SCRATCH-AI: A Tool to Predict Honey Wound Healing Properties
Abstract
1. Introduction
Novelty and Contributions Summary
- SCRATCH-AI is a graphical tool that unifies Bayesian networks and decision trees to classify honey samples into four wound-healing categories from biochemical markers and botanical origin. Unlike prior software focused on scratch-assay measurement from microscopy images (PyScratch v. 2 [12]), SCRATCH-AI provides two powerful but easily interpretable ML instruments to reveal how biological features interact and impact the four categories.
- The graphical user interface (GUI) supports real-time validation and expert feature weighting, aligning with laboratory workflows for non-programmers users.
- The work emphasizes understudied local honeys (Piedmont (Italy): chestnut, coriander, sunflower) and reports 10-fold cross-validated results with per-class metrics and confusion matrices, yielding actionable biological insights alongside accurate predictions.
2. Related Work
3. Methods
3.1. Data Collection
- Chemicals and reagents. All reagents, if not otherwise specified, were purchased from Merck (Milan, Italy).
- Cell Culture. L-glutamine (200 mM), 10% fetal bovine serum (FBS, Euroclone, Pero, Italy), and antibiotics (pen/strep) were added to the DMEM (high glucose, 4.5 g/L) used to cultivate the human spontaneously immortalized HaCaT keratinocytes (RRID:CVCL_0038) at 37 °C in a 5% humidified atmosphere.
- Honey Type. The honeys were obtained from “Davide e le sue Api” (https://www.ilgolosario.it/it/miele-davide-taverna, (accessed on 1 June 2025)), a local beekeeper, and collected in the Piedmont area (Northwest Italy). We utilized three different types of honey, i.e., chestnut, coriander, and sunflower. Honey samples were sourced during the summer 2024 harvest. All honey varieties (e.g., chestnut, sunflower, coriander) were collected from a distinct honey batch, defined as all honey from a specific floral source collected from a single apiary during the same harvest period. Upon collection, all samples were immediately transferred to opaque, airtight containers to protect them from light exposure. They were subsequently stored at 4 °C until analysis to preserve their physicochemical properties and prevent the degradation of bioactive compounds.
- Total Phenolic Content (TPC) assay. We used the colorimetric in vitro Folin–Ciocalteu method to determine the amount of phenol present (TPC, total phenolic content) in the honey samples under investigation [7,8,9]. 250 of Folin Ciocalteu (10% v/v) was mixed with 100 of honey solution (1 gr in 3 mL of ). 100 of 7.5% (w/v) was added after 5 min, and the mixture was incubated at 50 °C for 15 min. After that, samples were washed with 2.5 milliliters of water. With a plate reader (Infinite 200 Pro, Tecan), the absorbance was measured at 620 nm. The results were represented in milligrams of gallic acid equivalents ( 100 ), and the standard curve was defined by known quantities of gallic acid.
- Total Flavonoid Content (TFC) assay. The total flavonoid content (TFC) of the honey samples was determined using a spectrophotometric method [7,8,9] based on the formation of a flavonoid-aluminum complex. Briefly, a 1 mL aliquot of honey solution (0.5 g/mL) was combined with 1 mL of a 2% aluminum chloride () solution. The mixture was incubated at 25 °C for 10 min. The absorbance was then measured at 415 nm using a plate reader (Infinite 200 Pro, Tecan). A calibration curve was established using quercetin as the standard, and the results were expressed as milligrams of quercetin equivalent () per 0.5 g of honey.DPPH assay. The antioxidant capacity of honeys was evaluated by the free radical scavenging ability of 2,2-Diphenyl-1-picrylhydrazyl (DPPH). The assay was performed by adding 1 mM DPPH solution to honeys (1 gr in 3 mL of ) followed by a 10-min incubation. The absorbance was determined at 512 nm in a plate reader (Infinite 200 Pro, Tecan). The radical scavenging activity was compared to water.
