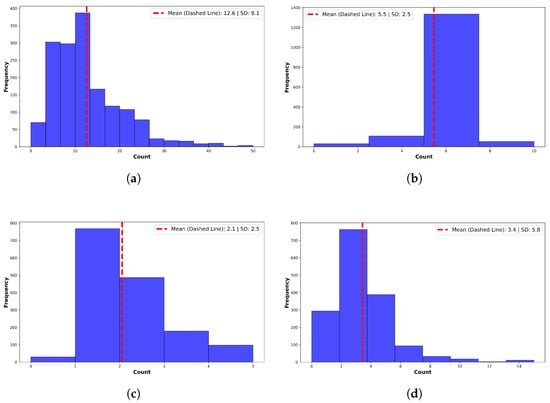
Figure 3.
Summary of corrections made by GPT-4o. (a) Acronyms and abbreviations. Preprocessing expanded a mean of per note. (b) Grammatical errors. Preprocessing corrected a mean of grammatical errors per note. (c) Slang, Jargon, and Non-Standard Terms. GPT-4o corrected non-standard terms per note. Example: “feeling blue” → “symptoms of depression”. (d) Spelling Errors. GPT-4o corrected a mean of errors per note. High variance reflects many outlier notes.
3.2. Preprocessing Improves HPO Term Extraction by Doc2Hpo
To evaluate whether preprocessing improves phenotype term extraction, we compared three parallel data flows:
- Raw Notes → Doc2Hpo → Raw Terms
- Preprocessed Notes → Doc2Hpo → Preprocessed Terms
- GPT-4o Extracted Phrases → Doc2Hpo → Ground Truth Terms
All extracted terms were mapped to the Human Phenotype Ontology (HPO) using Doc2Hpo. The ground truth set was defined as the union of HPO terms extracted by Doc2Hpo from GPT-4o-supplied candidate phrases. For each note, term sets from the raw and preprocessed notes were compared against the ground truth using standard classification metrics: precision, recall, accuracy, and F1 score.
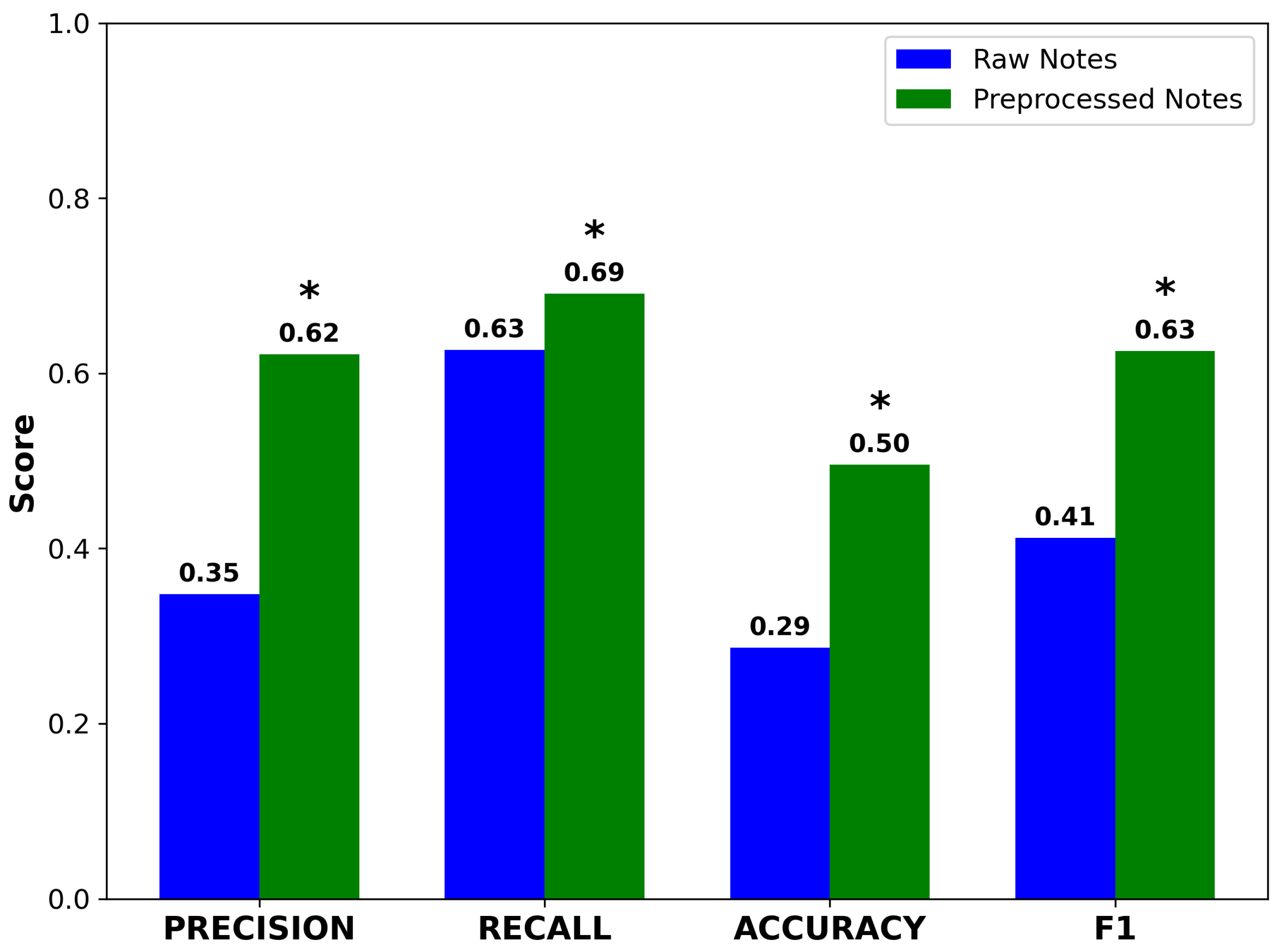
Preprocessed notes yielded higher precision and accuracy compared to raw notes. Recall also improved, rising from 0.61 to 0.68. Although modest in absolute terms, this gain was statistically significant due to the large sample size (n = 1618) and resulted in a meaningful improvement in F1 score (Figure 4). The concurrent improvement in both precision and recall suggests that GPT-4o neither introduced spurious concepts nor omitted clinically important terms. These results support the hypothesis that preprocessing enhances the performance of NLP pipelines performing ontology-based concept extraction.
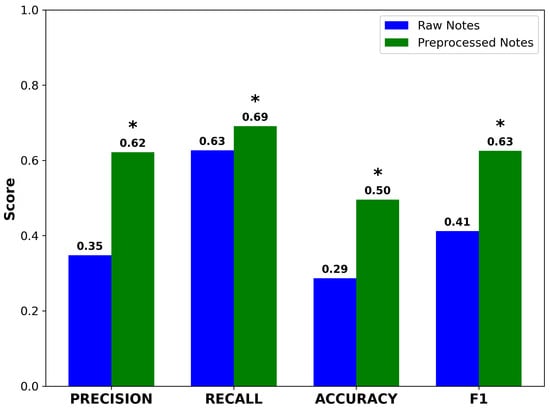
Figure 4.
