A Comparative Study of Machine Learning Classifiers for Enhancing Knee Osteoarthritis Diagnosis
Abstract
1. Introduction
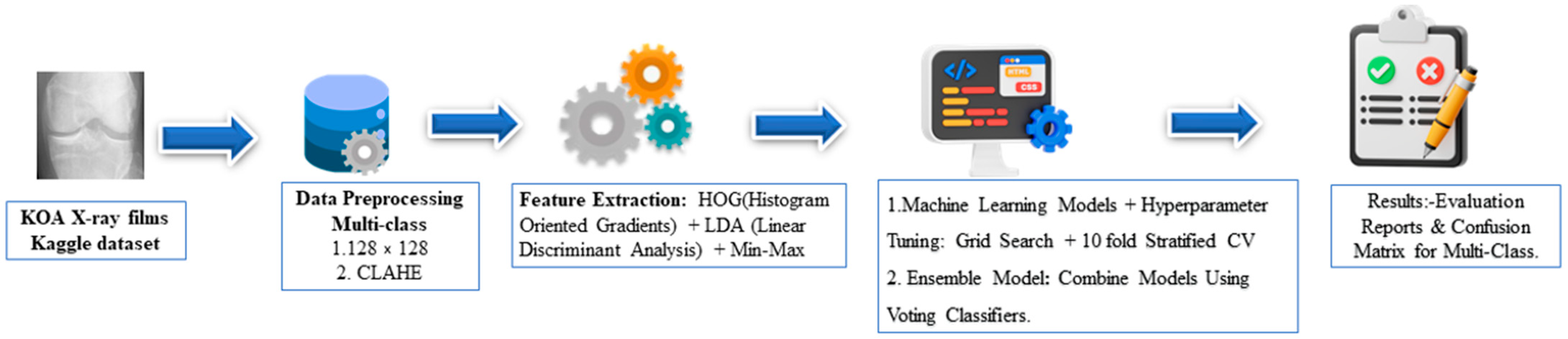
2. Materials and Methods
2.1. KOA X-ray Images Dataset
2.2. Data Pre-Processing
2.3. Feature Extraction