- Reactive oxygen species (ROS) content. As previously reported by [13,14], xylenol orange, a colorimetric test, was used to measure the amount of produced by honeys. In short, a solution comprising 25 mM ammonium ferrous sulphate in 0.25 M sulfuric acid was mixed with a 1:100 solution of 125 xylenol orange and 150 mM sorbitol to create a working solution. Next, 20 of the honey sample (1 gr in 3 mL ) was combined with 200 of the working solution. The absorbance was measured using a 96-well microplate reader (Infinite 200 Pro, Tecan).
- SCRATCH-score wound healing assay. A popular and easy “in vitro” method for examining the migration and proliferation of two-dimensional cell cultures is SCRATCH-score wound healing assay. It is based on the finding that when a confluent cell monolayer is exposed to an artificial gap, or scratch, the cells on the gap’s edge will multiply and migrate in the direction of the opening, gradually sealing the scratch until the monolayer is restored. In many physiological or pathological contexts, such as tissue development or wound healing, the behavior of various cellular lines can be easily and effectively studied using this “in vitro” technique, which mimics the migration and proliferation of cells “in vivo”.
3.2. SCRATCH-AI: A Tool for Predicting Wound Healing Properties
4. Results
4.1. A BN Model for SCRATCH CATEGORY Classification
4.2. A DT Model for SCRATCH CATEGORY Classification
4.3. GUI Evaluation
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bonsignore, G.; Martinotti, S.; Ranzato, E. Honey Bioactive Molecules: There Is a World Beyond the Sugars. BioTech 2024, 13, 47. [Google Scholar] [CrossRef]