Performance metrics for Doc2Hpo in mapping terms from raw and preprocessed notes to their correct HPO terms and IDs. Accuracy, precision, recall, and F1 score all improve significantly after preprocessing. Asterisks indicate statistically significant differences between raw and preprocessed notes (* , , paired two-tailed t-test).
Major academic medical centers are already exploring applications of LLMs to free-text EHR data—both to power predictive analytics [70] and to improve documentation quality among house officers and early-career clinicians [71].
4. Limitations
Preprocessing of physician notes does not directly reduce documentation burden, which is fundamentally tied to EHR design, clinical workflows, and organizational factors [10,72,73,74]. Technologies such as ambient AI and real-time documentation coaching may provide more comprehensive burden relief [75]. Encouragingly, recent surveys suggest that AI tools are improving physician satisfaction with EHRs [76].
This study was conducted using 1618 de-identified neurology notes, most of which were from patients with multiple sclerosis. Broader validation across multiple specialties, note types, and care settings is needed. Ground truth terms were generated using GPT-4o phrase extraction followed by human review. While this approach enabled scalable benchmarking, future studies may benefit from full manual annotation by domain experts.
We did not formally assess computational cost or latency. In our setup, processing time was approximately 10 s per note and cost averaged $5 per 1000 notes. Actual costs will vary by vendor and system configuration. Despite these costs, many institutions are actively piloting or adopting LLM-based tools in production environments [71,77].
Although notes were de-identified, we did not obtain explicit consent from patients or physicians. Real-time deployment in clinical settings may require additional legal or regulatory safeguards. US HIPAA-compliant LLM offerings are available through major cloud providers, including Microsoft, Amazon, and Google.