2.4. Model Selection and Hyperparameter Tuning
2.5. Performance Evaluations
2.6. Ensemble Model
2.7. Computational Settings
3. Results and Discussions
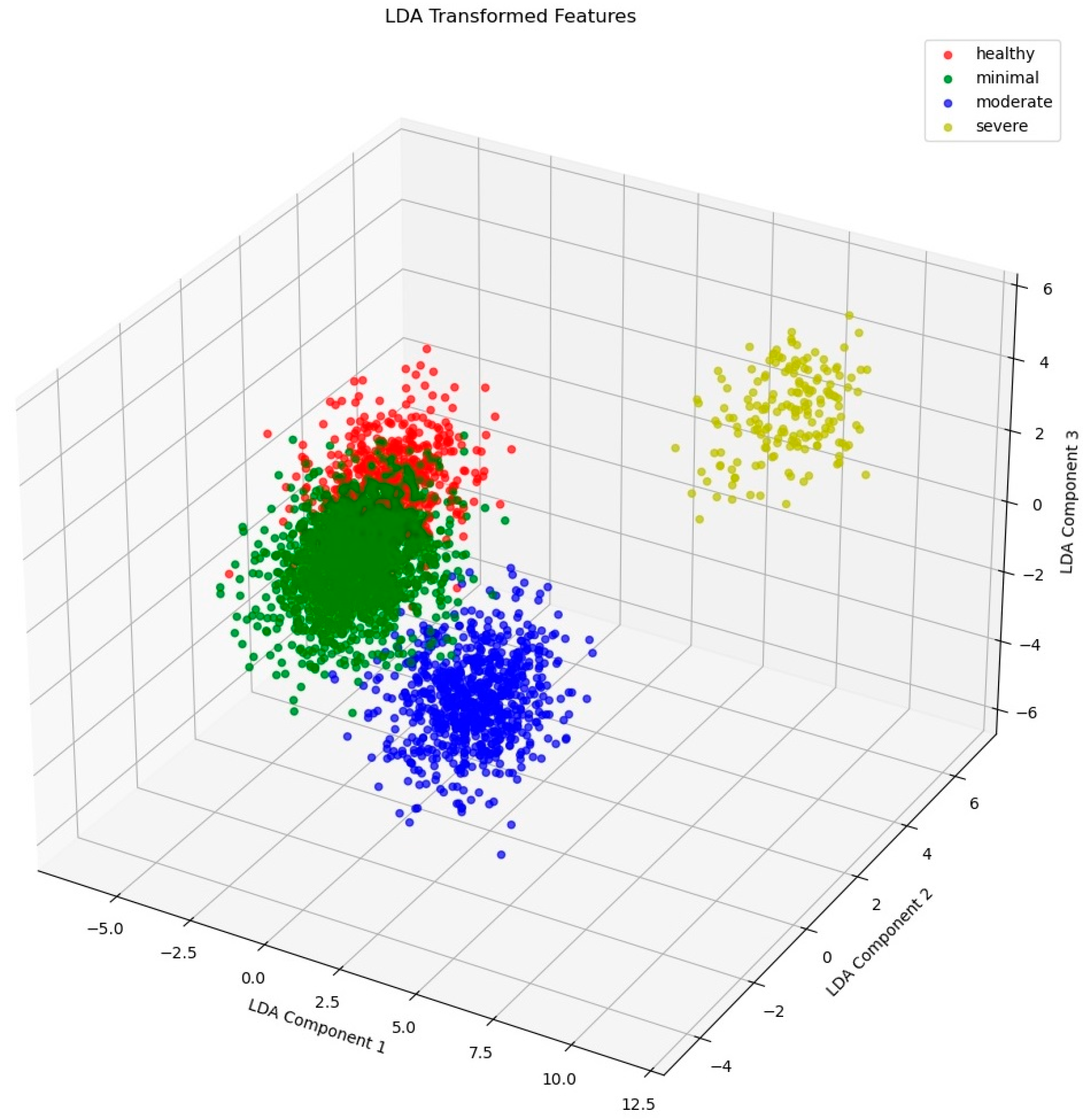
3.1. Feature Extraction Result
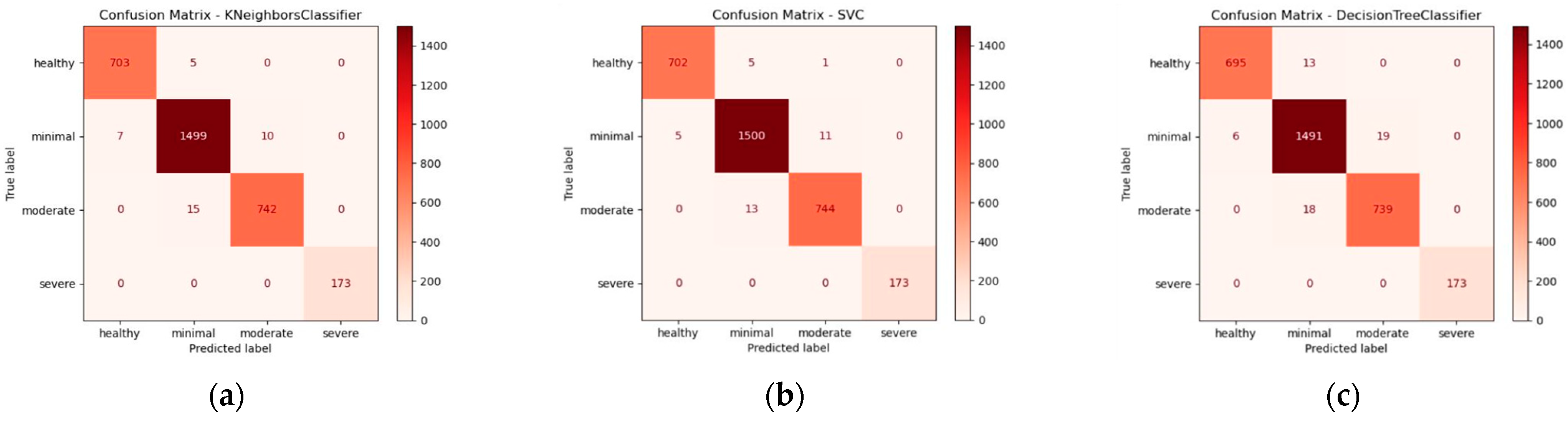
