Abstract
Machine learning approaches for pattern recognition are increasingly popular. However, the underlying algorithms are often not open source, may require substantial data for model training, and are not geared toward specific tasks. We used open-source software to build a green toad breeding call detection algorithm that will aid in field data analysis. We provide instructions on how to reproduce our approach for other animal sounds and research questions. Our approach using 34 green toad call sequences and 166 audio files without green toad sounds had an accuracy of 0.99 when split into training (70%) and testing (30%) datasets. The final algorithm was applied to amphibian sounds newly collected by citizen scientists. Our function used three categories: “Green toad(s) detected”, “No green toad(s) detected”, and “Double check”. Ninety percent of files containing green toad calls were classified as “Green toad(s) detected”, and the remaining 10% as “Double check”. Eighty-nine percent of files not containing green toad calls were classified as “No green toad(s) detected”, and the remaining 11% as “Double check”. Hence, none of the files were classified in the wrong category. We conclude that it is feasible for researchers to build their own efficient pattern recognition algorithm.
1. Introduction
Machine learning (ML) approaches as part of artificial intelligence systems for visual and sound pattern detection are receiving increasing popularity because of their high efficiency and comparatively simple application [1,2]. For instance, machine learning algorithms have been used to detect lesions in medical images [3], leukemia in blood cell count data patterns [4], movement and behavior of livestock [5], individual pig recognition [6], as well as face and speech recognition [7,8]. In the natural sciences, some identification applications of plants and animals (e.g., iNaturalist, observation.org) are assisted by artificial intelligence [9], but the algorithms are usually not open source or freely available. Most ML algorithms were developed by hiring costly programmers and need considerable model training data for acceptable performance. Hence, for biologists with specialized (‘niche’) research questions, artificial intelligence systems such as those described above might be out of reach.
In addition, automated call detection and species identification of amphibians are less widely available compared to other taxa (e.g., birds and bats). The available tools are based on specific call characteristics such as pulse rate and frequency [10], hence they might lack specificity in cases of species with similar calls, high levels of background noise, simultaneously calling individuals, or more specific research questions [11]. Furthermore, manual or semi-automatic preprocessing steps of the audio files are often needed to improve detection performances [12]. Thus, researchers would benefit from an easily adaptable small-scale approach, where they use familiar toolsets such as software R [13] to implement ML into their data analysis.
Earlier research has shown that algorithms based on decision trees might be a promising avenue for anuran call recognition [14]. A state-of-the-art candidate machine learning algorithm is the eXtreme Gradient boosting algorithm (XGboost) [15]. It is a further development of the gradient boosting approach and has consistently performed very well in multiple competitions hosted by the Kaggle AI/ML community platform [16]. Importantly, XGboost can be applied using all standard programming languages for statistical computing used in natural sciences, including R (via the xgboost package [17]).
While many species sampling efforts in field biology are based on visual detection, some taxa can be easily identified by the sounds they produce. For instance, species-specific vocalizations can be used to monitor birds [18] and frogs [19]. Especially for citizen science, acoustic detection is already used to collect vast datasets of species distributions [20,21]. However, anuran species acoustic identification is hardly used in citizen science compared to bird or bat species [22,23,24]. Recently the automated recording of soundscapes became a promising tool for efficient acoustic monitoring, even going beyond species identification [25].
Despite the advances in digitalization and automatization, a lot of the call identification of recorded audio still requires time-consuming effort by researchers [26,27], in particular for citizen science projects that collect very large datasets [28]. In part, utilizing existing ML algorithms or creating new ones might not be feasible (e.g., high costs); furthermore, the scope of existing algorithms might not fit the needs of a particular research question.
In the current study, we show that open-source tools can be easily used to develop a highly efficient call detection algorithm, at least for a single species, using the European green toad Bufotes viridis as an example. Our study is not a comparison between different possible detection algorithms, and there might be other equally or better performing solutions. However, for our approach, we provide a step-by-step instruction to reproduce our approach, with example sound files and annotated code including explanations on how it could be customized for other species. We expect our work to be of use to biologists working with sound data, extending the available tools in wildlife research.
