Optimized Deep Learning for Mammography: Augmentation and Tailored Architectures
Abstract
:1. Introduction
RelatedWorks
- Data preparation: Obtaining mammographic images from the mammographic dataset and performing pre-processing operations that include cleaning, normalization, and augmentation;
- Model architecture: Using pre-trained CNN models such as EfficientNet and ResNet, with customized changes for breast tissue categorization;
- Training and optimization: Using transfer learning, regularization, and learning rate modifications to improve the model performance;
- Reliable categorization: Applying evaluation measures such as accuracy, precision, recall, and F1 score;
- Interpretability: Using visualization techniques such as Grad-CAM to yield insights into the model’s decision-making while guaranteeing its clinical applicability.
2. Breast Cancer Detection
2.1. The Dataset
2.2. Cleaning the Dataset and Creating the Labels
2.3. Class Balancing
2.4. Image Augmentation
2.5. Performance Measures
2.6. Convolutional Neural Network Models
- In order to make the output of the base model’s convolutional layers ready for the dense layers, it is first converted into a one-dimensional vector. The next step after every completely linked layer is batch normalization, which speeds up and stabilizes the training process [23].
- By initializing the weights of the layers using ReLU activation, the He uniform initializer helps the network converge more quickly. Two dense layers, each with 512 and 256 units, are introduced. A dropout layer with a 0.3 dropout rate is added, and the ReLU activation function is employed in order to prevent overfitting by randomly deactivating a subset of the neurons during training [24].
- One of the three groups is identified using a dense layer with softmax activation to categorize the images [25]. The network can recognize particular textures, forms, or patterns that are important for diagnosing breast cancer thanks to the completely linked layers, which assist the network in making judgements based on the data taken from the photos.
- During training, the weights of the pre-trained network’s convolutional layers are kept constant, as they are frozen. This method makes use of these models’ strong feature extraction capabilities without requiring the time or computing resources to retrain them on the new dataset. In addition to preventing overfitting, freezing the foundation layers is also helpful when dealing with very limited datasets, as medical imaging frequently involves.
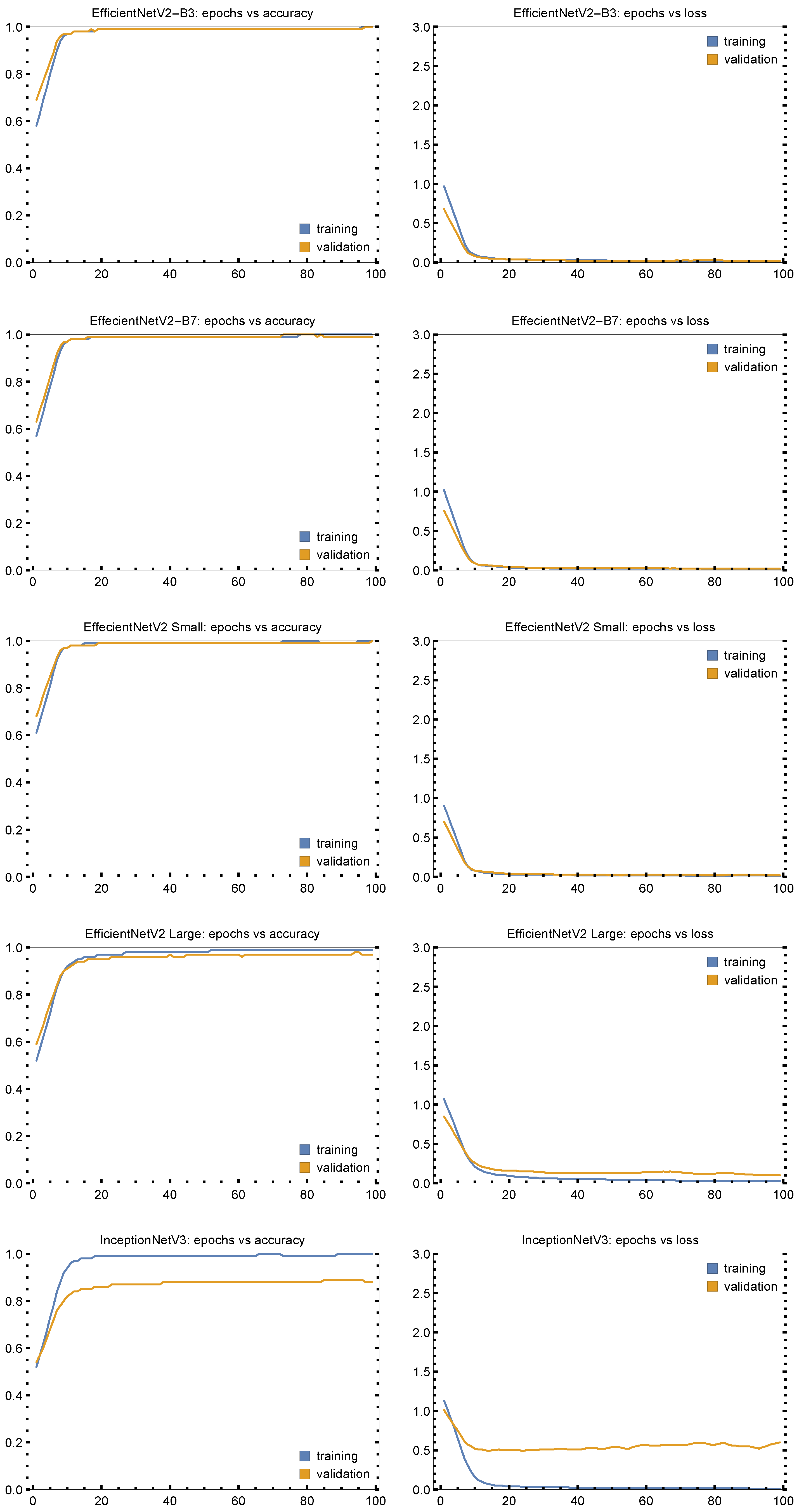