This cross-sectional study does not capture two key dynamics: (1) model performance drift—as clinical language evolves, periodic re-tuning may be needed to maintain accuracy; and (2) behavioral adaptation—as physicians grow accustomed to GPT-based editing, their writing may change in ways that affect both the editor’s output and downstream data use. Longitudinal studies should examine these phenomena using periodic F1 audits, user surveys, and dynamic prompts.
5. Conclusions
We demonstrate that LLM-based preprocessing—focused on correcting errors, expanding abbreviations, and standardizing terminology—enhances the clarity of clinical notes without altering their meaning or introducing hallucinated content. Across 1618 de-identified neurology notes, this approach yielded a statistically significant improvement in F1 score for HPO concept extraction, rising from 0.40 to 0.61.
Preprocessing is a lightweight, non-intrusive intervention that improves note quality without requiring any workflow changes from clinicians. Editing can be implemented transparently, and the resulting improvements strengthen the EHR as input for downstream applications, including precision medicine, decision support, quality improvement, and clinical research.
Looking ahead, LLM-powered preprocessing may serve as a practical on-ramp for broader reuse of EHR free text in real-time applications—such as quality monitoring, error correction, ontology mapping, cross-lingual translation, and generation of FHIR-compatible structured data.
Author Contributions
Conceptualization, All authors; Methodology, D.B.H. and T.O.-A.; Software, D.B.H.; Validation, all authors; Formal analysis, D.B.H., S.K.P. and T.O.-A.; Investigation and data acquisition, M.A.C.; Data curation, D.B.H. and M.A.C.; Writing—original draft preparation, D.B.H. and S.K.P.; Writing—review and editing, all authors; Visualization, D.B.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the National Science Foundation under Award Number 2423235.
Institutional Review Board Statement
The use of EHR clinical notes for research was approved by the IRB of the University of Illinois (Protocol 2017-0520Z).
Informed Consent Statement
Informed consent was obtained from all participants as part of enrollment in the UIC Neuroimmunology Biobank.
Data Availability Statement
Data and code supporting this study are available from the corresponding author on request.
Acknowledgments
We thank the UIC Neuroimmunology BioBank Team for their support.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| API | Application Programming Interface |
| ASCII | American Standard Code for Information Interchange |
| EHR | Electronic Health Record |
| FHIR | Fast Healthcare Interoperability Resources |
| HIPAA | Health Insurance Portability and Accountability Act |
| HPO | Human Phenotype Ontology |
| ICD | International Classification of Diseases |
| IRB | Institutional Review Board |
| JSON | JavaScript Object Notation |
| LLM | Large Language Model |
| LOINC | Logical Observation Identifiers Names and Codes |
| NLP | Natural Language Processing |
| REDCap | Research Electronic Data Capture |
| RxNorm | Standardized Nomenclature for Clinical Drugs |
| TP/FN/FP/TN | True Positive/False Negative/False Positive/True Negative |
Appendix A. Prompt to GPT-4o to Preprocess a Physician Note
You are a highly skilled medical terminologist
specializing in clinical note editing.
Your task is to edit the note using the following rules:
1. Expand abbreviations (e.g., BP ’ blood pressure),
retaining common abbreviations in parentheses.
2. Correct spelling and grammar while preserving meaning.
3. Reorganize content under the following headings:
History, Vital Signs, Examination, Labs, Radiology,
Impression, and Plan.
4. Replace non-standard terms with standard clinical terminology.
Appendix B. Note Format Used by GPT-4o for Preprocessed Notes
{
"HISTORY": {
"Chief Complaint": "...",
"Interim History": "..."
},
"VITAL SIGNS": {
"Blood Pressure": "...",
"Pulse": "...",
"Temperature": "...",
"Weight": "..."
},
"EXAMINATION": {
"Mental Status": "...",
"Cranial Nerves": "...",
"Motor": "...",
"Sensory": "...",
"Reflexes": "...",
"Coordination": "...",
"Gait and Station": "..."
},
"LABS": "...",
"RADIOLOGY": "...",
"IMPRESSION": {
"Assessment": "..."
},
"PLAN": {
"Testing": "...",
"Education Provided": {
"Instructions": "...",
"Barriers to Learning": "...",
"Content": "...",
"Outcome": "..."
},
"Return Visit": "..."
},
"Metrics": {
"Grammatical Errors": n,
"Abbreviations and Acronyms Expanded": ["..."],
"Spelling Errors": ["..."],
"Non-Standard Terms Corrected": ["..."]