2. Materials and Methods
2.1. Example Species and Call Characteristics
We used the breeding call of the European green toad (B. viridis Laurenti, 1768) as an example. The green toad is widely distributed in Europe and Asia [29], however, its populations are often threatened locally [30]. Its call is a trill with a fundamental frequency of about 1200 to 1400 Hz with about 20 pulses per s, with note lengths of about 4 s [31,32]. As the calls of different Bufotes species and subspecies may vary [29], in our study we focused on the nominate subspecies B. viridis viridis and only trained and tested on calls recorded in its range.
A previous study using passive acoustic monitoring has demonstrated the need for a green toad sound detection and identification algorithm [33]. In addition, it is the target species in the ongoing citizen science project AmphiBiom (https://amphi.at; accessed on 20 August 2024), with one of its goals being to use breeding calls submitted by volunteers to identify green toad occurrences; an automated algorithm could greatly increase the efficiency of related analyses.
2.2. Data Sources
To train the ML algorithm, we obtained audio files with and without calls of green toads from public platforms, provided by colleagues, or recorded by us (see details in Supplementary Tables S1 and S2). Recordings without green toad calls included other anuran calls, crickets, birds, and background noise. Files were recorded using different methods and in different audio formats, with professional microphones or mobile phone microphones.
The field collected data to be analyzed comprised 111 sound files recorded as MP3 audio files and uploaded by citizen scientists using the dedicated mobile phone application AmphiApp (https://www.amphi.at/en/app; downloaded on 2 May 2024). Each observation was identified by two of the amphibian experts in the team (Y.V.K. and S.B.).
2.3. Data Preparation
Before analysis, all files were converted to the WAV audio format if needed using the av_audio_convert function in the package av in software R (av::av_audio_convert) [34]. To train/test the ML algorithm, we extracted the specific parts with green toad calls using the R packages tuneR and seewave [35,36]. For non-green toad calls, we randomly selected 5 min long sections of each recording for algorithm training (to reduce the computation time); for samples below 5 min in length, the entire audio recording was used. We converted the audio files to data matrices. For this, we first resampled the wave to meet our target sampling rate (44,100 Hz). Then we filtered the frequency range between 1150 and 1650 Hz (function seewave::ffilter) and created a spectrogram object function seewave::spectro). For generating the spectrogram, we used the default parameters including the 512 window length, which resulted in a time difference between two time points of 0.0116 s. The amplitude values were then extracted and transposed, resulting in a matrix with frequency as columns and time as rows. To also represent previous and following frequency patterns in each time point, we added the row values of the 18 previous and 18 following rows to each row as additional columns (representing +/−0.2 s), hence, increasing the number of columns in the data set. As the columns were always arranged in the same way, each time point had the information about the amplitude per frequency for the current time point as well as the 18 previous and 18 following time points. Therefore, the algorithm has the possibility to be trained to the combination of amplitudes per frequency along a time period of about 0.4 s. The resulting matrix was then duplicated and scaled (function base::scale). Hence, the final matrix consisted of scaled and raw amplitude values for each row (time point). In addition, only for algorithm training and testing, we added a column indicating if the data are from a green toad call or not.
2.4. Call Detection Algorithm Training and Testing
We used the extreme gradient boost machine learning algorithm (function xgboost in the package xgboost [17]) for our green toad detection approach. Throughout we used a maximum tree depth (max.depth) of 100, an eta (i.e., learning rate) of 1 and 100 boosting rounds (nrounds); for all other parameters, we used the default values. The learning task of the algorithm (objective) was defined as a logistic regression for binary classification (binary:logistic).
First, we trained the algorithm on randomly selected 70% of the data (training data) and then compared the performance on the remaining 30% (test data). We used the confusionMatrix function of the caret package [37] to test the performance of the xgboost predictions (function predict) on the test data.