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anandhi, S.; Mahure, S.J.; Royal, T.Y.; Nikhil, V.V.; Viswanadh, U. Mammography scans for breast cancer detection using CNN. In Computer Science Engineering; CRC Press: Boca Raton, FL, USA, 2024; pp. 3–8. [Google Scholar]
- Aguerchi, K.; Jabrane, Y.; Habba, M.; El Hassani, A.H. A CNN Hyperparameters Optimization Based on Particle Swarm Optimization for Mammography Breast Cancer Classification. J. Imaging 2024, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Vijetha, K.J.; Priya, S.S.S. A Comparative Analysis of CNN Architectures and Regularization Techniques for Breast Cancer Classification in Mammograms. Ing. Syst. d’Inf. 2024, 29, 2433. [Google Scholar] [CrossRef]
- Wahed, M.A.; Alqaraleh, M.; Alzboon, M.S.; Al-Batah, M.S. Evaluating AI and Machine Learning Models in Breast Cancer Detection: A Review of Convolutional Neural Networks (CNN) and Global Research Trends. LatIA 2025, 3, 117. [Google Scholar] [CrossRef]
- Hussain, S.I.; Toscano, E. An extensive investigation into the use of machine learning tools and deep neural networks for the recognition of skin cancer: Challenges, future directions, and a comprehensive review. Symmetry 2024, 16, 366. [Google Scholar] [CrossRef]
- Şahin, Z.; Kalkan, Ö.; Aktaş, O. The physiology of laughter: Understanding laughter-related structures from brain lesions. J. Health Life 2022, 4, 242–251. [Google Scholar]
- Rautela, K.; Kumar, D.; Kumar, V. A systematic review on breast cancer detection using deep learning techniques. Arch. Comput. Methods Eng. 2022, 29, 4599–4629. [Google Scholar] [CrossRef]
- Liu, H.; Cui, G.; Luo, Y.; Guo, Y.; Zhao, L.; Wang, Y.; Subasi, A.; Dogan, S.; Tuncer, T. Artificial intelligence-based breast cancer diagnosis using ultrasound images and grid-based deep feature generator. Int. J. Gen. Med. 2022, 15, 2271–2282. [Google Scholar] [CrossRef]
- Huang, Z.; Shao, W.; Han, Z.; Alkashash, A.M.; De la Sancha, C.; Parwani, A.V.; Nitta, H.; Hou, Y.; Wang, T.; Salama, P.; et al. Artificial intelligence reveals features associated with breast cancer neoadjuvant chemotherapy responses from multi-stain histopathologic images. NPJ Precis. Oncol. 2023, 7, 14. [Google Scholar] [CrossRef]
- Marinovich, M.L.; Wylie, E.; Lotter, W.; Lund, H.; Waddell, A.; Madeley, C.; Pereira, G.; Houssami, N. Artificial intelligence (AI) for breast cancer screening: BreastScreen population-based cohort study of cancer detection. EBioMedicine 2023, 90, 104498. [Google Scholar] [CrossRef]
- Tan, T.Z.; Miow, Q.H.; Huang, R.Y.J.; Wong, M.K.; Ye, J.; Lau, J.A.; Wu, M.C.; Bin Abdul Hadi, L.H.; Soong, R.; Choolani, M.; et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol. Med. 2013, 5, 1051–1066. [Google Scholar] [CrossRef]
