MRF-Mixer: A Simulation-Based Deep Learning Framework for Accelerated and Accurate Magnetic Resonance Fingerprinting Reconstruction
Abstract
1. Introduction
- Our work is fully simulated, data-driven and employs an end-to-end training pipeline, starting with realistic tissue property maps of the brain and culminating in simulated MRF k-space data that incorporate undersampling patterns.
- A complex-valued neural network that preserves and models the inter-relationship between the real and imaginary components of the complex-valued MRF signal evolution.
- A spatio-temporal network architecture combining a voxel-based fully connected network with a patch-based multi-branch convolutional neural network.
2. Methods
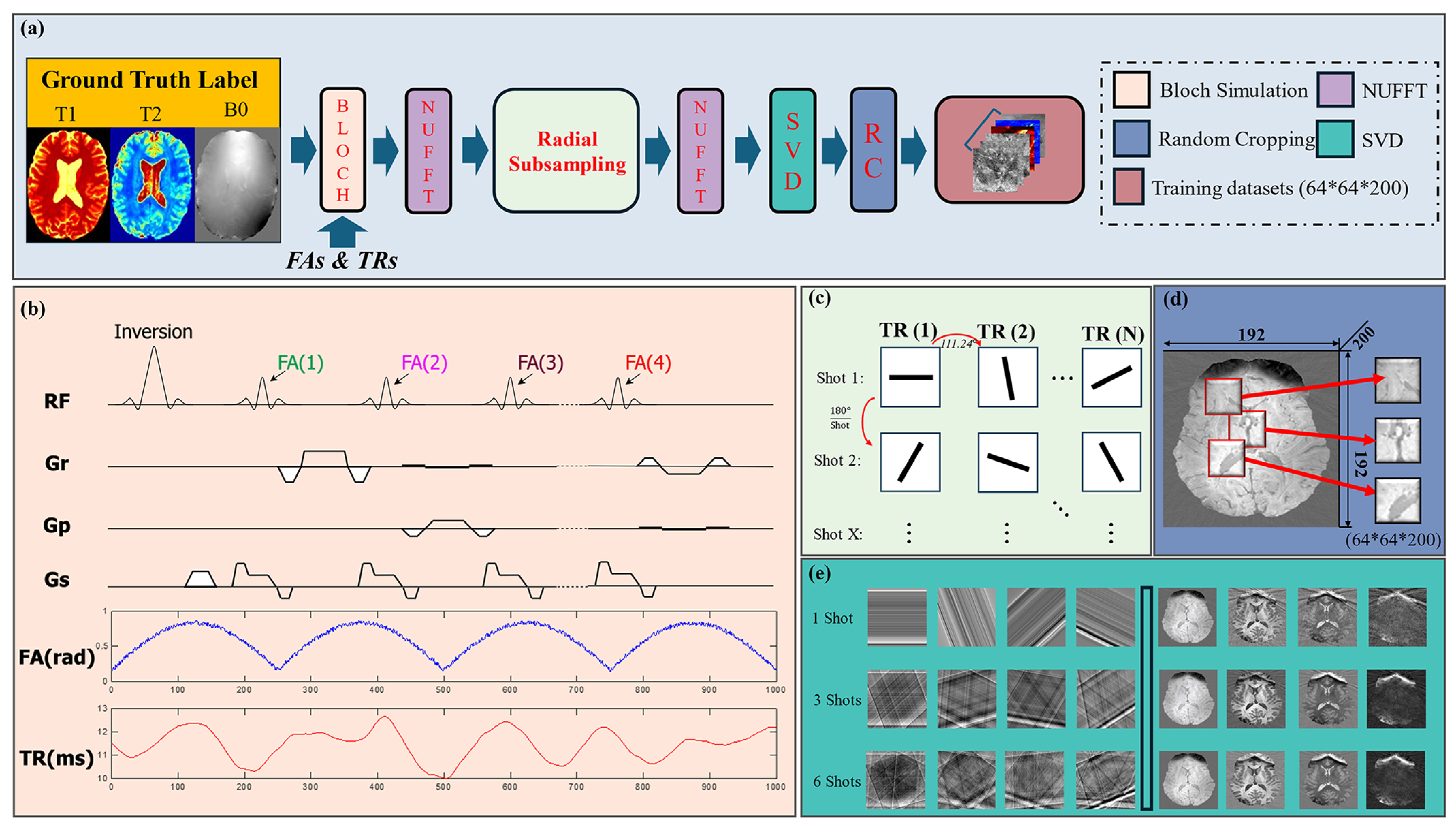
2.1. MRF Pulse Sequences and Simulations
2.2. Undersampling and Aliasing Effects
2.3. Synthetic Training Dataset
2.4. Time Series Dimension Reduction
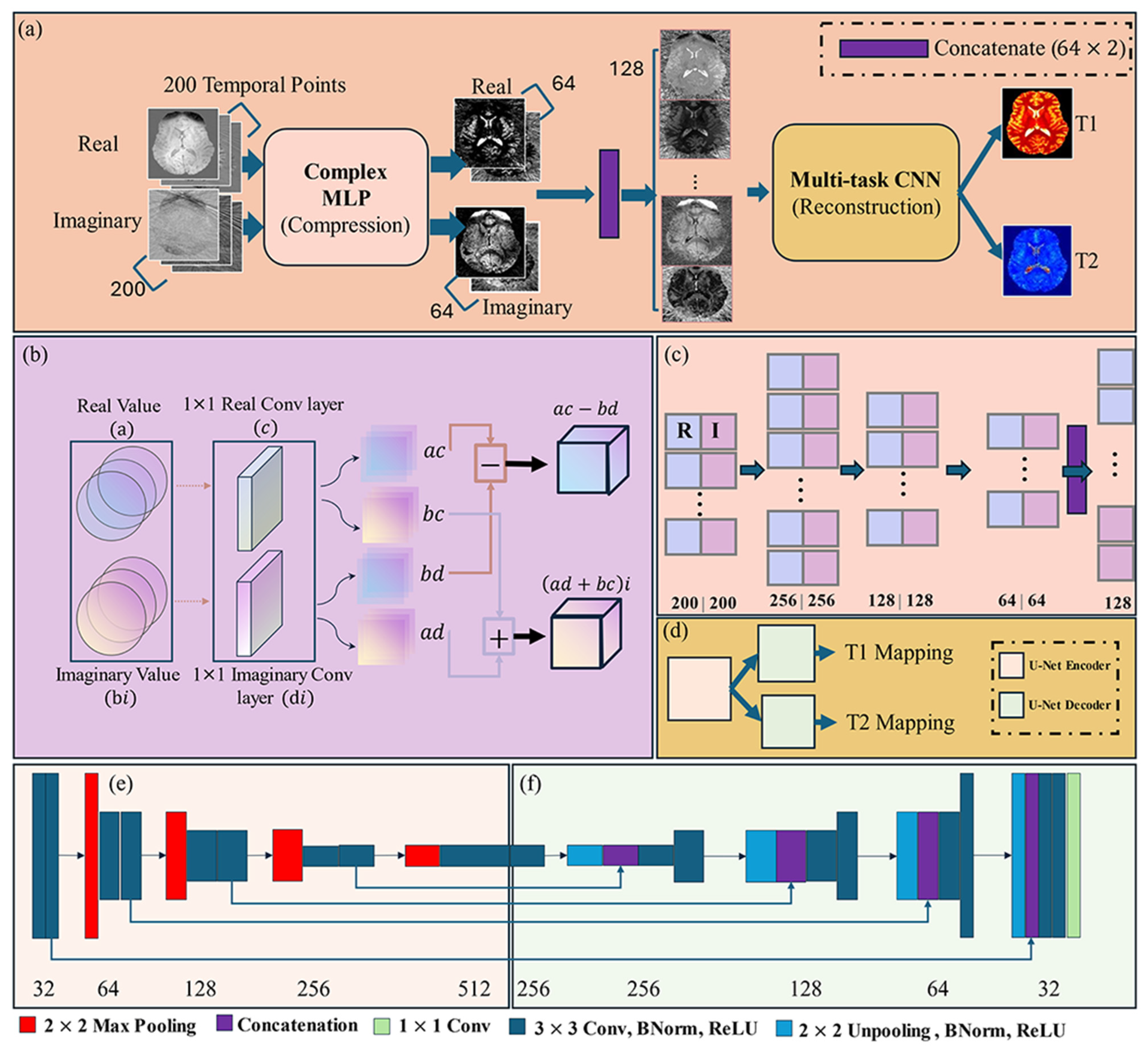
2.5. MRF-Mixer Neural Network
2.5.1. Complex-Valued Multi-Layer Perceptron (cMLP)
| Algorithm 1 MRF-Mixer Complex-MLP Block | |||
| Input: | |||
| Hyperparameters: | |||
| EncodingDepth = 2, | |||
| 1 | Step 1: Initial Complex Convolution | ||
| 2 | |||
| 3 | for to EncodingDepth do | ||
| 4 | |||
| 5 | Step 2: Complex Conv Layer with output channels | ||
| 6 | |||
| 7 | Step 3: Residual Connection | ||
| 8 | |||
| 9 | end for | ||
| Output: | |||
| Algorithm 2 Complex Convolution (CCovnLayer) with Kernel Size | |||
| Input: Complex feature map | |||
| Trainable Parameters: | |||
| Complex convolution weights | |||
| 1 | Step 1: Complex Convolution | ||
| 2 | Compute | ||
| 3 | ➢ denotes standard real-valued convolution | ||
| 4 | Compute | ||
| 5 | ➢ denotes standard real-valued convolution | ||
| 6 | Step 2: Batch Normalization | ||
| 7 | |||
| 8 | |||
| Output: | |||
2.5.2. Multi-Task CNN (U-Net)
| Algorithm 3 Multi-Task U-Net | ||||
| Input: | ➢ Output from cMLP | |||
| Hyperparameters: | ||||
| EncodingDepth = 4, | ||||
| 1 | unet Unet (EncodingDepth, In channels, Out channels) | |||
| 2 | Step 1: Encoding in U-Net: | |||
| 3 | ||||
| 4 | Step 2: Multi Decoders for T1, T2, B0 & PVs: | |||