Then we combined all data (the training and testing data from above) and repeated the algorithm training steps to make use of all the available data. The resulting algorithm was then applied on the field collected data, returning an assessment for each file (“Green toad(s) detected” if ≥10 consecutive time points are identified as containing a green toad call, “No green toad(s) detected” if less than two consecutive time points contain green toad calls, or “Double check” otherwise).
Following the application of the algorithm, we did a blinded manual identification of the audio files to identify files with and without green toad calls.
2.5. Example Protocol for Customized Call Recognition
For the example protocol to build customized detection algorithms for calls, we provided a suggestion for folder structure, an overview script (0_overview.R) that calls the scripts needed for data preparation (1_global_constants.R to centralize setup, 2_convert_audio_to_WAV.R, 3_target_species_sound_cleaning.R, 4_create_sound_matrices.R); a file for algorithm training (5_MLA_training.R), and the machine learning prediction function together with customized helper functions (functions_sound_identification.R). We also shared an application for this function using sound files not used for training. We worked through our example ensuring an easy workflow and general acceptable performance of this small-scale example. In the comments of the script together with the information in a help file (“Compiling a machine learning algorithm to detect target animal sounds.docx”), we provided suggestions for ways to adapt the algorithm for other sounds (files can be found in S1: Call_Example_Procedure.zip).
3. Results
We developed a highly efficient machine learning call detection algorithm, at least for a single species that can easily be adapted to other study cases. These results were obtained even if the field data audio recordings varied significantly, both in terms of what was recorded (e.g., no amphibians at all) and in the audio quality (e.g., including background noise and echo-like artifacts).
3.1. Call Recognition Performance
Within the training and testing datasets, the detection algorithm had an accuracy of 0.9948 (95% CI: 0.9946–0.9951). After training the algorithm on the combined data set (i.e., testing data set combined with training data set), we tested the resulting detection function (function: green_detection in Supplementary File: functions_calls_recogn_Aug2024.R; for the most up-to-date function and algorithm please visit the following GitHub page: https://github.com/lukasla/Amph_sounds) on 111 files downloaded from the AmphiApp back-end on 31 May 2024. Of 111 files to be analyzed, we removed 12 as they were <2 s long or empty; 29 files contained B. viridis calls (see example Figure 1A). The remaining 70 files contained no green toad calls. Of the 29 audios containing green toad calls, 90% (n = 26) were correctly classified as “Green toad(s) detected”, and 3 as “Double check”, with no misidentifications. Of the 70 files not containing green toad calls, 89% (n = 62) were correctly identified as “No green toad(s) detected”, and 8 as “Double check”. When only considering two categories, green toads present or not present, the detection function missed 3 out of 29 (i.e., 10%) green toad sound files (e.g., because of loud background noise, see Figure 1B). No files that did not contain green toads where misclassified. When double-checking files classified as “Double check”, the success rate would be 100% in both cases and it would reduce the workload by a factor of about 10 (i.e., checking 11 files instead of 99). The application of the algorithm did not require highly specialized or difficult to access computing power. We carried the calculations using a high-end personal computer (memory: Kingston 128 GB DDR4 3200 MHz, processor: AMD Ryzen Threadripper Pro 24 cores 3.80 GHz, GPU: AMD Radeon PRO W6400 4 GB).
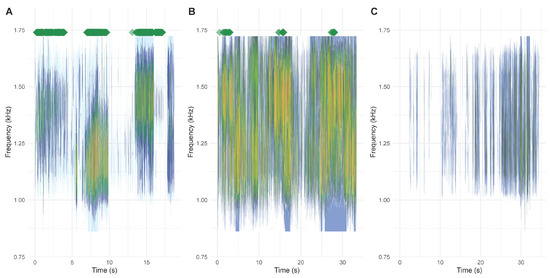
Figure 1.