3.2. Confusion Matrix and Classification Report of the Cross-Validation
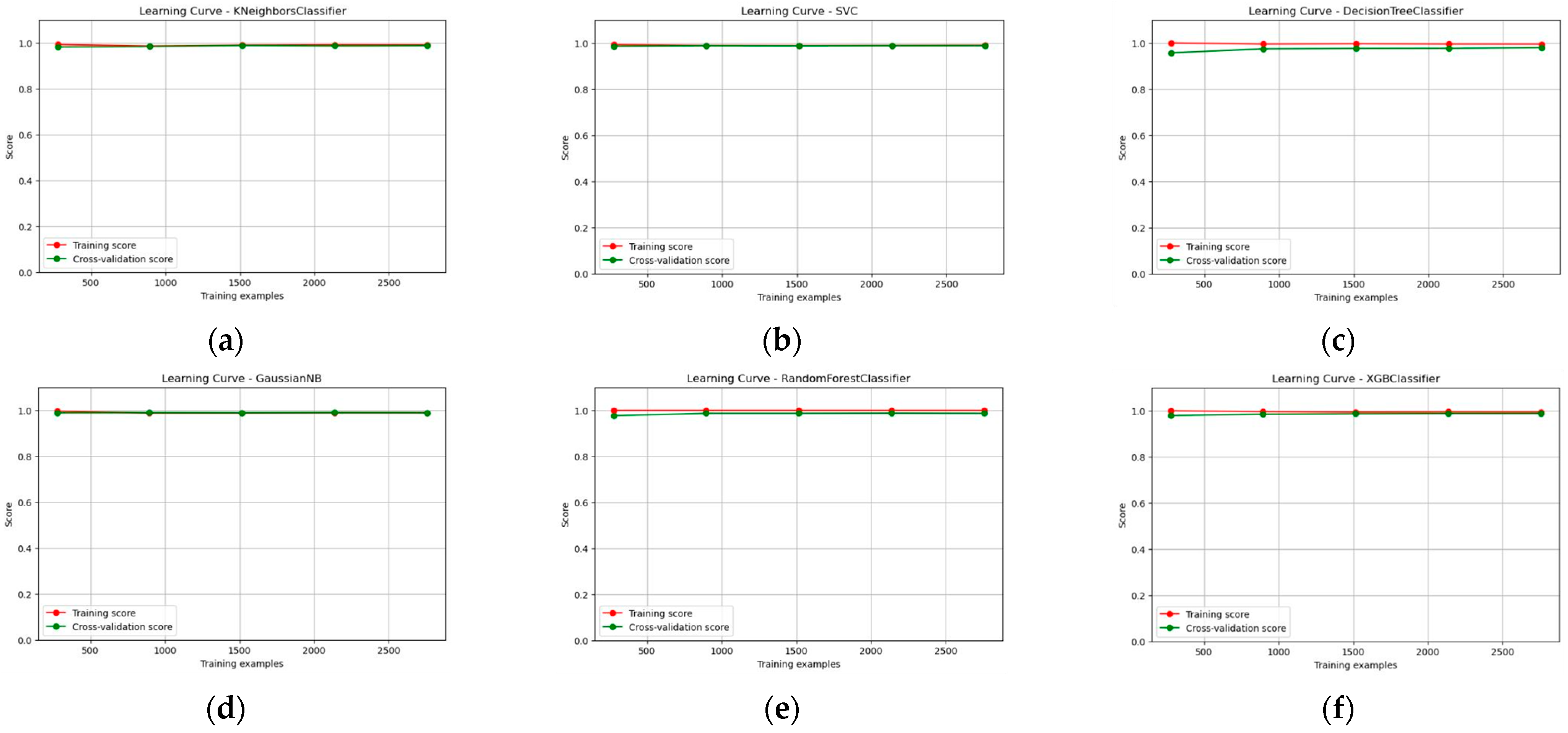
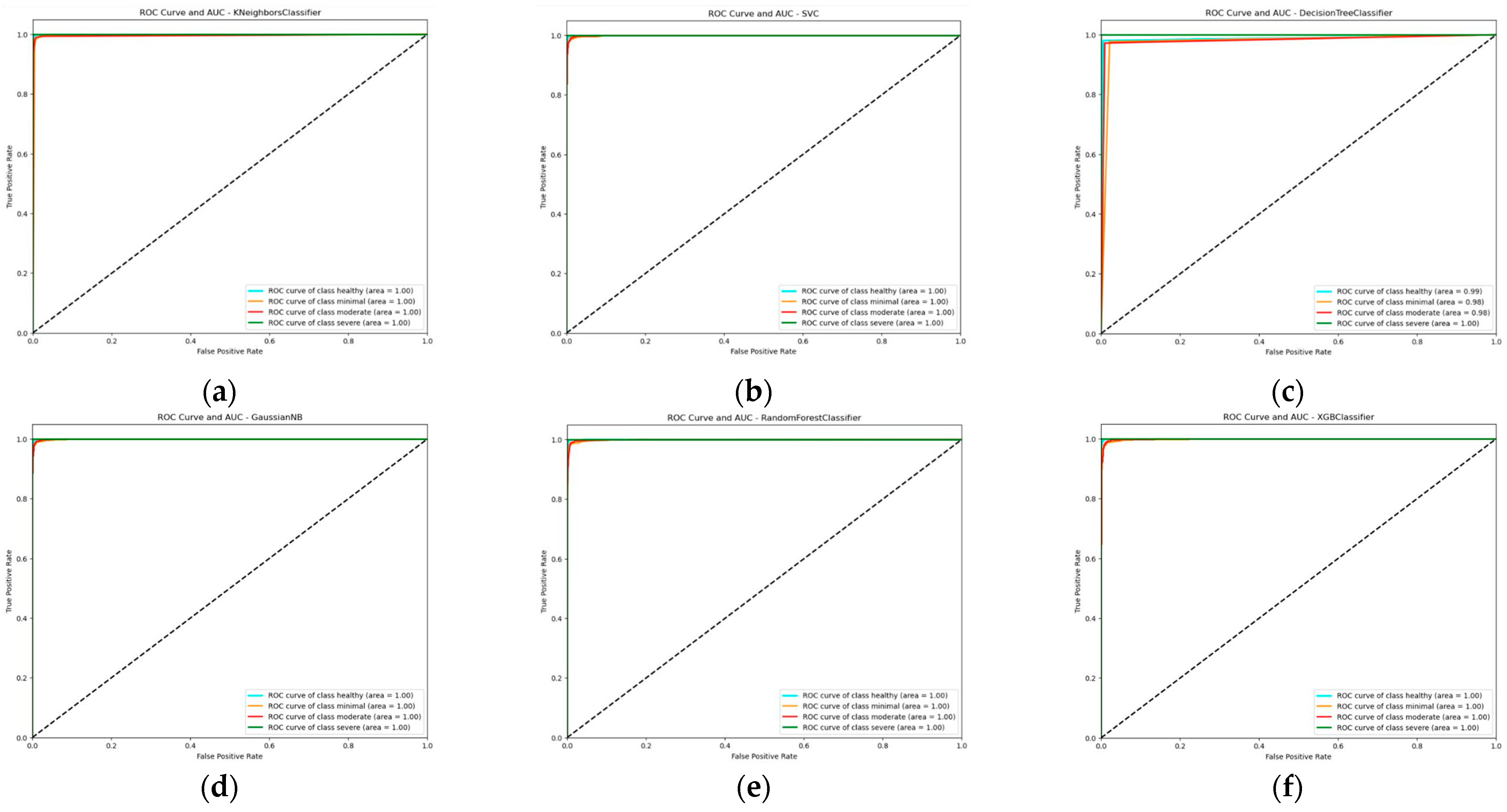
3.3. Learning Curve, ROC Curve, and AUC Score Report of the Multi-Model Classifier
3.4. Ensemble Model
3.5. Comparison with Available Literature
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hsu, H.; Siwiec, R.M. Knee Osteoarthritis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024; Available online: http://www.ncbi.nlm.nih.gov/books/NBK507884/ (accessed on 1 February 2024).