- Charan, G.; Yuvaraj, R. Estimating the effectiveness of alexnet in classifying tumor in comparison with resnet. In Proceedings of the AIP Conference Proceedings, Contemporary Innovations in Engineering and Management, Nandyal, India, 22–23 April 2022; AIP Publishing: Melville, NY, USA, 2023; Volume 2821. [Google Scholar]
- Ismail, N.S.; Sovuthy, C. Breast cancer detection based on deep learning technique. In Proceedings of the 2019 International UNIMAS STEM 12th engineering conference (EnCon), Kuching, Malaysia, 28–29 August 2019; pp. 89–92. [Google Scholar]
- Ibrahim, M.; Yadav, S.; Ogunleye, F.; Zakalik, D. Male BRCA mutation carriers: Clinical characteristics and cancer spectrum. BMC Cancer 2018, 18, 179. [Google Scholar] [CrossRef] [PubMed]
- Charan, S.; Khan, M.J.; Khurshid, K. Breast cancer detection in mammograms using convolutional neural network. In Proceedings of the 2018 International Conference on Computing, Mathematics and Engineering Technologies (iCoMET), Sukkur, Pakistan, 3–4 March 2018; pp. 1–5. [Google Scholar]
- Ebrahim, M.; Alsmirat, M.; Al-Ayyoub, M. Performance study of augmentation techniques for hep2 cnn classification. In Proceedings of the 2018 9th International Conference on Information and Communication Systems (ICICS), Irbid, Jordan, 3–5 April 2018; pp. 163–168. [Google Scholar]
- O’Gara, S.; McGuinness, K. Comparing data augmentation strategies for deep image classification. In Proceedings of the Irish Machine Vision & Image Processing Conference (IMVIP), Dublin, Ireland, 28–30 August 2019; Technological University Dublin: Dublin, Ireland, 2019. [Google Scholar]
- Thakur, A.; Gupta, M.; Sinha, D.K.; Mishra, K.K.; Venkatesan, V.K.; Guluwadi, S. Transformative breast Cancer diagnosis using CNNs with optimized ReduceLROnPlateau and Early stopping Enhancements. Int. J. Comput. Intell. Syst. 2024, 17, 14. [Google Scholar]
- Chanda, D.; Onim, M.S.H.; Nyeem, H.; Ovi, T.B.; Naba, S.S. DCENSnet: A new deep convolutional ensemble network for skin cancer classification. Biomed. Signal Process. Control 2024, 89, 105757. [Google Scholar] [CrossRef]
- Rao, Y.; Lee, Y.; Jarjoura, D.; Ruppert, A.S.; Liu, C.g.; Hsu, J.C.; Hagan, J.P. A comparison of normalization techniques for microRNA microarray data. Stat. Appl. Genet. Mol. Biol. 2008, 7, 22. [Google Scholar] [CrossRef]
- Kumar, R.; Corvisieri, G.; Fici, T.; Hussain, S.; Tegolo, D.; Valenti, C. Transfer Learning for Facial Expression Recognition. Information 2025, 16, 320. [Google Scholar] [CrossRef]