- Erler, S.; Moritz, R.F.A. Pharmacophagy and pharmacophory: Mechanisms of self-medication and disease prevention in the honeybee colony (Apis mellifera). Apidologie 2016, 47, 389–411. [Google Scholar] [CrossRef]
- Palma-Morales, M.; Huertas, J.R.; Rodríguez-Pérez, C. A Comprehensive Review of the Effect of Honey on Human Health. Nutrients 2023, 15, 3056. [Google Scholar] [CrossRef] [PubMed]
- Martinotti, S.; Bonsignore, G.; Ranzato, E. Applications of Beehive Products for Wound Repair and Skin Care. Cosmetics 2023, 10, 127. [Google Scholar] [CrossRef]
- Oryan, A.; Alemzadeh, E.; Moshiri, A. Biological properties and therapeutic activities of honey in wound healing: A narrative review and meta-analysis. J. Tissue Viability 2016, 25, 98–118. [Google Scholar] [CrossRef] [PubMed]
- Yupanqui Mieles, J.; Vyas, C.; Aslan, E.; Humphreys, G.; Diver, C.; Bartolo, P. Honey: An Advanced Antimicrobial and Wound Healing Biomaterial for Tissue Engineering Applications. Pharmaceutics 2022, 14, 1663. [Google Scholar] [CrossRef]
- Martinotti, S.; Bucekova, M.; Majtan, J.; Ranzato, E. Honey: An Effective Regenerative Medicine Product in Wound Management. Curr. Med. Chem. 2019, 26, 5230–5240. [Google Scholar] [CrossRef] [PubMed]
- Alam, F.; Islam, M.A.; Gan, S.H.; Khalil, M.I. Honey: A Potential Therapeutic Agent for Managing Diabetic Wounds. Evid.-Based Complement. Altern. Med. 2014, 2014, 169130. [Google Scholar] [CrossRef] [PubMed]