| Output: | ➢ Only T1, T2 maps in this work | |||
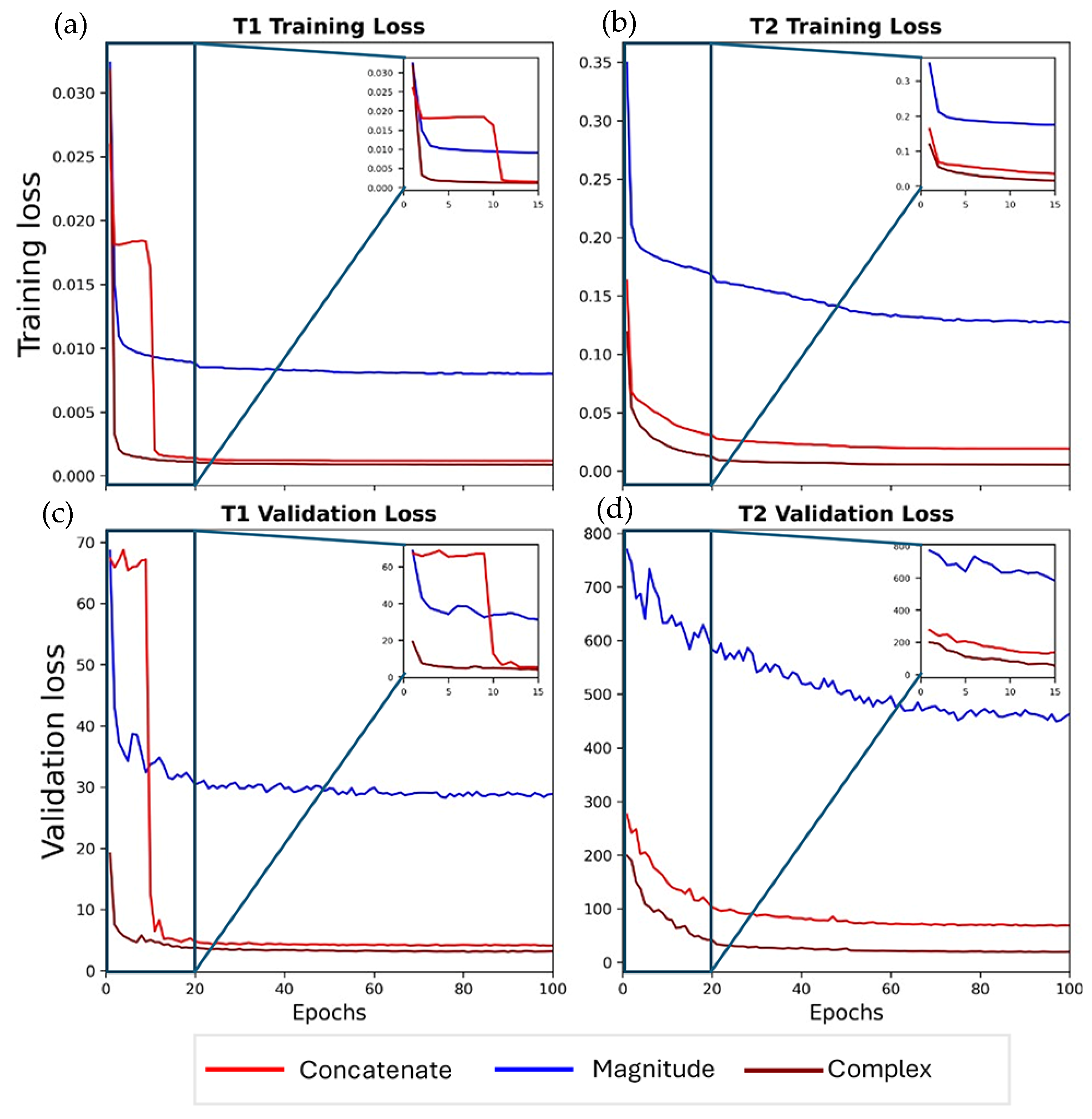
2.6. Network Training
2.7. Evaluation Experiments
2.7.1. Simulation Dataset
2.7.2. In Vivo Experiments
2.7.3. Comparison with Other Methods
2.8. Evaluation Metrics
3. Results
3.1. Ablation Study
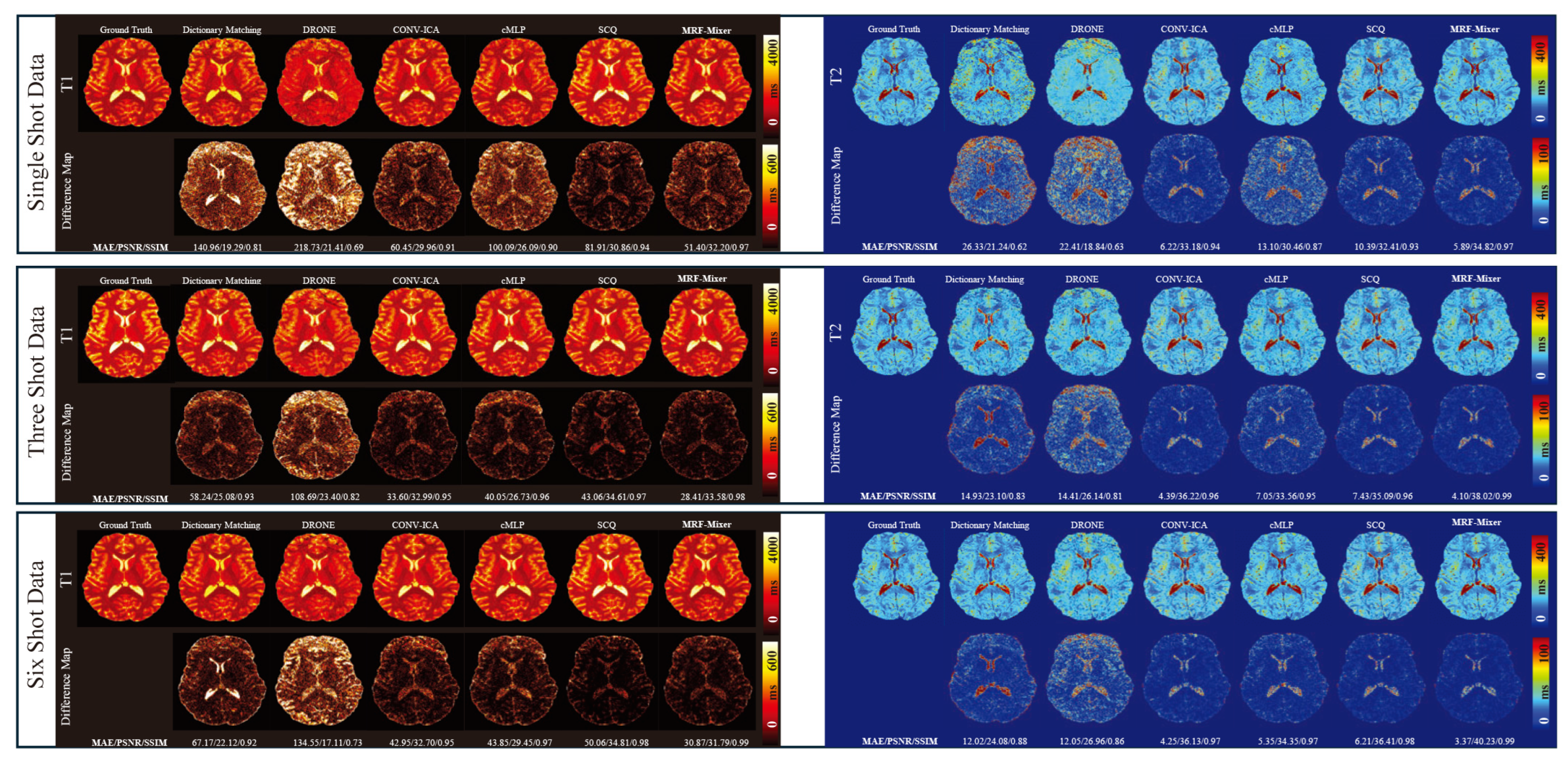
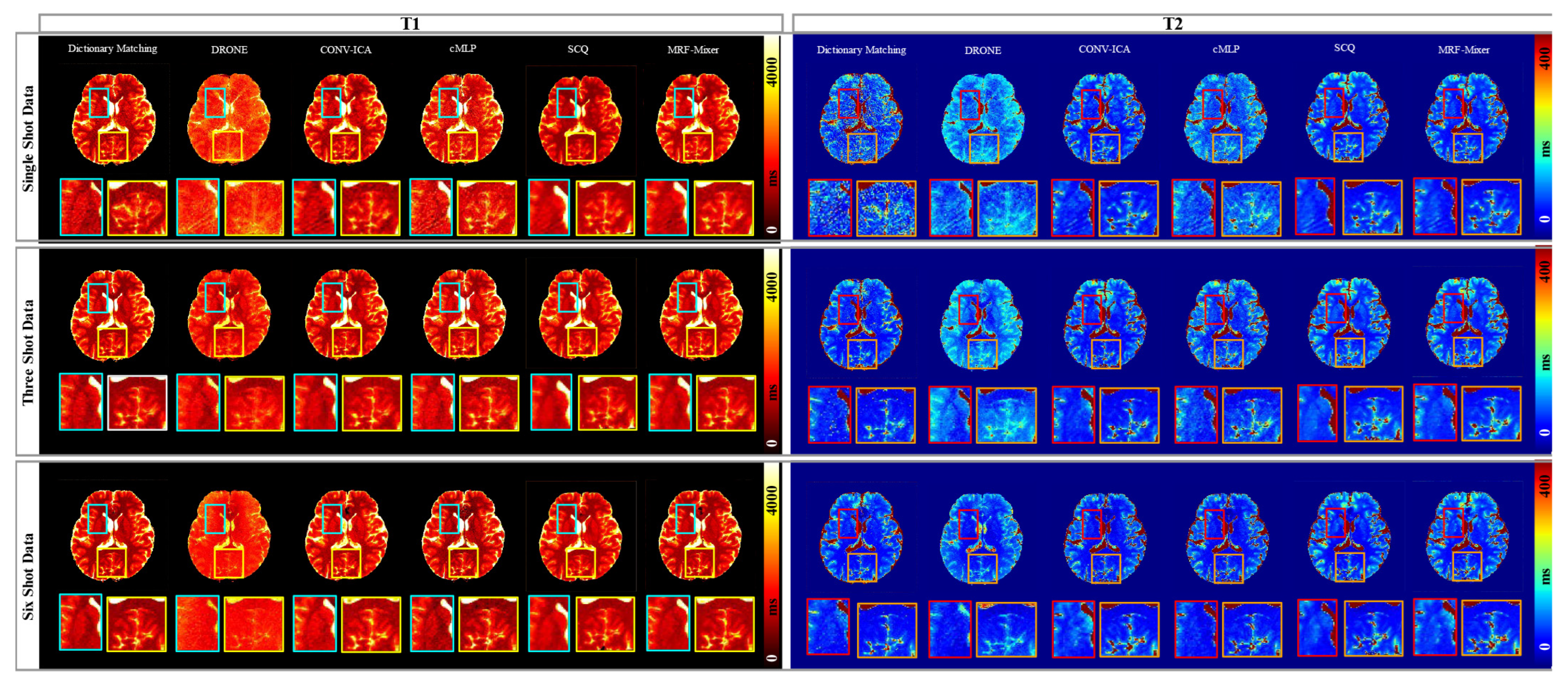
3.2. Simulation Results
3.3. In Vivo Experimental Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Cashmore, M.T.; McCann, A.J.; Wastling, S.J.; McGrath, C.; Thornton, J.; Hall, M.G. Clinical quantitative MRI and the need for metrology. Br. J. Radiol. 2021, 94, 20201215. [Google Scholar] [CrossRef] [PubMed]
- Gómez, P.A.; Cencini, M.; Golbabaee, M.; Schulte, R.F.; Pirkl, C.; Horvath, I.; Fallo, G.; Peretti, L.; Tosetti, M.; Menze, B.H.; et al. Rapid three-dimensional multiparametric MRI with quantitative transient-state imaging. Sci. Rep. 2020, 10, 13769. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Gulani, V.; Seiberlich, N.; Liu, K.; Sunshine, J.L.; Duerk, J.L.; Griswold, M.A. Magnetic resonance fingerprinting. Nature 2013, 495, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Deshmane, A.; McGivney, D.F.; Ma, D.; Jiang, Y.; Badve, C.; Gulani, V.; Seiberlich, N.; Griswold, M.A. Partial volume mapping using magnetic resonance fingerprinting. NMR Biomed. 2019, 32, e4082. [Google Scholar] [CrossRef]
- McGivney, D.F.; Boyacıoğlu, R.; Jiang, Y.; Poorman, M.E.; Seiberlich, N.; Gulani, V.; Keenan, K.E.; Griswold, M.A.; Ma, D. Magnetic resonance fingerprinting review part 2: Technique and directions. J. Magn. Reson. Imaging 2020, 51, 993–1007. [Google Scholar] [CrossRef]
- Cauley, S.F.; Setsompop, K.; Ma, D.; Jiang, Y.; Ye, H.; Adalsteinsson, E.; Griswold, M.A.; Wald, L.L. Fast group matching for MR fingerprinting reconstruction. Magn. Reson. Med. 2015, 74, 523–528. [Google Scholar] [CrossRef]
- McGivney, D.F.; Pierre, E.; Ma, D.; Jiang, Y.; Saybasili, H.; Gulani, V.; Griswold, M.A. SVD Compression for Magnetic Resonance Fingerprinting in the Time Domain. IEEE Trans. Med Imaging 2014, 33, 2311–2322. [Google Scholar] [CrossRef]