- Hayashi, D.; Roemer, F.W.; Jarraya, M.; Guermazi, A. Imaging in Osteoarthritis. Radiol. Clin. N. Am. 2017, 55, 1085–1102. [Google Scholar] [CrossRef]
- Schiphof, D.; Boers, M.; Bierma-Zeinstra, S.M.A. Differences in descriptions of Kellgren and Lawrence grades of knee osteoarthritis. Ann. Rheum. Dis. 2008, 67, 1034–1036. [Google Scholar] [CrossRef] [PubMed]
- Kellgren, J.H.; Lawrence, J.S. Radiological Assessment of Osteo-Arthrosis. Ann. Rheum. Dis. 1957, 16, 494–502. [Google Scholar] [CrossRef]
- Chen, P.; Gao, L.; Shi, X.; Allen, K.; Yang, L. Fully automatic knee osteoarthritis severity grading using deep neural networks with a novel ordinal loss. Comput. Med. Imaging Graph. 2019, 75, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Tiulpin, A.; Thevenot, J.; Rahtu, E.; Lehenkari, P.; Saarakkala, S. Automatic Knee Osteoarthritis Diagnosis from Plain Radiographs: A Deep Learning-Based Approach. Sci. Rep. 2018, 8, 1727. [Google Scholar] [CrossRef] [PubMed]
- Cui, A.; Li, H.; Wang, D.; Zhong, J.; Chen, Y.; Lu, H. Global, regional prevalence, incidence and risk factors of knee osteoarthritis in population-based studies. eClinicalMedicine 2020, 29–30, 100587. [Google Scholar] [CrossRef] [PubMed]
- Cross, M.; Smith, E.; Hoy, D.; Nolte, S.; Ackerman, I.; Fransen, M.; Bridgett, L.; Williams, S.; Guillemin, F.; Hill, C.L.; et al. The global burden of hip and knee osteoarthritis: Estimates from the Global Burden of Disease 2010 study. Ann. Rheum. Dis. 2014, 73, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Tiulpin, A.; Saarakkala, S. Automatic Grading of Individual Knee Osteoarthritis Features in Plain Radiographs Using Deep Convolutional Neural Networks. Diagnostics 2020, 10, 932. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.S.M.; Holi, M.S. Knee joint cartilage visualization and quantification in normal and osteoarthritis. In Proceedings of the 2010 International Conference on Systems in Medicine and Biology, Kharagpur, India, 16–18 December 2010; IEEE: New York, NY, USA, 2010; pp. 138–142. [Google Scholar] [CrossRef]
- Li, L. Deep Residual Autoencoder with Multiscaling for Semantic Segmentation of Land-Use Images. Remote Sens. 2019, 11, 2142. [Google Scholar] [CrossRef]
- Saygılı, A. A new approach for computer-aided detection of coronavirus (COVID-19) from CT and X-ray images using machine learning methods. Appl. Soft Comput. 2021, 105, 107323. [Google Scholar] [CrossRef]
- Ha, M.-K.; Phan, T.-L.; Nguyen, D.H.H.; Quan, N.H.; Ha-Phan, N.-Q.; Ching, C.T.S.; Hieu, N.V. Comparative Analysis of Audio Processing Techniques on Doppler Radar Signature of Human Walking Motion Using CNN Models. Sensors 2023, 23, 8743. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, X.; Ma, S.; Lin, T.; Gong, J. Monitoring ecosystem service change in the City of Shenzhen by the use of high-resolution remotely sensed imagery and deep learning. Land Degrad. Dev. 2019, 30, 1490–1501. [Google Scholar] [CrossRef]