- Miguel, M.; Antunes, M.; Faleiro, M. Honey as a Complementary Medicine. Integr. Med. Insights 2017, 12, 1178633717702869. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A. From Machine Learning to Explainable AI. In Proceedings of the 2018 World Symposium on Digital Intelligence for Systems and Machines (DISA), Košice, Slovakia, 23–25 August 2018; pp. 55–66. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2021, 23, 18. [Google Scholar] [CrossRef]
- Garcia-Fossa, F.; Gaal, V.; de Jesus, M.B. PyScratch: An ease of use tool for analysis of scratch assays. Comput. Methods Programs Biomed. 2020, 193, 105476. [Google Scholar] [CrossRef]
- Martinotti, S.; Laforenza, U.; Patrone, M.; Moccia, F.; Ranzato, E. Honey-Mediated Wound Healing: H2O2 Entry through AQP3 Determines Extracellular Ca2+ Influx. Int. J. Mol. Sci. 2019, 20, 764. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jin, Y.; Wei, Q.; Hu, Y.; Liu, L.; Feng, Y.; Jin, Y.; Jiang, Y. The relationship between aquaporins and skin diseases. Eur. J. Dermatol. 2023, 33, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Martinotti, S.; Bonsignore, G.; Patrone, M.; Ranzato, E. Correlation between Honey Parameters and Wound Healing Properties: The Case of Piedmont (Italy) Samples. Curr. Pharm. Biotechnol. 2025, 26, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Cianciosi, D.; Forbes-Hernández, T.Y.; Afrin, S.; Gasparrini, M.; Reboredo-Rodriguez, P.; Manna, P.P.; Zhang, J.; Bravo Lamas, L.; Martínez Flórez, S.; Agudo Toyos, P.; et al. Phenolic Compounds in Honey and Their Associated Health Benefits: A Review. Molecules 2018, 23, 2322. [Google Scholar] [CrossRef]