- Zhao, B.; Setsompop, K.; Adalsteinsson, E.; Gagoski, B.; Ye, H.; Ma, D.; Jiang, Y.; Ellen Grant, P.; Griswold, M.A.; Wald, L.L. Improved magnetic resonance fingerprinting reconstruction with low-rank and subspace modeling. Magn. Reason. Med. 2018, 79, 933–942. [Google Scholar] [CrossRef]
- Assländer, J.; Cloos, M.A.; Knoll, F.; Sodickson, D.K.; Hennig, J.; Lattanzi, R. Low rank alternating direction method of multipliers reconstruction for MR fingerprinting. Magn. Reson. Med. 2018, 79, 83–96. [Google Scholar] [CrossRef]
- Yang, M.; Ma, D.; Jiang, Y.; Hamilton, J.; Seiberlich, N.; Griswold, M.A.; McGivney, D. Low rank approximation methods for MR fingerprinting with large scale dictionaries. Magn. Reson. Med. 2018, 79, 2392–2400. [Google Scholar] [CrossRef]
- Cohen, O.; Zhu, B.; Rosen, M.S. MR fingerprinting deep reconstruction network (DRONE). Magn. Reason. Med. 2018, 80, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Chen, Y.; Liu, M.; Xiang, L.; Zhang, Q.; Wang, Q.; Lin, W.; Shen, D. Deep Learning for Fast and Spatially Constrained Tissue Quantification From Highly Accelerated Data in Magnetic Resonance Fingerprinting. IEEE Trans. Med. Imaging 2019, 38, 2364–2374. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, M.; Brizi, L.; Giampieri, E.; Solera, F.; Manners, D.N.; Castellani, G.; Testa, C.; Remondini, D. A deep learning approach for magnetic resonance fingerprinting: Scaling capabilities and good training practices investigated by simulations. Phys. Med. 2021, 89, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fang, Z.; Hung, S.-C.; Chang, W.-T.; Shen, D.; Lin, W. High-resolution 3D MR Fingerprinting using parallel imaging and deep learning. NeuroImage 2020, 206, 116329. [Google Scholar] [CrossRef]
- Golbabaee, M.; Chen, D.; Gomez, P.A.; Menzel, M.I.; Davies, M.E. Geometry of Deep Learning for Magnetic Resonance Fingerprinting. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 7825–7829. [Google Scholar]