- Al-Kababji, A.; Bensaali, F.; Dakua, S.P. Scheduling techniques for liver segmentation: Reducelronplateau vs onecyclelr. In Proceedings of the Second International Conference on Intelligent Systems and Pattern Recognition, Hammamet, Tunisia, 24–26 March 2022; Springer: Cham, Switzerland, 2022; Volume 1589, pp. 204–212. [Google Scholar]
- Chen, G.; Chen, P.; Shi, Y.; Hsieh, C.Y.; Liao, B.; Zhang, S. Rethinking the usage of batch normalization and dropout in the training of deep neural networks. arXiv 2019, arXiv:1905.05928. [Google Scholar]
- Huang, R.; Wu, H. Skin cancer severity analysis and prediction framework based on deep learning. In Proceedings of the 2024 3rd International Conference on Artificial Intelligence and Intelligent Information Processing, Tianjin China, 25–27 October 2024; pp. 192–198. [Google Scholar]
- Joseph, A.A.; Abdullahi, M.; Junaidu, S.B.; Ibrahim, H.H.; Chiroma, H. Improved multi-classification of breast cancer histopathological images using handcrafted features and deep neural network (dense layer). Intell. Syst. Appl. 2022, 14, 200066. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Gupta, K.; Prasad, P. Skin Lesion Analyser: An Efficient Seven-Way Multi-class Skin Cancer Classification Using MobileNet. In Proceedings of the Advanced Machine Learning Technologies and Applications, Jaipur, India, 13–15 February 2020; Springer: Singapore, 2020; Volume 1141. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. Lect. Notes Comput. Sci. 2016, 9908, 630–645. [Google Scholar]
- Huang, H.; Hsu, B.; Lee, C.; Tseng, V. Development of a light-weight deep learning model for cloud applications and remote diagnosis of skin cancers. J. Dermatol. 2021, 48, 310–316. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K. Densely Connected Convolutional Networks. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- El-Nouby, A.; Touvron, H.; Caron, M.; Bojanowski, P.; Douze, M.; Joulin, A.; Laptev, I.; Neverova, N.; Synnaeve, G.; Verbeek, J. XCiT: Cross-Covariance Image Transformers. arXiv 2021, arXiv:2106.09681. [Google Scholar]
- Xie, S.; Girshick, R.; Dollar, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning. Scientific Research, Long Beach, CA, USA, 9–15 June 2019. [Google Scholar]
- Pacal, I.; Ozdemir, B.; Zeynalov, J.; Gasimov, H.; Pacal, N. A novel CNN-ViT-based deep learning model for early skin cancer diagnosis. Biomed. Signal Process. Control 2025, 104, 107627. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for mobileNetV3. In Proceedings of the International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019. [Google Scholar]