Example results of the call detection algorithm using sound files from the AmphiBiom project, recorded by citizen scientists. Spectrograms of the audio files are in the kHz range of interest. Green diamonds above the spectrogram indicate positive detections of green toad calls (within 0.2 s time point samples). Examples show a “clean” green toad call correctly identified by the algorithm (A), a green toad calling along European tree frogs Hyla arborea, classified as “Double check” (B), and red-bellied toads Bombina bombina calling, correctly classified as “No green toad(s) detected” (C). Videos, including the spectrogram and associated soundtracks, are provided in the Supplementary Materials (Supplementary File: Videos_for_Fig1.zip). Results can be replicated by using the “Usage_of_green_toad_detection_function.R” together with the files in “Audios_Usage_Example.zip”.
3.2. Example Protocol for Customized Call Recognition Algorithm
We provide an easy-to-use procedure to implement your own species-specific call detection algorithm using freely available packages in R (Supplementary Zip File: Call_Example_Procedure.zip). Possibilities to tweak the algorithm, especially regarding the preparations of the sound file information are described in the Supplementary Materials. The R code and sound files we provide are restricted to some AmphiBiom sound files and a few additional files (see list in the working example folders) therefore do not encompass the database we used for the detection algorithm. Hence, one should not use the obtained “example algorithm” as a substitute for the green toad detection function described above. However, we provide these files to show the stepwise procedure to build the algorithm, which should enable other researchers to establish their own detection algorithm. We provide eight test files from our AmphiBiom project, not used in model training, four of which contain green toad calls. The resulting algorithm used inside our target species detection function identified all four green toad files correctly, the remaining files were either classified as “Double check” or “Target species not detected”.
4. Discussion
We demonstrate that customized, semi-automated, and highly accurate anuran call detection algorithms can be easily built and implemented using open-source software, if there are sufficient suitable reference vocalizations available. Our example protocol can be utilized by biologists of all areas working with sounds to build their own customized detection algorithm with little effort. Furthermore, we provide the tools in an environment (R) that should be familiar to a wide swath of researchers, running on major operating systems (Windows, Mac, Linux), and does not depend on external software companies or specialized programmers. However, like detection algorithms for bird songs (for example, the Merlin Bird ID application by The Cornell Lab of Ornithology), such tools may be integrated with available citizen science mobile phone applications such as iNaturalist or observation.org.
We show that the detection algorithm we built was robust and specific. It detected green toad calls when other amphibians were present, even with high background noise and low sound quality. However, great care should be taken in preparing the training and testing data sets to include sounds that are like the target species vocalizations, as well as common background noise. The best results are expected to be obtained when the training data cover the range of possible sound files to be evaluated by the algorithm. Hence, for a citizen science application, it would make sense to use the initial data collection period to obtain a training data set and then continue within the same framework but add the algorithm to speed up call identification. The size of the data set required likely varies a lot between research questions, target sound type, and file qualities.
Importantly, we prepared the algorithm to detect one specific call, the green toad breeding call. In cases of animals with several sound types (e.g., mating, warning, territorial calls), it might be possible to include all of them in one algorithm training procedure. However, in this case, the different calls could not be separated afterwards other than manually. Therefore, we would suggest training as many algorithms as there are sounds that should be detected and include all of them in one detection function. In this instance, one could differentiate not only if their target species is present, but also their behavioral state. The advantage of training several algorithms is that they can detect different sound types (e.g., warning versus mating) even when they are happening at the same time point. Similarly, one could train several algorithms for several different species and detect all of them in a single audio file, if all the algorithms are included in the detection function. Moreover, animal sounds can change from younger to older [38] and smaller to larger individuals [39] of the same species, potentially enabling researchers to estimate age/size structure through audio surveys.
We advise against training the data set on calls from one specific region and limited time frame in the season and then using the algorithms in a different area or/and different seasons. Small variations in call characteristics, but also background/non-target sounds between populations or regions (e.g., [40]) that could be missed by human ears, might decrease the accuracy of algorithms trained and applied on sound files obtained from different locations. We noticed during testing that the training files for such algorithms should be within the range of files that you expect to obtain. For instance, training the algorithm on files of natural sounds but then testing files containing music (e.g., piano) will likely lead to false results.