- Lundervold, S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Hoppe, E.; Körzdörfer, G.; Würfl, T.; Wetzl, J.; Lugauer, F.; Pfeuffer, J.; Maier, A. Deep learning for magnetic resonance fingerprinting: A new approach for predicting quantitative parameter values from time series. In German Medical Data Sciences: Visions and Bridges; IOS Press: Oldenburg, Germany, 2017; Volume 243, pp. 202–206. [Google Scholar] [CrossRef]
- Oksuz, I.; Cruz, G.; Clough, J.; Bustin, A.; Fuin, N.; Botnar, R.M.; King, A.P.; Schnabel, J.A. Magnetic Resonance Fingerprinting Using Recurrent Neural Networks. In Proceedings of the IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1537–1540. [Google Scholar]
- Soyak, R.; Navruz, E.; Ersoy, E.O.; Cruz, G.; Prieto, C.; King, A.P.; Unay, D.; Oksuz, I. Channel Attention Networks for Robust MR Fingerprint Matching. IEEE Trans. Biomed. Eng. 2022, 69, 1398–1405. [Google Scholar] [CrossRef]
- Balsiger, F.; Konar, A.S.; Chikop, S.; Chandran, V.; Scheidegger, O.; Geethanath, S.; Reyes, M. Magnetic resonance fingerprinting reconstruction via spatiotemporal convolutional neural networks. In Machine Learning for Medical Image Reconstruction; Springer: Cham, Switzerland, 2018; pp. 39–46. [Google Scholar]
- Cao, P.; Cui, D.; Vardhanabhuti, V.; Hui, E.S. Development of fast deep learning quantification for magnetic resonance fingerprinting in vivo. Magn. Reson. Imaging 2020, 70, 81–90. [Google Scholar] [CrossRef]
- Song, P.; Eldar, Y.C.; Mazor, G.; Rodrigues, M.R.D. Magnetic Resonance Fingerprinting Using a Residual Convolutional Neural Network. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1040–1044. [Google Scholar]
- Li, P.; Hu, Y. Deep magnetic resonance fingerprinting based on Local and Global Vision Transformer. Med. Image Anal. 2024, 95, 103198. [Google Scholar] [CrossRef]
- Li, P.; Hu, Y. Deep graph embedding based on Laplacian eigenmaps for MR fingerprinting reconstruction. Med. Image Anal. 2025, 101, 103481. [Google Scholar] [CrossRef]
- Gao, Y.; Ding, T.; Cloos, M.; Sun, H. MRF-mixer: A self-supervised deep learning MRF framework. In Proceedings of the 2023 ISMRM & ISMRT Annual Meeting, Toronto, ON, Canada, 3–8 June 2023. [Google Scholar]
- Ding, T.; Gao, Y.; Xiong, Z.; Cloos, M.; Sun, H. Multi Complex-valued Spatio-temporal Fusion Networks for Robust MRF Reconstruction. In Proceedings of the 2024 ISMRM & ISMRT Annual Meeting, Singapore, 4–9 May 2024. [Google Scholar]