- Hapsari, Y.; Syamsuryadi. Weather Classification Based on Hybrid Cloud Image Using Principal Component Analysis (PCA) and Linear Discriminant Analysis (LDA). J. Phys. Conf. Ser. 2019, 1167, 012064. [Google Scholar] [CrossRef]
- Anifah, L.; Purnama, I.K.E.; Hariadi, M.; Purnomo, M.H. Osteoarthritis Classification Using Self Organizing Map Based on Gabor Kernel and Contrast-Limited Adaptive Histogram Equalization. Open Biomed. Eng. J. 2013, 7, 18–28. [Google Scholar] [CrossRef]
- Wahyuningrum, R.T.; Anifah, L.; Purnama, I.K.E.; Purnomo, M.H. A novel hybrid of S2DPCA and SVM for knee osteoarthritis classification. In Proceedings of the 2016 IEEE International Conference on Computational Intelligence and Virtual Environments for Measurement Systems and Applications (CIVEMSA), Budapest, Hungary, 27–28 June 2016; IEEE: New York, NY, USA, 2016; pp. 1–5. [Google Scholar] [CrossRef]
- Mohammed, A.S.; Hasanaath, A.A.; Latif, G.; Bashar, A. Knee Osteoarthritis Detection and Severity Classification Using Residual Neural Networks on Preprocessed X-ray Images. Diagnostics 2023, 13, 1380. [Google Scholar] [CrossRef]
- Kotti, M.; Duffell, L.D.; Faisal, A.A.; McGregor, A.H. Detecting knee osteoarthritis and its discriminating parameters using random forests. Med. Eng. Phys. 2017, 43, 19–29. [Google Scholar] [CrossRef]
- Kokkotis, C.; Moustakidis, S.; Papageorgiou, E.; Giakas, G.; Tsaopoulos, D.E. Machine learning in knee osteoarthritis: A review. Osteoarthr. Cartil. Open 2020, 2, 100069. [Google Scholar] [CrossRef] [PubMed]
- Gornale, S.S.; Patravali, P.U.; Manza, R.R. Detection of Osteoarthritis Using Knee X-ray Image Analyses: A Machine Vision based Approach. Int. J. Comput. Appl. 2016, 145, 20–26. [Google Scholar]
- Brahim, A.; Jennane, R.; Riad, R.; Janvier, T.; Khedher, L.; Toumi, H.; Lespessailles, E. A decision support tool for early detection of knee OsteoArthritis using X-ray imaging and machine learning: Data from the OsteoArthritis Initiative. Comput. Med. Imaging Graph. 2019, 73, 11–18. [Google Scholar] [CrossRef]
- Mehta, S.; Gaur, A.; Sarathi, M.P. A Simplified Method of Detection and Predicting the Severity of Knee Osteoarthritis. In Proceedings of the 2023 14th International Conference on Computing Communication and Networking Technologies (ICCCNT), Delhi, India, 6–8 July 2023; IEEE: New York, NY, USA, 2023; pp. 1–7. [Google Scholar] [CrossRef]
- Mahum, R.; Rehman, S.U.; Meraj, T.; Rauf, H.T.; Irtaza, A.; El-Sherbeeny, A.M.; El-Meligy, M.A. A Novel Hybrid Approach Based on Deep CNN Features to Detect Knee Osteoarthritis. Sensors 2021, 21, 6189. [Google Scholar] [CrossRef]
- Bayramoglu, N.; Tiulpin, A.; Hirvasniemi, J.; Nieminen, M.T.; Saarakkala, S. Adaptive segmentation of knee radiographs for selecting the optimal ROI in texture analysis. Osteoarthr. Cartil. 2020, 28, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Belete, D.M.; Huchaiah, M.D. Grid search in hyperparameter optimization of machine learning models for prediction of HIV/AIDS test results. Int. J. Comput. Appl. 2022, 44, 875–886. [Google Scholar] [CrossRef]