- Touvron, H.; Bojanowski, P.; Caron, M.; Cord, M.; El-Nouby, A.; Grave, E.; Izacard, G.; Joulin, A.; Synnaeve, G.; Verbeek, J.; et al. ResMLP: Feedforward Networks for Image Classification With Data-Efficient Training. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 5314–5321. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhou, P.; Yan, S.; Wang, X. InceptionNeXt: When Inception Meets ConvNeXt. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 17–22 June 2024; pp. 5672–5683. [Google Scholar]
- Tang, Y.; Han, K.; Guo, J.; Xu, C.; Xu, C.; Wang, Y. GhostNetV2: Enhance Cheap Operation with Long-Range Attention. Adv. Neural Inf. Process. Syst. 2022, 35, 9969–9982. [Google Scholar]
- Shahin, A.; Kamal, A.; Elattar, M. Deep Ensemble Learning for Skin Lesion Classification from Dermoscopic Images. In Proceedings of the 9th Cairo International Biomedical Engineering Conference, Cairo, Egypt, 20–22 December 2018. [Google Scholar]
- Carcagnì, P.; Leo, M.; Cuna, A.; Mazzeo, P.; Spagnolo, P.; Celeste, G.; Distante, C. Classification of Skin Lesions by Combining Multilevel Learnings in a DenseNet Architecture. In Proceedings of the 20th International Conference, Image Analysis and Processing, Trento, Italy, 9–13 September 2019; Springer: Cham, Switzerland, 2019; Volume 11751. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. PVT v2: Improved baselines with Pyramid Vision Transformer. Comp. Vis. Media 2022, 8, 415–424. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Volume 139, pp. 10347–10357. [Google Scholar]
- Han, D.; Yun, S.; Heo, B.; Yoo, Y. Rethinking Channel Dimensions for Efficient Model Design. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021. [Google Scholar]
- Wang, A.; Chen, H.; Lin, Z.; Han, J.; Ding, G. RepViT: Revisiting Mobile CNN From ViT Perspective. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024. [Google Scholar]
- Wu, K.; Zhang, J.; Peng, H.; Liu, M.; Xiao, B.; Fu, J.; Yuan, L. TinyViT: Fast Pretraining Distillation for Small Vision Transformers. In Proceedings of the 17th European Conference, Tel Aviv, Israel, 23–27 October 2022; Springer: Cham, Switzerland, 2022; Volume 13681. [Google Scholar]
- Hangbo, B.; Li, D.; Songhao, P.; Furu, W. BEiT: BERT Pre-Training of Image Transformers. arXiv 2021, arXiv:2106.08254. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021. [Google Scholar]
- Hatamizadeh, A.; Yin, H.; Heinrich, G.; Kautz, J.; Molchanov, P. Global Context Vision Transformers. In Proceedings of the 40th International Conference on Machine Learning, Honolulu, HI, USA, 23–29 July 2023. [Google Scholar]
- Chaturvedi, S.; Tembhurne, J.; Diwan, T. A multi-class skin Cancer classification using deep convolutional neural networks. Multimed. Tools Appl. 2020, 79, 28477–28498. [Google Scholar] [CrossRef]