Importantly, the majority of the sounds we analyzed were likely exclusively being collected with built-in smartphone microphones through the AmphiApp and directly saved in MP3 format. Thus, the sound quality might vary based on the specific hardware and software characteristics of individual devices and their ability to record and convert sounds in real time, the distance from the recording device to the calling toad, and/or background noise [22,41,42]. Furthermore, the MP3 format is widely used for audio recording as it generates relatively small-size files but achieves this by using lossy data compression consisting of inexact approximations and the partial discarding of data (including reducing the accuracy of certain components of sound that are beyond the hearing capabilities of most humans). Therefore, care needs to be taken that key characteristics of the animal calls recorded and analyzed are not lost during recording/conversion and subsequently unavailable for analysis and recognition by the algorithm. Training data should thus include a range of sounds with varying recording quality. However, considering these limitations, at least in our case, the algorithm was robust and accurate even when analyzing recordings with quality deemed poor to the human listeners.
Using machine learning approaches for automated species detection in addition to professional (human) assessments could improve data quality for auditory surveys [43,44]. Such auditory surveys are common in amphibian ecology [45] and can help monitor species distribution and to some extent even population trends. New developments in automated sound recording (e.g., soundscape recordings) could easily boost our understanding of the activity and distribution of calling amphibians, as it is already transforming marine acoustic studies [46]. As most anuran calls are specifically related to reproductive efforts [47], acoustic information is highly valuable, because it relates to a reproductively active population. In the future, such calls might also be used to assess the status of populations, as the calls are related to the size [39] and potentially health status of individuals.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/info15100610/s1, Table S1: Calls_collection_Aug_20_2024.xlsx, Table S2: Other_collection_Aug_20_2024.xlsx, Scripts S1: Call_Example_Procedure.zip; Scripts S2: Usage_of_green_toad_detection_function.R, Audio Files S1: Audios_Usage_Example.zip and Video S1: Videos_for_Fig1.zip.
Author Contributions
Conceptualization, L.L., F.H. and D.D.; methodology, L.L., Y.V.K., S.B., M.K., M.S. and J.S.; software, L.L. and Y.V.K.; validation, Y.V.K., S.B. and J.S.; formal analysis, L.L.; investigation, L.L., Y.V.K., S.B., M.K., M.S. and J.S.; data curation, L.L., Y.V.K., S.B., M.K., M.S. and J.S.; writing—original draft preparation, L.L.; writing—review and editing, L.L., Y.V.K., S.B., M.S., J.S., F.H. and D.D.; visualization, L.L.; funding acquisition: L.L., F.H. and D.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Biodiversity Fund of the Federal Ministry of Austria for Climate Action, Environment, Energy, Mobility, Innovation and Technology and Next Generation EU (project no. C321025).
Data Availability Statement
The detection algorithm, R scripts and data to reproduce the approach are available in the Supplementary Information and on GitHub.