- Tashkandi, H. Honey in wound healing: An updated review. Open Life Sci. 2021, 16, 1091–1100. [Google Scholar] [CrossRef]
- López, B.; Latorre, M.; Fernández, M.; García, M.; García, S.; Herreroa, C. Chemometric classification of honeys according to their type based on quality control data. Food Chem. 1996, 55, 281–287. [Google Scholar] [CrossRef]
- Martinotti, S.; Pellavio, G.; Laforenza, U.; Ranzato, E. Propolis Induces AQP3 Expression: A Possible Way of Action in Wound Healing. Molecules 2019, 24, 1544. [Google Scholar] [CrossRef]
- Balko, S.; Kerr, E.; Buchel, E.; Logsetty, S.; Raouf, A. A Robust and Standardized Approach to Quantify Wound Closure Using the Scratch Assay. Methods Protoc. 2023, 6, 87. [Google Scholar] [CrossRef]
- Walter, M.N.M.; Wright, K.T.; Fuller, H.R.; MacNeil, S.; Johnson, W.E.B. Mesenchymal stem cell-conditioned medium accelerates skin wound healing: An in vitro study of fibroblast and keratinocyte scratch assays. Exp. Cell Res. 2010, 316, 1271–1281. [Google Scholar] [CrossRef]
- Charatan, Q.; Kans, A. Python Graphics with Tkinter. In Programming in Two Semesters: Using Python and Java; Springer International Publishing: Cham, Swizerland, 2022; pp. 211–254. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Kramer, O. Scikit-Learn. In Machine Learning for Evolution Strategies; Springer International Publishing: Cham, Swizerland, 2016; pp. 45–53. [Google Scholar] [CrossRef]
- Lucas, P.J.F.; van der Gaag, L.C.; Abu-Hanna, A. Bayesian networks in biomedicine and health-care. Artif. Intell. Med. 2004, 30, 201–214. [Google Scholar] [CrossRef]