}
}
Appendix C. Example of Corrections Made by GPT-4o
"Abbreviations Expanded": [
"BP", "IVIG", "MRI", "EMG", "PT", "OTC", "OT", "CSF",
"WBC", "RBC", "HSV", "PCR", "CIDP", "INCAT", "BPD",
"CBD", "BSA", "FPL", "EHL", "FN", "PA"
],
"Spelling Errors Corrected": [
"wreight", "materal", "unknwon", "schizphernia", "tjhan"
],
"Non-Standard Terms Mapped": [
"heart attack -> myocardial infarction"
]
Appendix D. Prompt to GPT-4o to Identify Ground Truth Terms
You are an expert medical coder with expertise in medical
terminologies such as the Human Phenotype Ontology (HPO).
From the note text {note_text}, extract all potential HPO terms
the patient may have. Return a JSON object with a list of
extracted terms under the key "hpo_terms".
Use this exact format:
{
"hpo_terms": ["term1", "term2", "term3"]
}
Appendix E. Sample Neurological Examinations: Before and After Preprocessing
Neurological Examination Prior to Preprocessing. Expanded terms are highlighted in blue.
Neurologic:
Mental status : awake, alert, oriented to person,
place, and time. Follows commands briskly,
including 2-step commands. Naming and repetition intact.
Fluent speech with no dysarthria.
Cranial nerves : PERRL, no rAPD,
unable to perform full L
lateral gaze but otherwise EOMI, facial sensation full
and symmetric, smile full and symmetric, palate and
uvula elevate symmetrically, shoulder shrug intact,
tongue midline
...continues....
Neurological Examination After Preprocessing by GPT-4o.
EXAMINATION:
Mental Status: Awake, alert, oriented to person, place,
and time. Follows commands briskly, including
two-step commands. Naming and repetition intact.
Fluent speech with no dysarthria.
Cranial Nerves: Pupils equal, round, and reactive
to light, no relative afferent pupillary
defect, unable to perform full left lateral gaze but
otherwise extraocular movements are intact,
facial sensation full and symmetric,
smile full and symmetric, palate and uvula elevate
symmetrically, shoulder shrug intact, tongue midline
...continues....
References
- Menachemi, N.; Collum, T.H. Benefits and drawbacks of electronic health record systems. Risk Manag. Healthc. Policy 2011, 4, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Bruner, A.; Kasdan, M.L. Handwriting errors: Harmful, wasteful and preventable. J.-Ky. Med Assoc. 2001, 99, 189–192. [Google Scholar]
- Kozak, E.A.; Dittus, R.S.; Smith, W.R.; Fitzgerald, J.F.; Langfeld, C.D. Deciphering the physician note. J. Gen. Intern. Med. 1994, 9, 52–54. [Google Scholar] [CrossRef]
- Rodríguez-Vera, F.J.; Marin, Y.; Sanchez, A.; Borrachero, C.; Pujol, E. Illegible handwriting in medical records. J. R. Soc. Med. 2002, 95, 545–546. [Google Scholar] [CrossRef]
- Holmgren, A.J.; Hendrix, N.; Maisel, N.; Everson, J.; Bazemore, A.; Rotenstein, L.; Phillips, R.L.; Adler-Milstein, J. Electronic health record usability, satisfaction, and burnout for family physicians. JAMA Netw. Open 2024, 7, e2426956. [Google Scholar] [CrossRef]
- Muhiyaddin, R.; Elfadl, A.; Mohamed, E.; Shah, Z.; Alam, T.; Abd-Alrazaq, A.; Househ, M. Electronic health records and physician burnout: A scoping review. In Informatics and Technology in Clinical Care and Public Health; IOS Press: Amsterdam, The Netherlands, 2022; pp. 481–484. [Google Scholar]
- Downing, N.L.; Bates, D.W.; Longhurst, C.A. Physician burnout in the electronic health record era: Are we ignoring the real cause? Ann. Intern. Med. 2018, 169, 50–51. [Google Scholar] [CrossRef]
- Elliott, M.; Padua, M.; Schwenk, T.L. Electronic health records, medical practice problems, and physician distress. Int. J. Behav. Med. 2022, 29, 387–392. [Google Scholar] [CrossRef]
- Carroll, A.E. How health information technology is failing to achieve its full potential. JAMA Pediatr. 2015, 169, 201–202. [Google Scholar] [CrossRef][Green Version]