Acknowledgments
We thank Peter Bader who recorded the audio files in the Austrian Danubian floodplains which were used as part of the algorithm training data set (without green toads). These recordings were obtained during the RIMECO project funded by the Austrian Science Fund (FWF, No. I5006-B). Audio files used in model training were recorded by multiple people (see Tables S1 and S2) including unreleased recordings from Jasmin Proksch, Nadine Lehofer, Luise Aiglsperger and Hendrik Walcher.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Asht, S.; Dass, R. Pattern Recognition Techniques: A Review. Int. J. Comput. Sci. Telecommun. 2012, 3, 25–29. [Google Scholar]
- Sukthankar, R.; Ke, Y.; Hoiem, D. Semantic Learning for Audio Applications: A Computer Vision Approach. In Proceedings of the 2006 Conference on Computer Vision and Pattern Recognition Workshop (CVPRW’06), New York, NY, USA, 17–22 June 2006; p. 112. [Google Scholar]
- Jiang, H.; Diao, Z.; Shi, T.; Zhou, Y.; Wang, F.; Hu, W.; Zhu, X.; Luo, S.; Tong, G.; Yao, Y.-D. A Review of Deep Learning-Based Multiple-Lesion Recognition from Medical Images: Classification, Detection and Segmentation. Comput. Biol. Med. 2023, 157, 106726. [Google Scholar] [CrossRef]
- Jiwani, N.; Gupta, K.; Pau, G.; Alibakhshikenari, M. Pattern Recognition of Acute Lymphoblastic Leukemia (ALL) Using Computational Deep Learning. IEEE Access 2023, 11, 29541–29553. [Google Scholar] [CrossRef]
- Myat Noe, S.; Zin, T.T.; Tin, P.; Kobayashi, I. Comparing State-of-the-Art Deep Learning Algorithms for the Automated Detection and Tracking of Black Cattle. Sensors 2023, 23, 532. [Google Scholar] [CrossRef]
- Marsot, M.; Mei, J.; Shan, X.; Ye, L.; Feng, P.; Yan, X.; Li, C.; Zhao, Y. An Adaptive Pig Face Recognition Approach Using Convolutional Neural Networks. Comput. Electron. Agric. 2020, 173, 105386. [Google Scholar] [CrossRef]
- Nassif, A.B.; Shahin, I.; Attili, I.; Azzeh, M.; Shaalan, K. Speech Recognition Using Deep Neural Networks: A Systematic Review. IEEE Access 2019, 7, 19143–19165. [Google Scholar] [CrossRef]
- Adjabi, I.; Ouahabi, A.; Benzaoui, A.; Taleb-Ahmed, A. Past, Present, and Future of Face Recognition: A Review. Electronics 2020, 9, 1188. [Google Scholar] [CrossRef]
- Campbell, C.J.; Barve, V.; Belitz, M.W.; Doby, J.R.; White, E.; Seltzer, C.; Di Cecco, G.; Hurlbert, A.H.; Guralnick, R. Identifying the Identifiers: How iNaturalist Facilitates Collaborative, Research-Relevant Data Generation and Why It Matters for Biodiversity Science. BioScience 2023, 73, 533–541. [Google Scholar] [CrossRef]
- Lapp, S.; Wu, T.; Richards-Zawacki, C.; Voyles, J.; Rodriguez, K.M.; Shamon, H.; Kitzes, J. Automated Detection of Frog Calls and Choruses by Pulse Repetition Rate. Conserv. Biol. 2021, 35, 1659–1668. [Google Scholar] [CrossRef]
- Xie, J.; Towsey, M.; Zhang, J.; Roe, P. Frog Call Classification: A Survey. Artif. Intell. Rev. 2018, 49, 375–391. [Google Scholar] [CrossRef]
- Alonso, J.B.; Cabrera, J.; Shyamnani, R.; Travieso, C.M.; Bolaños, F.; García, A.; Villegas, A.; Wainwright, M. Automatic Anuran Identification Using Noise Removal and Audio Activity Detection. Expert Syst. Appl. 2017, 72, 83–92. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Luque, A.; Romero-Lemos, J.; Carrasco, A.; Barbancho, J. Non-Sequential Automatic Classification of Anuran Sounds for the Estimation of Climate-Change Indicators. Expert Syst. Appl. 2018, 95, 248–260. [Google Scholar] [CrossRef]