- Ozdemir, B.; Pacal, I. A robust deep learning framework for multiclass skin cancer classification. Sci. Rep. 2025, 15, 4938. [Google Scholar] [CrossRef]
- Alsunaidi, S.; Almuhaideb, A.; Ibrahim, N. Applications of Big Data Analytics to Control COVID-19 Pandemic. Sensors 2021, 21, 2282. [Google Scholar] [CrossRef]
- Aladhadh, S.; Alsanea, M.; Aloraini, M.; Khan, T.; Habib, S.; Islam, M. An Effective Skin Cancer Classification Mechanism via Medical Vision Transformer. Sensors 2022, 22, 4008. [Google Scholar] [CrossRef]
- Thwin, S.; Hyun-Seok Park, H.; Seo, S. A Trustworthy Framework for Skin Cancer Detection Using a CNN with a Modified Attention Mechanism. Appl. Sci. 2025, 15, 1067. [Google Scholar] [CrossRef]





| Class | Original | Augmented | Train (70%) | Val (15%) | Test (15%) |
|---|---|---|---|---|---|
| Images | Samples | ||||
| Normal | 209 | 12,540 | 8760 | 1890 | 1890 |
| Benign | 62 | 3720 | 2580 | 570 | 570 |
| Malignant | 51 | 3060 | 2160 | 450 | 450 |
| Total | 322 | 19,320 | 13,500 | 2910 | 2910 |
| CNN Models | Accuracy | Precision | Recall | F1 Score | Kappa Score |
|---|---|---|---|---|---|
| VGG-19 | 0.9557 | 0.9557 | 0.9557 | 0.9554 | 0.9332 |
| MobileNet-V3 Large | 0.9215 | 0.9221 | 0.9215 | 0.9216 | 0.8814 |
| DenseNet-201 | 0.7545 | 0.7557 | 0.7545 | 0.7548 | 0.6290 |
| XceptionNet | 0.7887 | 0.7886 | 0.7887 | 0.7884 | 0.6806 |
| MobileNet-V2 | 0.8028 | 0.8034 | 0.8028 | 0.8029 | 0.7022 |
| DenseNet-121 | 0.8672 | 0.8698 | 0.8672 | 0.8668 | 0.7989 |
| DenseNet-169 | 0.8692 | 0.8706 | 0.8692 | 0.8690 | 0.8019 |
| Resnet-50 | 0.9416 | 0.9427 | 0.9416 | 0.9415 | 0.9118 |
| Resnet-101 | 0.9135 | 0.9147 | 0.9135 | 0.9137 | 0.8691 |
| Resnet-152 | 0.9427 | 0.9477 | 0.9477 | 0.9477 | 0.9211 |
| EfficientNetV2-B3 | 0.9970 | 0.9970 | 0.9970 | 0.9997 | 0.9954 |
| NASNet | 0.7903 | 0.7904 | 0.7903 | 0.7903 | 0.6832 |
| EfficientNetV2 Large | 0.9774 | 0.9774 | 0.9774 | 0.9774 | 0.9658 |
| CONVNet | 0.5445 | 0.5817 | 0.5445 | 0.5201 | 0.2923 |
| EfficientNetV2 Small | 1.000 | 0.9991 | 0.9990 | 0.9974 | 0.9989 |
| EfficientNetV2 B7 | 0.9985 | 0.9985 | 0.9985 | 0.9985 | 0.9977 |
| Inception-ResnetV2 | 0.8612 | 0.8647 | 0.8612 | 0.8616 | 0.7901 |
| Resnet101V2 | 0.7692 | 0.7696 | 0.7692 | 0.7688 | 0.6504 |
| NasNet Large | 0.8778 | 0.8785 | 0.8778 | 0.8780 | 0.8155 |
| InceptionNet V3 | 0.8643 | 0.8646 | 0.8643 | 0.8644 | 0.7946 |
| CNN Models | Accuracy | Precision | Recall | F1 Score | Kappa Score |
|---|---|---|---|---|---|
| VGG-19 | 0.9437 | 0.9439 | 0.9437 | 0.9436 | 0.9147 |