- Cheng, J.; Greiner, R. Comparing Bayesian Network Classifiers. arXiv 2013, arXiv:1301.6684. [Google Scholar] [CrossRef]
- Friedman, N.; Geiger, D.; Goldszmidt, M. Bayesian Network Classifiers. Mach. Learn. 1997, 29, 131–163. [Google Scholar] [CrossRef]
- Heckerman, D.; Geiger, D.; Chickering, D.M. Learning Bayesian Networks: The Combination of Knowledge and Statistical Data. Mach. Learn. 1995, 20, 197–243. [Google Scholar] [CrossRef]
- Mantovani, R.G.; Horvath, T.; Cerri, R.; Vanschoren, J.; de Carvalho, A.C. Hyper-Parameter Tuning of a Decision Tree Induction Algorithm. In Proceedings of the 2016 5th Brazilian Conference on Intelligent Systems (BRACIS), Recife, Brazil, 9–12 October 2016; pp. 37–42. [Google Scholar] [CrossRef]
- Salzberg, S. Book Review: C4.5: Programs for Machine Learning by J. Ross Quinlan. Morgan Kaufmann Publishers, Inc., 1993. Mach. Learn. 1994, 16, 235–240. [Google Scholar] [CrossRef]
- Brooke, J. SUS—A quick and dirty usability scale. In Usability Evaluation in Industry; CRC Press: London, UK; Bristol, PA, USA, 1996; ISBN 9780748404605. [Google Scholar]
- Skobelev, D.O.; Zaytseva, T.M.; Kozlov, A.D.; Perepelitsa, V.L.; Makarova, A.S. Laboratory information management systems in the work of the analytic laboratory. Meas. Tech. 2011, 53, 1182–1189. [Google Scholar] [CrossRef]
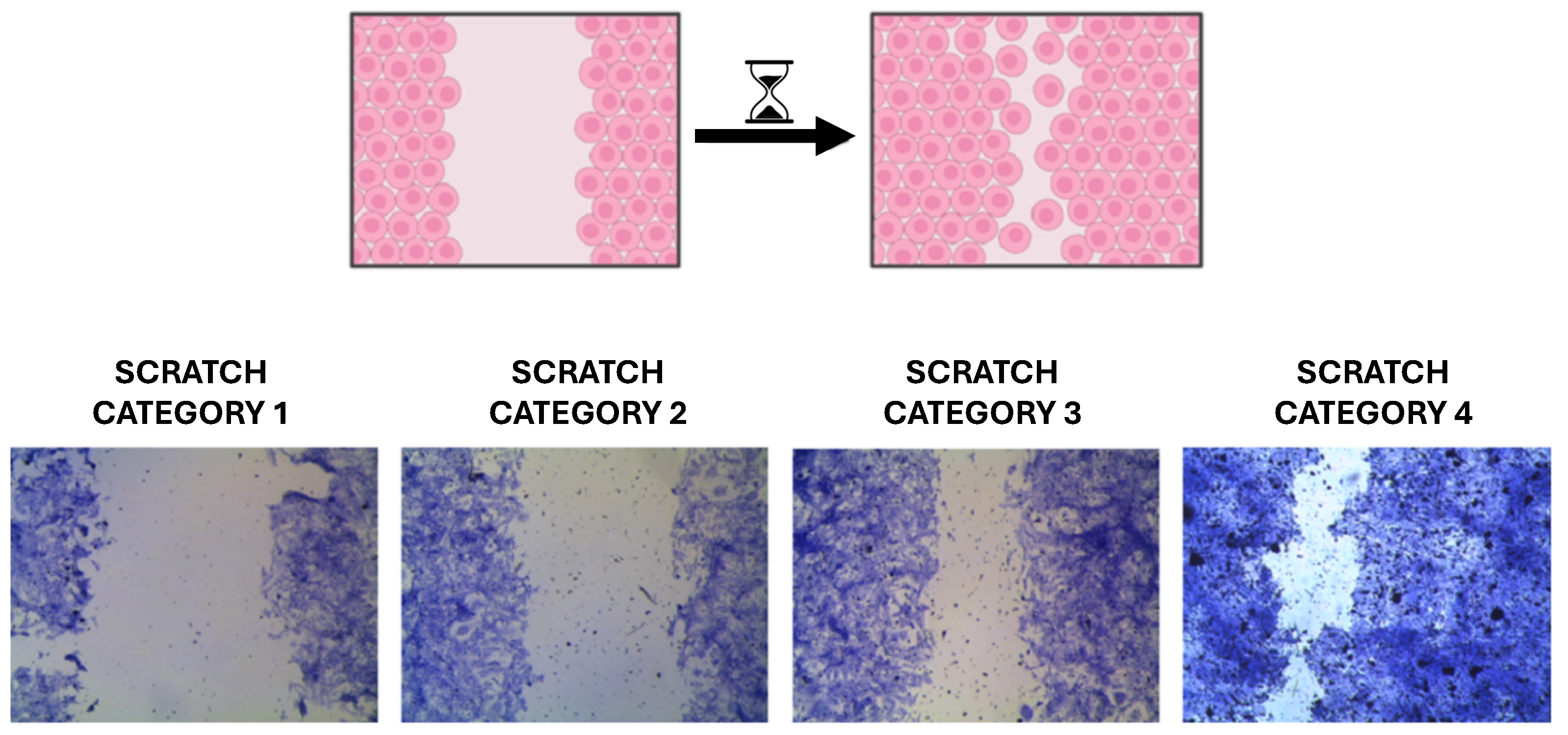

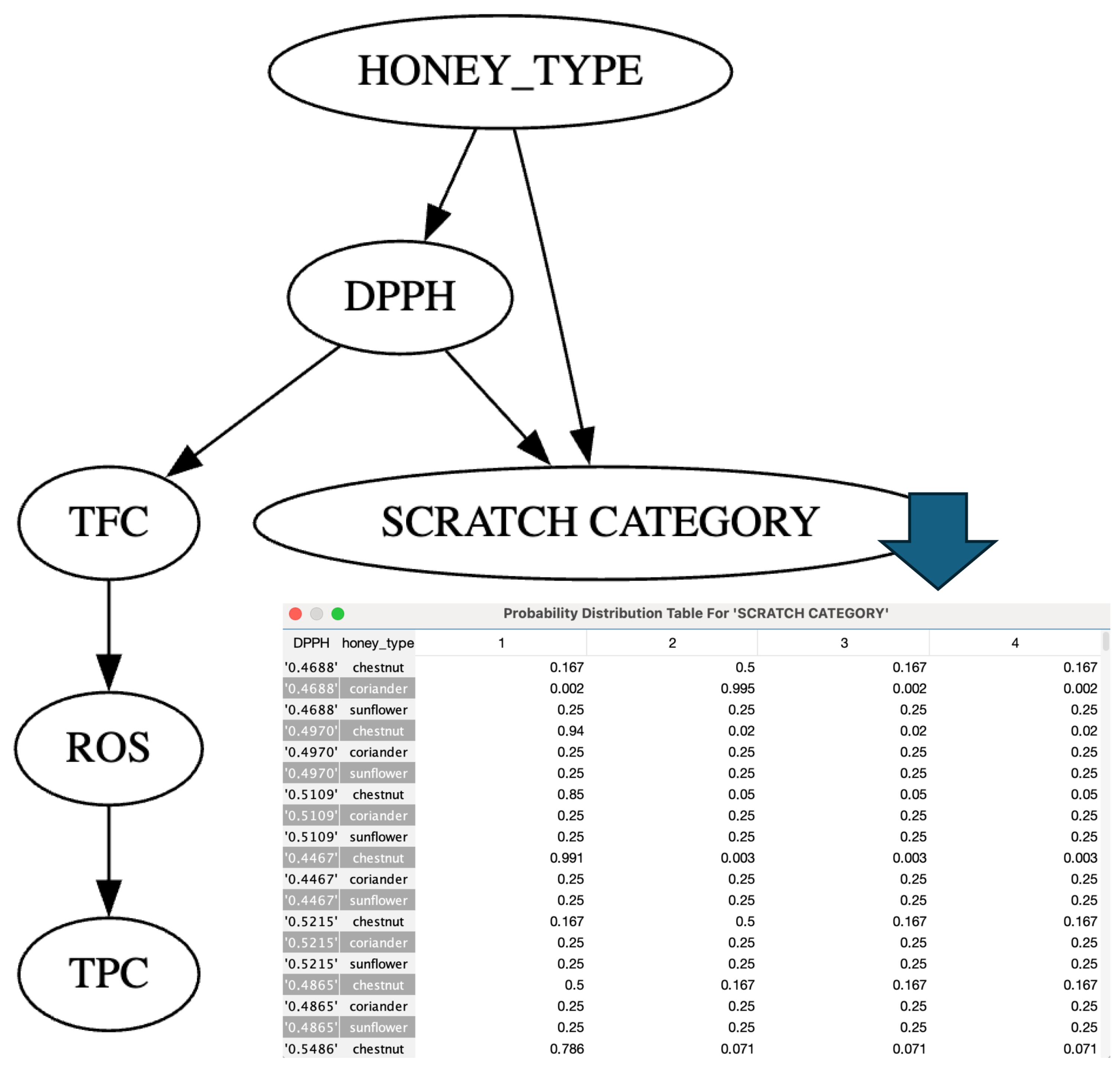
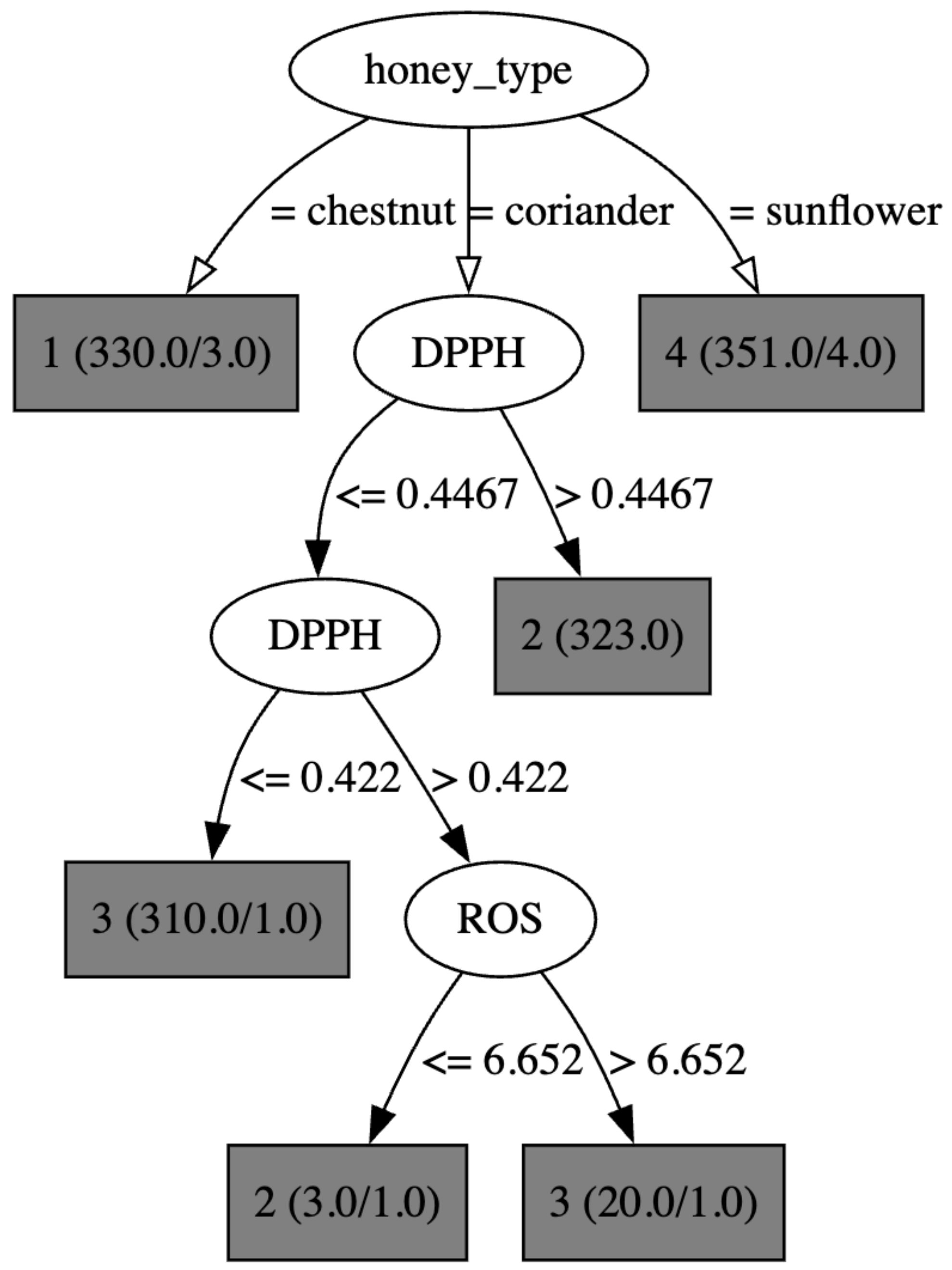
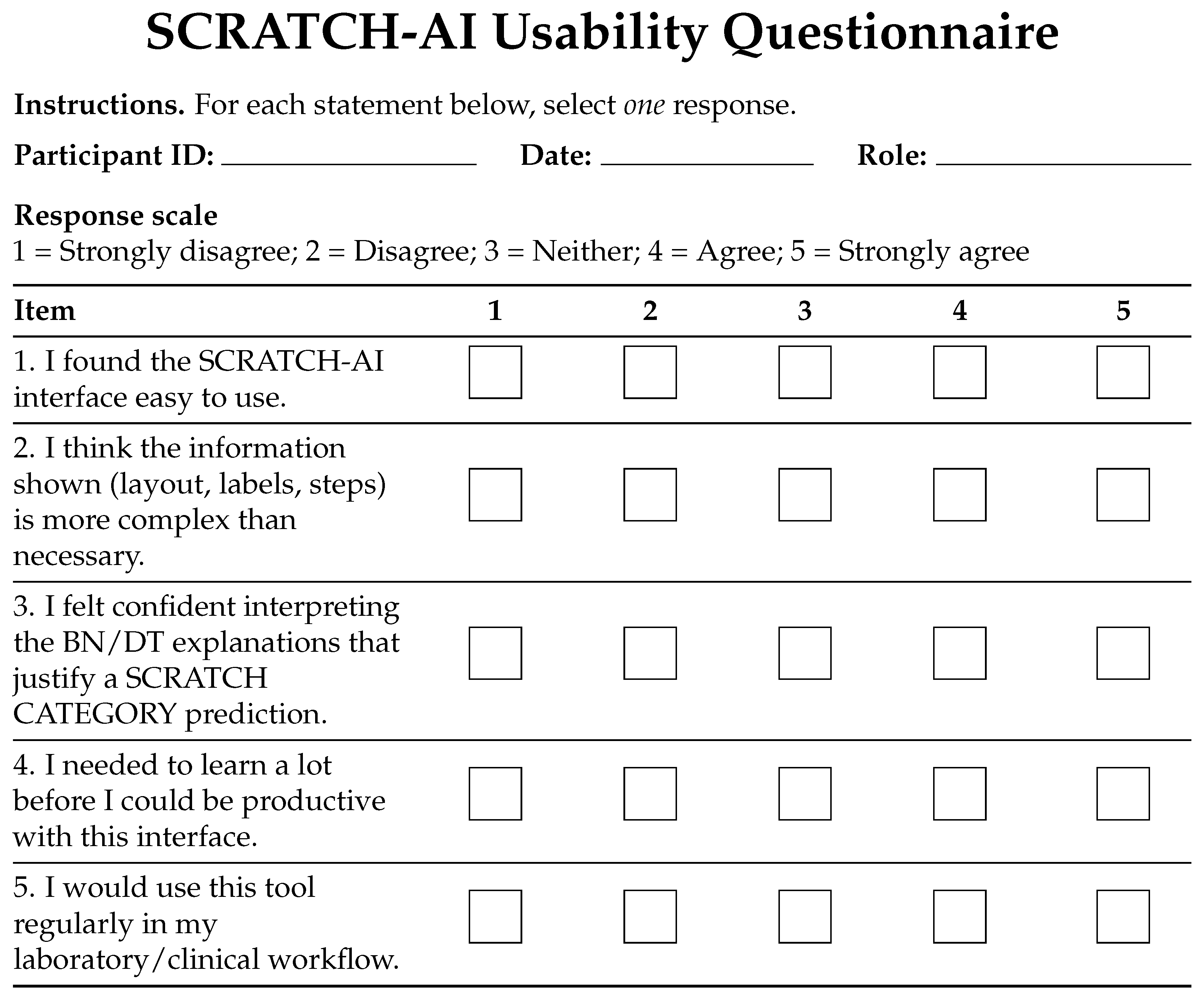
| Dimension | PyScratch ([12]) | SCRATCH-AI (This Work) |
|---|---|---|
| Primary objective | Automate scratch-closure measurement from images | Classify honey samples into wound-healing categories |