- Cloos, M.A.; Assländer, J.; Abbas, B.; Fishbaugh, J.; Babb, J.S.; Gerig, G.; Lattanzi, R. Rapid Radial T1 and T2 Mapping of the Hip Articular Cartilage with Magnetic Resonance Fingerprinting. J. Magn. Reson. Imaging 2019, 50, 810–815. [Google Scholar] [CrossRef]
- Sun, H.; Cleary, J.O.; Glarin, R.; Kolbe, S.C.; Ordidge, R.J.; Moffat, B.A.; Pike, G.B. Extracting more for less: Multi-echo MP2RAGE for simultaneous T1-weighted imaging, T1 mapping, mapping, SWI, and QSM from a single acquisition. Magn. Reson. Med. 2020, 83, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- El-Rewaidy, H.; Neisius, U.; Mancio, J.; Kucukseymen, S.; Rodriguez, J.; Paskavitz, A.; Menze, B.; Nezafat, R. Deep complex convolutional network for fast reconstruction of 3D late gadolinium enhancement cardiac MRI. NMR Biomed. 2020, 33, e4312. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cheng, H.; Ying, L.; Xiao, T.; Ke, Z.; Zheng, H.; Liang, D. DeepcomplexMRI: Exploiting deep residual network for fast parallel MR imaging with complex convolution. Magn. Reson. Imaging 2020, 68, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cloos, M.; Liu, F.; Crozier, S.; Pike, G.B.; Sun, H. Accelerating quantitative susceptibility and R2* mapping using incoherent undersampling and deep neural network reconstruction. NeuroImage 2021, 240, 118404. [Google Scholar] [CrossRef]
- Ma, S.; Wang, N.; Fan, Z.; Kaisey, M.; Sicotte, N.L.; Christodoulou, A.G.; Li, D. Three-dimensional whole-brain simultaneous T1, T2, and T1ρ quantification using MR Multitasking: Method and initial clinical experience in tissue characterization of multiple sclerosis. Magn. Reson. Med. 2021, 85, 1938–1952. [Google Scholar] [CrossRef]
- Deoni, S.C. High-resolution T1 mapping of the brain at 3T with driven equilibrium single pulse observation of T1 with high-speed incorporation of RF field inhomogeneities (DESPOT1-HIFI). J. Magn. Reson. Imaging 2007, 26, 1106–1111. [Google Scholar] [CrossRef]
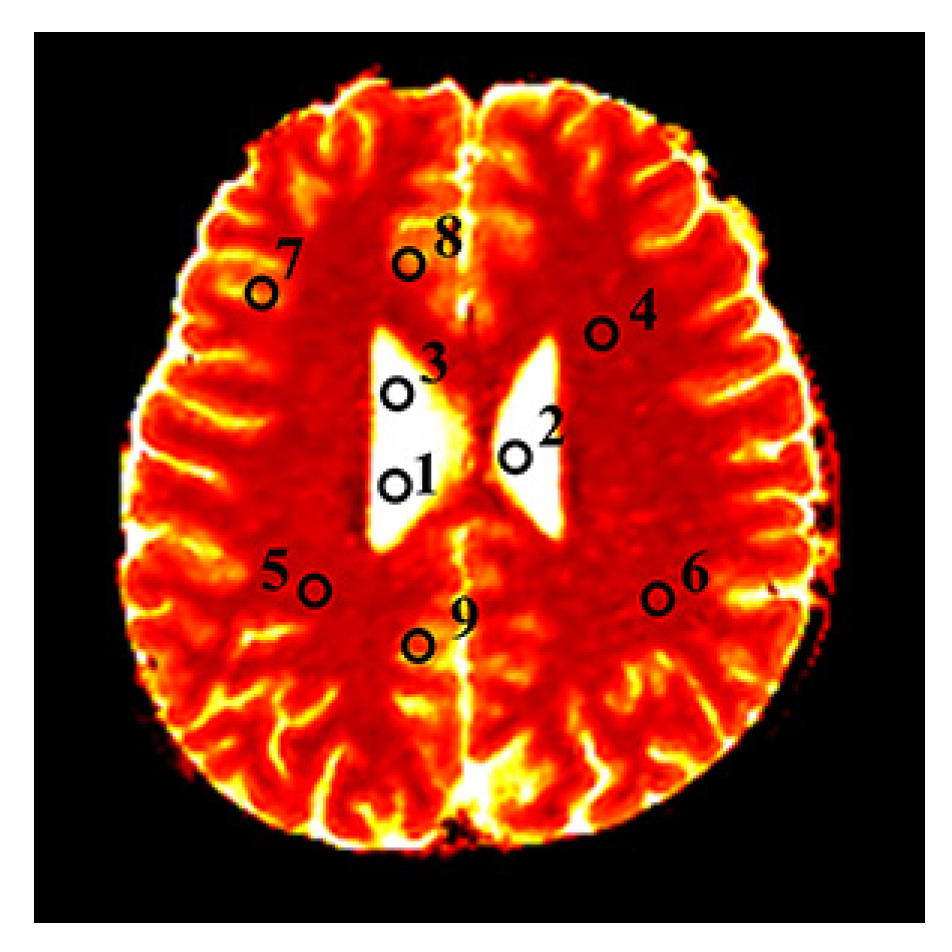
| Model | Property | MAE () | PSNR (dB) () | SSIM () | RMSE () |