- Tariq, T.; Suhail, Z.; Nawaz, Z. Machine Learning Approaches for the Classification of Knee Osteoarthritis. In Proceedings of the 2023 3rd International Conference on Electrical, Computer, Communications and Mechatronics Engineering (ICECCME), Tenerife, Canary Islands, Spain, 19–21 July 2023; IEEE: New York, NY, USA, 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Thornton, C.; Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Auto-WEKA: Combined selection and hyperparameter optimization of classification algorithms. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; ACM: New York, NY, USA, 2013; pp. 847–855. [Google Scholar] [CrossRef]
- DeCastro-García, N.; Castañeda, Á.L.M.; García, D.E.; Carriegos, M.V. Effect of the Sampling of a Dataset in the Hyperparameter Optimization Phase over the Efficiency of a Machine Learning Algorithm. Complexity 2019, 2019, 6278908. [Google Scholar] [CrossRef]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Wang, R. Efficient kNN Classification with Different Numbers of Nearest Neighbors. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- Chapelle, O.; Vapnik, V.; Bousquet, O.; Mukherjee, S. Choosing Multiple Parameters for Support Vector Machines. Mach. Learn. 2002, 46, 131–159. [Google Scholar] [CrossRef]
- Probst, P.; Wright, M.; Boulesteix, A.-L. Hyperparameters and Tuning Strategies for Random Forest. arXiv 2018, arXiv:1804.03515. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Ntakolia, C.; Kokkotis, C.; Moustakidis, S.; Tsaopoulos, D. A machine learning pipeline for predicting joint space narrowing in knee osteoarthritis patients. In Proceedings of the 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, OH, USA, 26–28 October 2020; pp. 934–941. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Chen, P. Knee Osteoarthritis Severity Grading Dataset. Mendeley 2018. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of Oriented Gradients for Human Detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; IEEE: New York, NY, USA, 2005; pp. 886–893. [Google Scholar] [CrossRef]
- Gornale, S.S.; Patravali, P.U.; Marathe, K.S.; Hiremath, P.S. Determination of Osteoarthritis Using Histogram of Oriented Gradients and Multiclass SVM. Int. J. Image Graph. Signal Process. 2017, 9, 41–49. [Google Scholar] [CrossRef]
- Zöller, M.-A.; Huber, M.F. Benchmark and Survey of Automated Machine Learning Frameworks. arXiv 2019, arXiv:1904.12054. [Google Scholar] [CrossRef]
- Hutter, F.; Kotthoff, L.; Vanschoren, J. (Eds.) Automated Machine Learning: Methods, Systems, Challenges. In The Springer Series on Challenges in Machine Learning; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]


| Classifier | Hyperparameters | Tuning Value |
|---|---|---|
| K Nearest Neighbors classifier (KNN) | n_neighbors | {3, 5, 7, 9, 11, 13} |
| Support Vector Machine (SVM) | C | {1, 10, 50, 70, 100] |
| gamma | {‘scale’, ‘auto’} | |
| kernel | {‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’} | |