- Rodríguez-Fernández, J.M.; Loeb, J.A.; Hier, D.B. It’s time to change our documentation philosophy: Writing better neurology notes without the burnout. Front. Digit. Health 2022, 4, 1063141. [Google Scholar] [CrossRef] [PubMed]
- Koopman, R.J.; Steege, L.M.B.; Moore, J.L.; Clarke, M.A.; Canfield, S.M.; Kim, M.S.; Belden, J.L. Physician information needs and electronic health records (EHRs): Time to reengineer the clinic note. J. Am. Board Fam. Med. 2015, 28, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Budd, J. Burnout related to electronic health record use in primary care. J. Prim. Care Community Health 2023, 14, 21501319231166921. [Google Scholar] [CrossRef]
- Kugic, A.; Schulz, S.; Kreuzthaler, M. Disambiguation of acronyms in clinical narratives with large language models. J. Am. Med. Inform. Assoc. 2024, 31, 2040–2046. [Google Scholar] [CrossRef]
- Aronson, J.K. When I use a word…Medical slang: A taxonomy. BMJ 2023, 382, 1516. [Google Scholar] [CrossRef]
- Lee, E.H.; Patel, J.P.; VI, A.H.F. Patient-centric medical notes: Identifying areas for improvement in the age of open medical records. Patient Educ. Couns. 2017, 100, 1608–1611. [Google Scholar] [CrossRef]
- Castro, C.M.; Wilson, C.; Wang, F.; Schillinger, D. Babel babble: Physicians’ use of unclarified medical jargon with patients. Am. J. Health Behav. 2007, 31, S85–S95. [Google Scholar] [CrossRef]
- Pitt, M.B.; Hendrickson, M.A. Eradicating jargon-oblivion—a proposed classification system of medical jargon. J. Gen. Intern. Med. 2020, 35, 1861–1864. [Google Scholar] [CrossRef]
- Workman, T.E.; Shao, Y.; Divita, G.; Zeng-Treitler, Q. An efficient prototype method to identify and correct misspellings in clinical text. BMC Res. Notes 2019, 12, 42. [Google Scholar] [CrossRef]
- Hamiel, U.; Hecht, I.; Nemet, A.; Pe’er, L.; Man, V.; Hilely, A.; Achiron, A. Frequency, comprehension and attitudes of physicians towards abbreviations in the medical record. Postgrad. Med J. 2018, 94, 254–258. [Google Scholar] [CrossRef]
- Myers, J.S.; Gojraty, S.; Yang, W.; Linsky, A.; Airan-Javia, S.; Polomano, R.C. A randomized-controlled trial of computerized alerts to reduce unapproved medication abbreviation use. J. Am. Med. Inform. Assoc. 2011, 18, 17–23. [Google Scholar] [CrossRef]
- Horon, K.; Hayek, K.; Montgomery, C. Prohibited abbreviations: Seeking to educate, not enforce. Can. J. Hosp. Pharm. 2012, 65, 294. [Google Scholar] [CrossRef]
- Cheung, S.; Hoi, S.; Fernandes, O.; Huh, J.; Kynicos, S.; Murphy, L.; Lowe, D. Audit on the use of dangerous abbreviations, symbols, and dose designations in paper compared to electronic medication orders: A multicenter study. Ann. Pharmacother. 2018, 52, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Shultz, J.; Strosher, L.; Nathoo, S.N.; Manley, J. Avoiding potential medication errors associated with non-intuitive medication abbreviations. Can. J. Hosp. Pharm. 2011, 64, 246. [Google Scholar] [CrossRef]
- Baker, D.E. Campaign to Eliminate Use of Error-Prone Abbreviations. Hosp. Pharm. 2006, 41, 809–810. [Google Scholar] [CrossRef]
- American Hospital Association; American Society of Health-System Pharmacists. Medication safety issue brief. Eliminating dangerous abbreviations, acronyms and symbols. Hosp. Health Networks 2005, 79, 41–42. [Google Scholar]
- Hultman, G.M.; Marquard, J.L.; Lindemann, E.; Arsoniadis, E.; Pakhomov, S.; Melton, G.B. Challenges and opportunities to improve the clinician experience reviewing electronic progress notes. Appl. Clin. Inform. 2019, 10, 446–453. [Google Scholar] [CrossRef]
- Wrenn, J.O.; Stein, D.M.; Bakken, S.; Stetson, P.D. Quantifying clinical narrative redundancy in an electronic health record. J. Am. Med Inform. Assoc. 2010, 17, 49–53. [Google Scholar] [CrossRef]