| MobileNet-V3 Large | 0.9416 | 0.943 | 0.9416 | 0.9417 | 0.9116 |
| DNS-201 | 0.7686 | 0.7694 | 0.7686 | 0.7683 | 0.6491 |
| XceptionNet | 0.8451 | 0.8450 | 0.8451 | 0.8448 | 0.7653 |
| MobileNet-V2 | 0.8390 | 0.8398 | 0.8390 | 0.8391 | 0.756 |
| DenseNet-121 | 0.9115 | 0.9124 | 0.9115 | 0.9117 | 0.8661 |
| DenseNet-169 | 0.8531 | 0.8550 | 0.8531 | 0.8532 | 0.7769 |
| Resnet-50 | 0.9557 | 0.9560 | 0.9557 | 0.9557 | 0.9329 |
| Resnet-101 | 0.9316 | 0.9321 | 0.9316 | 0.9315 | 0.8962 |
| Resnet-152 | 0.9457 | 0.9458 | 0.9457 | 0.9457 | 0.9178 |
| EffecientNetV2-B3 | 0.9970 | 0.9970 | 0.9970 | 0.9998 | 0.9956 |
| NASNet | 0.7949 | 0.7967 | 0.7949 | 0.7954 | 0.6919 |
| EffecientNetV2 Large | 0.9955 | 0.9955 | 0.9955 | 0.9955 | 0.9932 |
| CONVNet | 0.5716 | 0.5845 | 0.5716 | 0.5617 | 0.3492 |
| EffecientNetV2 Small | 0.9970 | 0.9997 | 0.9997 | 0.9997 | 0.9955 |
| EffecientNetV2 B7 | 0.9955 | 0.9955 | 0.9955 | 0.9955 | 0.9932 |
| Inception-ResnetV2 | 0.8959 | 0.8960 | 0.8959 | 0.8959 | 0.8434 |
| Resnet101V2 | 0.8009 | 0.8025 | 0.8009 | 0.8010 | 0.7002 |
| NasNet Large | 0.8748 | 0.8749 | 0.8748 | 0.8746 | 0.8116 |
| InceptionNet V3 | 0.8974 | 0.8991 | 0.8974 | 0.8977 | 0.8459 |
| CNN Models | Accuracy | Precision | Recall | F1 Score | Kappa Score |
|---|---|---|---|---|---|
| VGG-19 | 0.8567 | 0.8571 | 0.8567 | 0.8568 | 0.7848 |
| MobileNet-V3 Large | 0.9020 | 0.9020 | 0.9020 | 0.9018 | 0.8526 |
| DenseNet-201 | 0.8944 | 0.8945 | 0.8944 | 0.8944 | 0.8412 |
| XceptionNet | 0.9578 | 0.9579 | 0.9578 | 0.9577 | 0.9365 |
| MobileNet-V2 | 0.9457 | 0.9462 | 0.9457 | 0.9455 | 0.9183 |
| DenseNet-121 | 0.9668 | 0.9670 | 0.9668 | 0.9668 | 0.9501 |
| DenseNet-169 | 0.9502 | 0.9503 | 0.9502 | 0.9502 | 0.9252 |
| Resnet-50 | 0.9744 | 0.9744 | 0.9744 | 0.9743 | 0.9615 |
| Resnet-101 | 0.9985 | 0.9985 | 0.9985 | 0.9985 | 0.9977 |
| Resnet-152 | 0.9834 | 0.9934 | 0.9834 | 0.9834 | 0.9751 |
| EfficientNetV2-B3 | 1.0000 | 0.9993 | 0.9997 | 0.9998 | 0.9994 |
| NASNet | 0.8115 | 0.8115 | 0.8113 | 0.8115 | 0.7139 |
| EfficientNetV2 Large | 0.9894 | 0.9895 | 0.9894 | 0.9895 | 0.9840 |
| CONVNet | 0.5445 | 0.5684 | 0.5445 | 0.5271 | 0.2897 |
| EfficientNetV2 Small | 1.0000 | 0.9998 | 1.0000 | 0.9998 | 0.9995 |
| EfficientNetV2 B7 | 1.0000 | 0.9995 | 0.9995 | 0.9995 | 0.9991 |
| Inception-ResnetV2 | 0.9351 | 0.9351 | 0.9351 | 0.9350 | 0.9015 |
| Resnet101V2 | 0.7783 | 0.7785 | 0.7783 | 0.7781 | 0.6637 |
| NasNet Large | 0.8658 | 0.8659 | 0.8658 | 0.8657 | 0.7963 |
| InceptionNet V3 | 0.9005 | 0.9005 | 0.9005 | 0.9003 | 0.8487 |
| Approach | Accuracy | Precision | Recall | F1 Score | Kappa Score |
|---|---|---|---|---|---|
| MobileNet [26] | 0.8310 | 0.8900 | 0.8300 | 0.8300 | — |