| Input modality | Microscopy images of scratch assays | Biochemical markers (DPPH, TFC, TPC, ROS) + HONEY TYPE |
| Core methods | Image processing by using OpenCV python library | Bayesian Networks (BN) + Decision Trees (DT) in one pipeline |
| Main outputs | Quantitative closure metrics (e.g., % wound area over time) | Predicted class (4 levels) + BN graph of dependencies + DT rules |
| GUI/usability | Interface tailored to image analysis workflow | Desktop GUI for non-programmers: real-time input checks, “show model” toggle, expert feature-weighting (0–1) |
| Domain emphasis | General scratch-assay image pipelines | Under-studied local honeys (e.g., chestnut, coriander, sunflower; beyond Manuka) |
| Evaluation focus | Accuracy/quality of closure measurement | Stratified 10-fold CV with per-class metrics, confusion matrices |
| Limitation addressed | Manual/subjectiveimage measurement | White-box ML: provides transparent dependencies and explainable information |
| Class | TP Rate | FP Rate | Precision | Recall | F-Measure | MCC | ROC Area | PRC Area | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.963 | 0.035 | 0.900 | 0.963 | 0.931 | 0.908 | 0.997 | 0.991 | |
| 2 | 0.976 | 0.010 | 0.970 | 0.976 | 0.973 | 0.964 | 0.998 | 0.996 | |
| 3 | 0.895 | 0.017 | 0.946 | 0.895 | 0.920 | 0.894 | 0.994 | 0.986 | |
| 4 | 0.966 | 0.005 | 0.985 | 0.966 | 0.975 | 0.967 | 0.997 | 0.991 | |
| Wt. Avg. | 0.950 | 0.016 | 0.951 | 0.950 | 0.950 | 0.934 | 0.997 | 0.991 | 0.9498 |
| Actual ↓ | ← Predicted | ||||
|---|---|---|---|---|---|