- Moja, L.; Kwag, K.H.; Lytras, T.; Bertizzolo, L.; Brandt, L.; Pecoraro, V.; Rigon, G.; Vaona, A.; Ruggiero, F.; Mangia, M.; et al. Effectiveness of computerized decision support systems linked to electronic health records: A systematic review and meta-analysis. Am. J. Public Health 2014, 104, e12–e22. [Google Scholar] [CrossRef]
- Coorevits, P.; Sundgren, M.; Klein, G.O.; Bahr, A.; Claerhout, B.; Daniel, C.; Dugas, M.; Dupont, D.; Schmidt, A.; Singleton, P.; et al. Electronic health records: New opportunities for clinical research. J. Intern. Med. 2013, 274, 547–560. [Google Scholar] [CrossRef]
- Carr, L.H.; Christ, L.; Ferro, D.F. The electronic health record as a quality improvement tool: Exceptional potential with special considerations. Clin. Perinatol. 2023, 50, 473–488. [Google Scholar] [CrossRef]
- Vorisek, C.N.; Lehne, M.; Klopfenstein, S.A.I.; Mayer, P.J.; Bartschke, A.; Haese, T.; Thun, S. Fast healthcare interoperability resources (FHIR) for interoperability in health research: Systematic review. JMIR Med. Inform. 2022, 10, e35724. [Google Scholar] [CrossRef]
- Lehne, M.; Luijten, S.; Vom Felde Genannt Imbusch, P.; Thun, S. The use of FHIR in digital health–a review of the scientific literature. In German Medical Data Sciences: Shaping Change–Creative Solutions for Innovative Medicine; IOS Press: Amsterdam, The Netherlands, 2019; pp. 52–58. [Google Scholar]
- Sitapati, A.; Kim, H.; Berkovich, B.; Marmor, R.; Singh, S.; El-Kareh, R.; Clay, B.; Ohno-Machado, L. Integrated precision medicine: The role of electronic health records in delivering personalized treatment. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9, e1378. [Google Scholar] [CrossRef] [PubMed]
- Kruse, C.S.; Stein, A.; Thomas, H.; Kaur, H. The use of electronic health records to support population health: A systematic review of the literature. J. Med. Syst. 2018, 42, 214. [Google Scholar] [CrossRef]
- Li, I.; Pan, J.; Goldwasser, J.; Verma, N.; Wong, W.P.; Nuzumlalı, M.Y.; Rosand, B.; Li, Y.; Zhang, M.; Chang, D.; et al. Neural natural language processing for unstructured data in electronic health records: A review. Comput. Sci. Rev. 2022, 46, 100511. [Google Scholar] [CrossRef]
- Xiao, C.; Choi, E.; Sun, J. Opportunities and challenges in developing deep learning models using electronic health records data: A systematic review. J. Am. Med Inform. Assoc. 2018, 25, 1419–1428. [Google Scholar] [CrossRef]
- McDonald, C.J.; Huff, S.M.; Suico, J.G.; Hill, G.; Leavelle, D.; Aller, R.; Forrey, A.; Mercer, K.; DeMoor, G.; Hook, J.; et al. LOINC, a universal standard for identifying laboratory observations: A 5-year update. Clin. Chem. 2003, 49, 624–633. [Google Scholar] [CrossRef]
- Hanna, J.; Joseph, E.; Brochhausen, M.; Hogan, W.R. Building a drug ontology based on RxNorm and other sources. J. Biomed. Semant. 2013, 4, 1–9. [Google Scholar] [CrossRef]
- Zarei, J.; Golpira, R.; Hashemi, N.; Azadmanjir, Z.; Meidani, Z.; Vahedi, A.; Bakhshandeh, H.; Fakharian, E.; Sheikhtaheri, A. Comparison of the accuracy of inpatient morbidity coding with ICD-11 and ICD-10. Health Inf. Manag. J. 2025, 54, 14–24. [Google Scholar] [CrossRef]
- Lee, D.; de Keizer, N.; Lau, F.; Cornet, R. Literature review of SNOMED CT use. J. Am. Med Inform. Assoc. 2014, 21, e11–e19. [Google Scholar] [CrossRef]
- Köhler, S.; Vasilevsky, N.A.; Engelstad, M.; Foster, E.; McMurry, J.; Aymé, S.; Baynam, G.; Bello, S.M.; Boerkoel, C.F.; Boycott, K.M.; et al. The human phenotype ontology in 2017. Nucleic Acids Res. 2017, 45, D865–D876. [Google Scholar] [CrossRef]
- Al Hadidi, S.; Upadhaya, S.; Shastri, R.; Alamarat, Z. Use of dictation as a tool to decrease documentation errors in electronic health records. J. Community Hosp. Intern. Med. Perspect. 2017, 7, 282–286. [Google Scholar] [CrossRef]