| class_weight | {None, ‘balanced’} | |
| GaussianNB | var_smoothing | {10−11, 10−10, 10−9, 10−8} |
| Decision Tree classifier | max_depth | {None, 3, 5, 7, 10} |
| Random Forest classifier | n_estimators | {50, 100, 200, 300, 500} |
| warm_start | {True, False} | |
| n_jobs | {None, 1, 2, 3} | |
| XGBoost classifier | max_depth | {3, 5, 7} |
| learning_rate | {0.1, 0.01, 0.001} | |
| n_estimators | {50, 100, 200} | |
| gamma | {0, 1, 2} | |
| subsample | {0.8, 0.9, 1.0} |
| Model | Optimized Hyperparameter Value | Healthy (Precision, Recall, F1-Score) | Minimal (Precision, Recall, F1-Score) | Moderate (Precision, Recall, F1-Score) | Severe (Precision, Recall, F1-Score) | Cross Validation Accuracy (%) |
|---|---|---|---|---|---|---|
| K Nearest Neighbors (KNN) | n_neighbors: 3 | 0.99, 0.99, 0.99 | 0.99, 0.99, 0.99 | 0.99, 0.98, 0.98 | 1.00, 1.00, 1.00 | 98.83 |
| SVM | C: 100, kernel: rbf | 0.99, 0.99, 0.99 | 0.99, 0.99, 0.99 | 0.98, 0.98, 0.98 | 1.00, 1.00, 1.00 | 98.89 |
| GaussianNB | var_smoothing: 10−9 | 0.99, 0.99, 0.99 | 0.99, 0.99, 0.99 | 0.99, 0.98, 0.98 | 1.00, 1.00, 1.00 | 98.89 |
| Decision Tree | max_depth: 7 | 0.99, 0.98, 0.99 | 0.98, 0.98, 0.98 | 0.97, 0.98, 0.98 | 1.00, 1.00, 1.00 | 98.19 |
| Random Forest | n_estimators: 200, warm_start: false | 0.99, 0.99, 0.99 | 0.99, 0.99, 0.99 | 0.98, 0.98, 0.98 | 1.00, 1.00, 1.00 | 98.67 |
| XGBoost | gamma: 1, learning_rate: 0.1, max_depth: 5, n_estimators: 100, subsample: 1.0 | 0.99, 1.00, 0.99 | 0.99, 0.99, 0.99 | 0.98, 0.98, 0.98 | 1.00, 1.00, 1.00 | 98.90 |
| References | Methodology | Accuracy |
|---|---|---|
| [17] | SVM with Structural 2-dimensional Principle Component Analysis | 94.33% |
| [21] | SVM with HOG feature extraction | ~95% |
| [22] | RF and NB with Independent Component Analysis | 82.98% |
| [27] | LR with HOG + Haralick Features | 84.50% |
| This study | XGBoost Classifer with CLAHE, HOG + LDA feature extraction, Min–Max scaling | 98.90% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raza, A.; Phan, T.-L.; Li, H.-C.; Hieu, N.V.; Nghia, T.T.; Ching, C.T.S. A Comparative Study of Machine Learning Classifiers for Enhancing Knee Osteoarthritis Diagnosis. Information 2024, 15, 183. https://doi.org/10.3390/info15040183
Raza A, Phan T-L, Li H-C, Hieu NV, Nghia TT, Ching CTS. A Comparative Study of Machine Learning Classifiers for Enhancing Knee Osteoarthritis Diagnosis. Information. 2024; 15(4):183. https://doi.org/10.3390/info15040183
Chicago/Turabian StyleRaza, Aquib, Thien-Luan Phan, Hung-Chung Li, Nguyen Van Hieu, Tran Trung Nghia, and Congo Tak Shing Ching. 2024. "A Comparative Study of Machine Learning Classifiers for Enhancing Knee Osteoarthritis Diagnosis" Information 15, no. 4: 183. https://doi.org/10.3390/info15040183
APA StyleRaza, A., Phan, T.-L., Li, H.-C., Hieu, N. V., Nghia, T. T., & Ching, C. T. S. (2024). A Comparative Study of Machine Learning Classifiers for Enhancing Knee Osteoarthritis Diagnosis. Information, 15(4), 183. https://doi.org/10.3390/info15040183