|---|---|---|---|---|---|
| cMLP (without U-Net) | T1 | ||||
| T2 | |||||
| Single-Branch (single decoder U-Net) | T1 | *** | *** | *** | *** |
| T2 | |||||
| MRF-Mixer (multi decoder U-Net) | T1 | ||||
| T2 |
| Shot | Model | T1 Metrics | |||
|---|---|---|---|---|---|
| MAE () | PSNR () | SSIM () | RMSE () | ||
| 1-shot | MRF-Mixer | 48.63 5.03 | 30.81 1.35 | 0.96 0.00 | 102.01 9.63 |
| cMLP | 94.56 10.36 | 21.70 1.81 | 0.88 0.02 | 179.71 18.95 | |
| CONV-ICA | 58.04 8.12 *** | 27.5 2.49 *** | 0.81 0.06 *** | 116.97 13.75 *** | |
| DRONE | 221.86 33.19 | 16.57 1.65 | 0.41 0.06 | 404.07 50.95 | |
| DM | 105.96 13.37 | 23.31 0.65 | 0.85 0.01 | 260.29 25.64 | |
| SCQ | 76.34 | 30.13 0.69 | 0.92 0.01 | 120.87 13.00 | |
| 3-shot | MRF-Mixer | 33.42 4.86 | 33.01 1.91 | 0.98 0.00 | 78.68 9.53 |
| cMLP | 46.06 8.62 | 25.99 2.65 | 0.95 0.02 | 103.72 15.38 | |
| CONV-ICA | 39.92 6.27 *** | 29.59 2.45 *** | 0.97 0.01 *** | 88.27 10.95 *** | |
| DRONE | 126.09 21.20 | 16.90 2.02 | 0.53 0.02 | 275.60 42.21 | |
| DM | 61.64 9.26 | 25.95 0.92 | 0.93 0.01 | 182.62 23.60 | |
| SCQ | 52.41 9.73 | 33.21 1.03 | 0.96 0.01 | 85.22 12.90 | |
| 6-shot | MRF-Mixer | 28.80 4.51 | 33.48 1.83 | 0.98 0.01 | 72.90 9.19 |
| cMLP | 40.46 7.95 | 26.45 2.80 | 0.96 0.01 | 95.32 14.36 | |
| CONV-ICA | 35.98 6.02 *** | 30.56 2.35 *** | 0.83 0.08 *** | 83.93 10.87 *** | |
| DRONE | 117.64 28.02 | 18.10 1.70 | 0.55 0.04 | 239.19 43.61 | |
| DM | 55.10 8.25 | 26.79 0.97 | 0.94 0.01 | 162.76 21.18 | |
| SCQ | 44.47 8.36 | 34.33 1.01 | 0.97 0.01 | 74.84 11.01 | |
| Shot | Model | T2 Metrics | |||
|---|---|---|---|---|---|
| MAE () | PSNR () | SSIM () | RMSE () | ||
| 1-shot | MRF-Mixer | 9.25 ± 3.34 | 31.09 ± 2.61 | 0.95 ± 0.02 | 24.00 ± 7.97 |
| cMLP | 16.84 ± 3.90 | 27.65 ± 2.10 | 0.87 ± 0.02 | 35.68 ± 8.53 | |
| CONV-ICA | 9.04 ± 3.08 *** | 31.25 ± 2.07 | 0.90 ± 0.04 *** | 22.43 ± 6.94 *** | |
| DRONE | 28.31 ± 6.97 | 20.65 ± 1.35 | 0.42 ± 0.04 | 58.83 ± 15.35 | |
| DM | 32.60 ± 6.06 | 19.11 ± 0.67 | 0.65 ± 0.03 | 90.88 ± 8.11 | |
| SCQ | 14.49 ± 4.72 | 29.40 ± 2.40 | 0.93 ± 0.02 | 30.20 ± 9.06 | |