- Ash, J.S.; Corby, S.; Mohan, V.; Solberg, N.; Becton, J.; Bergstrom, R.; Orwoll, B.; Hoekstra, C.; Gold, J.A. Safe use of the EHR by medical scribes: A qualitative study. J. Am. Med Inform. Assoc. 2021, 28, 294–302. [Google Scholar] [CrossRef]
- Mehta, R.; Radhakrishnan, N.S.; Warring, C.D.; Jain, A.; Fuentes, J.; Dolganiuc, A.; Lourdes, L.S.; Busigin, J.; Leverence, R.R. The use of evidence-based, problem-oriented templates as a clinical decision support in an inpatient electronic health record system. Appl. Clin. Inform. 2016, 7, 790–802. [Google Scholar]
- Iannello, J.; Waheed, N.; Neilan, P. Template Design and Analysis: Integrating Informatics Solutions to Improve Clinical Documentation. Fed. Pract. 2020, 37, 527. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.H.; Topaz, M.; Goss, F.R.; Zhou, L. Automated misspelling detection and correction in clinical free-text records. J. Biomed. Inform. 2015, 55, 188–195. [Google Scholar] [CrossRef]
- Amosa, T.I.; Izhar, L.I.B.; Sebastian, P.; Ismail, I.B.; Ibrahim, O.; Ayinla, S.L. Clinical errors from acronym use in electronic health record: A review of NLP-based disambiguation techniques. IEEE Access 2023, 11, 59297–59316. [Google Scholar] [CrossRef]
- Baughman, D.J.; Botros, P.A.; Waheed, A. Technology in Medicine: Improving Clinical Documentation. FP Essentials 2024, 537, 26–38. [Google Scholar] [PubMed]
- NYU Langone Health. Artificial Intelligence Feedback on Physician Notes Improves Patient Care. 2024. Available online: https://nyulangone.org/news/artificial-intelligence-feedback-physician-notes-improves-patient-care (accessed on 8 May 2025).
- Balloch, J.; Sridharan, S.; Oldham, G.; Wray, J.; Gough, P.; Robinson, R.; Sebire, N.J.; Khalil, S.; Asgari, E.; Tan, C.; et al. Use of an ambient artificial intelligence tool to improve quality of clinical documentation. Future Healthc. J. 2024, 11, 100157. [Google Scholar] [CrossRef]
- Stults, C.D.; Deng, S.; Martinez, M.C.; Wilcox, J.; Szwerinski, N.; Chen, K.H.; Driscoll, S.; Washburn, J.; Jones, V.G. Evaluation of an Ambient Artificial Intelligence Documentation Platform for Clinicians. JAMA Netw. Open 2025, 8, e258614. [Google Scholar] [CrossRef]
- Mess, S.A.; Mackey, A.J.; Yarowsky, D.E. Artificial Intelligence Scribe and Large Language Model Technology in Healthcare Documentation: Advantages, Limitations, and Recommendations. Plast. Reconstr. Surg.-Open 2025, 13, e6450. [Google Scholar] [CrossRef] [PubMed]
- Avendano, J.P.; Gallagher, D.O.; Hawes, J.D.; Boyle, J.; Glasser, L.; Aryee, J.; Katt, B.M. Interfacing With the Electronic Health Record (EHR): A Comparative Review of Modes of Documentation. Cureus 2022, 14, e26330. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.S.; Plasek, J.M.; Xu, H.; Uzuner, Ö.; Cohen, T.; Yetisgen, M.; Liu, H.; Meystre, S.; Wang, Y. Large language models for biomedicine: Foundations, opportunities, challenges, and best practices. J. Am. Med Inform. Assoc. 2024, 31, 2114–2124. [Google Scholar] [CrossRef]
- Yan, C.; Ong, H.H.; Grabowska, M.E.; Krantz, M.S.; Su, W.C.; Dickson, A.L.; Peterson, J.F.; Feng, Q.; Roden, D.M.; Stein, C.M.; et al. Large language models facilitate the generation of electronic health record phenotyping algorithms. J. Am. Med. Inform. Assoc. 2024, 31, 1994–2001. [Google Scholar] [CrossRef] [PubMed]
- Munzir, S.I.; Hier, D.B.; Carrithers, M.D. High Throughput Phenotyping of Physician Notes with Large Language and Hybrid NLP Models. arXiv 2024, arXiv:2403.05920. [Google Scholar] [CrossRef]
- Omiye, J.A.; Gui, H.; Rezaei, S.J.; Zou, J.; Daneshjou, R. Large language models in medicine: The potentials and pitfalls. arXiv 2023, arXiv:2309.00087. [Google Scholar] [CrossRef]