| 315 | 0 | 12 | 0 | ||
| 3 | 322 | 4 | 1 | ||
| 21 | 10 | 297 | 4 | ||
| 11 | 0 | 1 | 366 |
| Class | TP Rate | FP Rate | Precision | Recall | F-Measure | MCC | ROC Area | PRC Area | Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.917 | 0.001 | 0.997 | 0.917 | 0.955 | 0.943 | 0.997 | 0.988 | |
| 2 | 0.976 | 0.002 | 0.994 | 0.976 | 0.985 | 0.980 | 0.992 | 0.983 | |
| 3 | 0.907 | 0.003 | 0.990 | 0.907 | 0.947 | 0.931 | 0.992 | 0.977 | |
| 4 | 0.986 | 0.066 | 0.841 | 0.986 | 0.907 | 0.877 | 0.990 | 0.972 | |
| Wt.Avg. | 0.947 | 0.019 | 0.954 | 0.947 | 0.948 | 0.932 | 0.993 | 0.980 | 0.9469 |
| Actual ↓ | ← Predicted | ||||
|---|---|---|---|---|---|
| 300 | 0 | 0 | 27 | ||
| 1 | 322 | 0 | 7 | ||
| 0 | 0 | 301 | 31 | ||
| 0 | 2 | 3 | 343 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinotti, S.; Montani, S.; Ranzato, E.; Striani, M. SCRATCH-AI: A Tool to Predict Honey Wound Healing Properties. Information 2025, 16, 827. https://doi.org/10.3390/info16100827
Martinotti S, Montani S, Ranzato E, Striani M. SCRATCH-AI: A Tool to Predict Honey Wound Healing Properties. Information. 2025; 16(10):827. https://doi.org/10.3390/info16100827
Chicago/Turabian StyleMartinotti, Simona, Stefania Montani, Elia Ranzato, and Manuel Striani. 2025. "SCRATCH-AI: A Tool to Predict Honey Wound Healing Properties" Information 16, no. 10: 827. https://doi.org/10.3390/info16100827
APA StyleMartinotti, S., Montani, S., Ranzato, E., & Striani, M. (2025). SCRATCH-AI: A Tool to Predict Honey Wound Healing Properties. Information, 16(10), 827. https://doi.org/10.3390/info16100827