| 3-shot | MRF-Mixer | 5.99 ± 2.25 | 34.47 ± 2.69 | 0.98 ± 0.01 | 16.24 ± 5.57 |
| cMLP | 8.67 ± 2.69 | 32.10 ± 1.95 | 0.96 ± 0.01 | 20.81 ± 5.69 | |
| CONV-ICA | 5.97 ± 2.03 | 34.63 ± 2.37 | 0.97 ± 0.03 *** | 15.28 ± 4.81 *** | |
| DRONE | 15.58 ± 4.38 | 22.32 ± 3.53 | 0.57 ± 0.06 | 34.79 ± 9.49 | |
| DM | 20.40 ± 6.42 | 21.22 ± 1.16 | 0.84 ± 0.01 | 74.59 ± 11.67 | |
| SCQ | 9.76 ± 3.20 | 32.66 ± 2.38 | 0.97 ± 0.01 | 20.76 ± 6.33 | |
| 6-shot | MRF-Mixer | 4.97 ± 1.87 | 35.90 ± 2.48 | 0.98 ± 0.02 | 13.67 ± 4.62 |
| cMLP | 6.90 ± 2.35 | 33.55 ± 2.33 | 0.97 ± 0.01 | 17.71 ± 5.34 | |
| CONV-ICA | 5.21 ± 1.79 *** | 35.33 ± 2.42 | 0.93 ± 0.04 *** | 13.59 ± 4.27 | |
| DRONE | 11.72 ± 3.14 | 25.00 ± 2.80 | 0.64 ± 0.05 | 26.33 ± 6.72 | |
| DM | 18.54 ± 6.53 | 21.96 ± 1.40 | 0.86 ± 0.01 | 69.71 ± 12.85 | |
| SCQ | 8.45 ± 2.89 | 33.75 ± 2.56 | 0.97 ± 0.01 | 18.42 ± 5.90 | |
| Method | DM | DRONE | CONV-ICA | cMLP | SCQ | MRF-Mixer | |
|---|---|---|---|---|---|---|---|
| CSF | 1-shot | 3988.89 ± 21.71 | 3158.63 ± 289.79 | 4393.15 ± 277.23 | 4363.95 ± 126.97 | 3957.03 ± 141.40 | 3961.73 ± 81.46 |
| 3-shot | 3983.33 ± 42.83 | 2615.96 ± 320.37 | 4009.75 ± 121.58 | 4251.35 ± 132.43 | 3859.29 ± 153.10 | 3938.06 ± 82.70 | |
| 6-shot | 4000.00 ± 0.00 | 3712.80 ± 283.22 | 4700.96 ± 113.69 | 4270.01 ± 113.10 | 4092.38 ± 48.87 | 3817.02 ± 51.93 | |
| GM | 1-shot | 1571.30 ± 196.88 | 1599.84 ± 85.38 | 1588.21 ± 138.15 | 1703.32 ± 219.84 | 1569.50 ± 111.99 | 1585.94 ± 116.21 |
| 3-shot | 1496.30 ± 106.71 | 1524.83 ± 90.97 | 1516.34 ± 98.45 | 1574.81 ± 104.45 | 1546.38 ± 90.60 | 1582.99 ± 103.90 | |
| 6-shot | 1436.11 ± 126.89 | 1613.06 ± 75.64 | 1459.22 ± 103.03 | 1440.22 ± 118.10 | 1463.40 ± 30.23 | 1486.05 ± 99.85 | |
| WM | 1-shot | 909.28 ± 134.35 | 1288.99 ± 175.66 | 912.35 ± 60.17 | 1157.50 ± 129.68 | 946.76 ± 19.12 | 969.52 ± 48.33 |
| 3-shot | 935.42 ± 71.13 | 1151.71 ± 54.82 | 963.98 ± 66.02 | 987.41 ± 110.88 | 965.79 ± 45.52 | 956.03 ± 38.14 | |
| 6-shot | 903.47 ± 66.37 | 1361.15 ± 91.60 | 882.75 ± 84.03 | 875.16 ± 71.46 | 927.48 ± 40.06 | 916.90 ± 38.26 |
| Method | DM | DRONE | CONV-ICA | cMLP | SCQ | MRF-Mixer | |
|---|---|---|---|---|---|---|---|