- Clusmann, J.; Kolbinger, F.R.; Muti, H.S.; Carrero, Z.I.; Eckardt, J.N.; Laleh, N.G.; Löffler, C.M.L.; Schwarzkopf, S.C.; Unger, M.; Veldhuizen, G.P.; et al. The future landscape of large language models in medicine. Commun. Med. 2023, 3, 141. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Yerebakan, H.; Shinagawa, Y.; Luo, Y. Enhancing Health Data Interoperability with Large Language Models: A FHIR Study. arXiv 2023, arXiv:2310.12989. [Google Scholar]
- Van Veen, D.; Van Uden, C.; Blankemeier, L.; Delbrouck, J.B.; Aali, A.; Bluethgen, C.; Pareek, A.; Polacin, M.; Reis, E.P.; Seehofnerova, A.; et al. Clinical text summarization: Adapting large language models can outperform human experts. Res. Sq. 2024, 30, 1134–1142. [Google Scholar]
- Tang, L.; Sun, Z.; Idnay, B.; Nestor, J.G.; Soroush, A.; Elias, P.A.; Xu, Z.; Ding, Y.; Durrett, G.; Rousseau, J.F.; et al. Evaluating large language models on medical evidence summarization. NPJ Digit. Med. 2023, 6, 158. [Google Scholar] [CrossRef]
- Zhou, W.; Bitterman, D.; Afshar, M.; Miller, T.A. Considerations for health care institutions training large language models on electronic health records. arXiv 2023, arXiv:2309.12339. [Google Scholar]
- Qiu, J.; Li, L.; Sun, J.; Peng, J.; Shi, P.; Zhang, R.; Dong, Y.; Lam, K.; Lo, F.P.W.; Xiao, B.; et al. Large ai models in health informatics: Applications, challenges, and the future. IEEE J. Biomed. Health Inform. 2023, 27, 6074–6078. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Petzold, L. Are Large Language Models Ready for Healthcare. A Comparative Study on Clinical Language Understanding. arXiv 2023, arXiv:2304.05368. [Google Scholar]
- Ficarra, B.J. Grammar and Medicine. Arch. Surg. 1981, 116, 251–252. [Google Scholar] [CrossRef]
- Goss, F.R.; Zhou, L.; Weiner, S.G. Incidence of speech recognition errors in the emergency department. Int. J. Med Inform. 2016, 93, 70–73. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Peres Kury, F.S.; Li, Z.; Ta, C.; Wang, K.; Weng, C. Doc2Hpo: A web application for efficient and accurate HPO concept curation. Nucleic Acids Res. 2019, 47, W566–W570. [Google Scholar] [CrossRef]
- Powers, D.M. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar]
- Jiang, L.Y.; Liu, X.C.; Nejatian, N.P.; Nasir-Moin, M.; Wang, D.; Abidin, A.; Eaton, K.; Riina, H.A.; Laufer, I.; Punjabi, P.; et al. Health system-scale language models are all-purpose prediction engines. Nature 2023, 619, 357–362. [Google Scholar] [CrossRef]
- Feldman, J.; Hochman, K.A.; Guzman, B.V.; Goodman, A.; Weisstuch, J.; Testa, P. Scaling note quality assessment across an academic medical center with AI and GPT-4. NEJM Catal. Innov. Care Deliv. 2024, 5, CAT–23. [Google Scholar] [CrossRef]
- Bakken, S. Can informatics innovation help mitigate clinician burnout? J. Am. Med. Inform. Assoc. 2019, 26, 93–94. [Google Scholar] [CrossRef]
- Kapoor, M. Physician burnout in the electronic health record era. Ann. Intern. Med. 2019, 170, 216. [Google Scholar] [CrossRef]
- Kang, C.; Sarkar, N. Interventions to reduce electronic health record-related burnout: A systematic review. Appl. Clin. Informatics 2024, 15, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.; Reilly, C.; Feeley, A.; Sheehan, E.; Merghani, K.; Feeley, I. Artificial Intelligence (AI)–Powered Documentation Systems in Healthcare: A Systematic Review. J. Med Syst. 2025, 49, 28. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J. Fewer Physicians Consider Leaving Medicine, Survey Finds. MedPage Today, 29 March 2025. [Google Scholar]
- Ji, Z.; Wei, Q.; Xu, H. Bert-based ranking for biomedical entity normalization. AMIA Summits Transl. Sci. Proc. 2020, 2020, 269. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).