| ResNetv250 [27] | 0.8493 | 0.7809 | 0.7406 | 0.7571 | — |
| Deep learning models [28] | 0.8580 | 0.7518 | — | — | — |
| DenseNet121 [29] | 0.8635 | 0.8126 | 0.7798 | 0.7932 | — |
| XCiT-Small-Patch16 [30] | 0.8785 | 0.8390 | 0.7921 | 0.8123 | — |
| Res2NeXt50 [31] | 0.8798 | 0.8401 | 0.8144 | 0.8255 | — |
| EfficientNet-B4 [32] | 0.8827 | 0.8210 | 0.8035 | 0.8110 | — |
| EfficientNetv2-small [33] | 0.8858 | 0.8580 | 0.8148 | 0.8324 | — |
| Xception [34] | 0.8858 | 0.8677 | 0.8094 | 0.8345 | — |
| MobileNetv3-large-075 [35] | 0.8877 | 0.8442 | 0.8077 | 0.8249 | — |
| ResMLP-24 [36] | 0.8885 | 0.8786 | 0.8171 | 0.8449 | — |
| InceptionNeXt-base [37] | 0.8929 | 0.8616 | 0.8308 | 0.8444 | — |
| GhostNetv2-100 [38] | 0.8982 | 0.8615 | 0.8337 | 0.8440 | — |
| ResNet-50 + Inception V3 [39] | 0.8990 | 0.8620 | 0.7960 | — | — |
| nseNet + SVM [40] | 0.9000 | 0.8800 | 0.7600 | 0.8200 | — |
| PvTv2-B2 [41] | 0.9029 | 0.8754 | 0.8399 | 0.8532 | — |
| DeiT-base [42] | 0.9034 | 0.8887 | 0.8359 | 0.8588 | — |
| RexNet200 [43] | 0.9040 | 0.8785 | 0.8596 | 0.8677 | — |
| RepViT-m2 [44] | 0.9061 | 0.8792 | 0.8669 | 0.8713 | — |
| Tiny-ViT-21 [45] | 0.9082 | 0.8740 | 0.8724 | 0.8720 | — |
| BeiTv2-base [46] | 0.9090 | 0.8775 | 0.8731 | 0.8741 | — |
| PiT-base [44] | 0.9092 | 0.8952 | 0.8456 | 0.8675 | — |
| Swinv-base [47] | 0.9179 | 0.9049 | 0.8757 | 0.8893 | — |
| GcViT-small [48] | 0.9213 | 0.9127 | 0.8742 | 0.8913 | — |
| ResNeXt-101 [49] | 0.9320 | 0.8800 | 0.8800 | — | — |
| ConvNeXtV2 + ViT [50] | 0.9348 | 0.9324 | 0.9070 | 0.9182 | — |
| Deep learning models [51] | 0.9580 | 0.9222 | 0.8420 | 0.8803 | — |
| MVT + MLP [52] | 0.9614 | 0.9600 | 0.9650 | 0.9700 | — |
| DCAN-Net [53] | 0.9757 | 0.9700 | 0.9757 | 0.9710 | — |
| Proposed methodology | 1.0000 | 0.9998 | 1.0000 | 0.9998 | 0.9995 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, S.I.; Toscano, E. Optimized Deep Learning for Mammography: Augmentation and Tailored Architectures. Information 2025, 16, 359. https://doi.org/10.3390/info16050359
Hussain SI, Toscano E. Optimized Deep Learning for Mammography: Augmentation and Tailored Architectures. Information. 2025; 16(5):359. https://doi.org/10.3390/info16050359
Chicago/Turabian StyleHussain, Syed Ibrar, and Elena Toscano. 2025. "Optimized Deep Learning for Mammography: Augmentation and Tailored Architectures" Information 16, no. 5: 359. https://doi.org/10.3390/info16050359
APA StyleHussain, S. I., & Toscano, E. (2025). Optimized Deep Learning for Mammography: Augmentation and Tailored Architectures. Information, 16(5), 359. https://doi.org/10.3390/info16050359