| CSF | 1-shot | 590.56 ± 98.67 | 509.70 ± 75.66 | 798.48 ± 40.98 | 646.65 ± 67.85 | 870.35 ± 37.90 | 773.82 ± 47.57 |
| 3-shot | 586.94 ± 45.24 | 450.44 ± 64.00 | 981.58 ± 55.45 | 774.48 ± 33.01 | 906.94 ± 26.82 | 883.86 ± 40.69 | |
| 6-shot | 565.28 ± 33.09 | 595.98 ± 54.46 | 1092.98 ± 37.41 | 774.48 ± 33.01 | 920.62 ± 20.78 | 993.97 ± 28.44 | |
| GM | 1-shot | 108.61 ± 47.19 | 111.58 ± 13.41 | 112.61 ± 18.16 | 124.27 ± 18.76 | 113.15 ± 17.76 | 120.60 ± 18.97 |
| 3-shot | 98.70 ± 21.00 | 137.77 ± 13.63 | 105.74 ± 15.46 | 120.07 ± 21.95 | 114.72 ± 13.99 | 117.19 ± 17.55 | |
| 6-shot | 83.61 ± 9.61 | 108.01 ± 14.87 | 100.14 ± 13.09 | 120.07 ± 21.95 | 101.18 ± 12.08 | 101.96 ± 12.69 | |
| WM | 1-shot | 76.64 ± 43.55 | 76.27 ± 16.43 | 56.76 ± 5.79 | 82.48 ± 15.61 | 59.30 ± 3.56 | 60.38 ± 5.54 |
| 3-shot | 56.35 ± 20.13 | 90.64 ± 6.51 | 55.38 ± 5.21 | 67.15 ± 10.06 | 61.81 ± 3.79 | 62.35 ± 4.17 | |
| 6-shot | 49.22 ± 7.34 | 76.55 ± 12.98 | 52.69 ± 5.80 | 67.15 ± 10.06 | 52.03 ± 3.04 | 52.99 ± 2.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, T.; Gao, Y.; Xiong, Z.; Liu, F.; Cloos, M.A.; Sun, H. MRF-Mixer: A Simulation-Based Deep Learning Framework for Accelerated and Accurate Magnetic Resonance Fingerprinting Reconstruction. Information 2025, 16, 218. https://doi.org/10.3390/info16030218
Ding T, Gao Y, Xiong Z, Liu F, Cloos MA, Sun H. MRF-Mixer: A Simulation-Based Deep Learning Framework for Accelerated and Accurate Magnetic Resonance Fingerprinting Reconstruction. Information. 2025; 16(3):218. https://doi.org/10.3390/info16030218
Chicago/Turabian StyleDing, Tianyi, Yang Gao, Zhuang Xiong, Feng Liu, Martijn A. Cloos, and Hongfu Sun. 2025. "MRF-Mixer: A Simulation-Based Deep Learning Framework for Accelerated and Accurate Magnetic Resonance Fingerprinting Reconstruction" Information 16, no. 3: 218. https://doi.org/10.3390/info16030218
APA StyleDing, T., Gao, Y., Xiong, Z., Liu, F., Cloos, M. A., & Sun, H. (2025). MRF-Mixer: A Simulation-Based Deep Learning Framework for Accelerated and Accurate Magnetic Resonance Fingerprinting Reconstruction. Information, 16(3), 218. https://doi.org/10.3390/info16030218