- Bentéjac, C.; Csörgő, A.; Martínez-Muñoz, G. A Comparative Analysis of Gradient Boosting Algorithms. Artif. Intell. Rev. 2021, 54, 1937–1967. [Google Scholar] [CrossRef]
- Nalluri, M.; Pentela, M.; Eluri, N.R. A Scalable Tree Boosting System: XG Boost. Int. J. Res. Stud. Sci. Eng. Technol. 2020, 12, 36–51. [Google Scholar]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. Xgboost: Extreme Gradient Boosting; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Budka, M.; Jobda, M.; Szałański, P.; Piórkowski, H. Acoustic Approach as an Alternative to Human-Based Survey in Bird Biodiversity Monitoring in Agricultural Meadows. PLoS ONE 2022, 17, e0266557. [Google Scholar] [CrossRef]
- Melo, I.; Llusia, D.; Bastos, R.P.; Signorelli, L. Active or Passive Acoustic Monitoring? Assessing Methods to Track Anuran Communities in Tropical Savanna Wetlands. Ecol. Indic. 2021, 132, 108305. [Google Scholar] [CrossRef]
- Rowley, J.J.L.; Callaghan, C.T. Tracking the Spread of the Eastern Dwarf Tree Frog (Litoria fallax) in Australia Using Citizen Science. Aust. J. Zool. 2023, 70, 204–210. [Google Scholar] [CrossRef]
- Farr, C.M.; Ngo, F.; Olsen, B. Evaluating Data Quality and Changes in Species Identification in a Citizen Science Bird Monitoring Project. Citiz. Sci. Theory Pract. 2023, 8, 24. [Google Scholar] [CrossRef]
- Rowley, J.J.L.; Callaghan, C.T. The FrogID Dataset: Expert-Validated Occurrence Records of Australia’s Frogs Collected by Citizen Scientists. ZooKeys 2020, 912, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Westgate, M.J.; Scheele, B.C.; Ikin, K.; Hoefer, A.M.; Beaty, R.M.; Evans, M.; Osborne, W.; Hunter, D.; Rayner, L.; Driscoll, D.A. Citizen Science Program Shows Urban Areas Have Lower Occurrence of Frog Species, but Not Accelerated Declines. PLoS ONE 2015, 10, e0140973. [Google Scholar] [CrossRef]
- Paracuellos, M.; Rodríguez-Caballero, E.; Villanueva, E.; Santa, M.; Alcalde, F.; Dionisio, M.Á.; Fernández Cardenete, J.R.; García, M.P.; Hernández, J.; Tapia, M.; et al. Citizen Science Reveals Broad-Scale Variation of Calling Activity of the Mediterranean Tree Frog (Hyla meridionalis) in Its Westernmost Range. Amphib.-Reptil. 2022, 43, 251–261. [Google Scholar] [CrossRef]
- Nieto-Mora, D.A.; Rodríguez-Buritica, S.; Rodríguez-Marín, P.; Martínez-Vargaz, J.D.; Isaza-Narváez, C. Systematic Review of Machine Learning Methods Applied to Ecoacoustics and Soundscape Monitoring. Heliyon 2023, 9, e20275. [Google Scholar] [CrossRef] [PubMed]
- Platenberg, R.J.; Raymore, M.; Primack, A.; Troutman, K. Monitoring Vocalizing Species by Engaging Community Volunteers Using Cell Phones. Wildl. Soc. Bull. 2020, 44, 782–789. [Google Scholar] [CrossRef]
- Čeirāns, A.; Pupina, A.; Pupins, M. A New Method for the Estimation of Minimum Adult Frog Density from a Large-Scale Audial Survey. Sci. Rep. 2020, 10, 8627. [Google Scholar] [CrossRef] [PubMed]
- Walters, C.L.; Collen, A.; Lucas, T.; Mroz, K.; Sayer, C.A.; Jones, K.E. Challenges of Using Bioacoustics to Globally Monitor Bats. In Bat Evolution, Ecology, and Conservation; Adams, R.A., Pedersen, S.C., Eds.; Springer: New York, NY, USA, 2013; pp. 479–499. ISBN 978-1-4614-7397-8. [Google Scholar]
- Dufresnes, C.; Mazepa, G.; Jablonski, D.; Oliveira, R.C.; Wenseleers, T.; Shabanov, D.A.; Auer, M.; Ernst, R.; Koch, C.; Ramírez-Chaves, H.E.; et al. Fifteen Shades of Green: The Evolution of Bufotes Toads Revisited. Mol. Phylogenet. Evol. 2019, 141, 106615. [Google Scholar] [CrossRef] [PubMed]
- Vences, M. Development of New Microsatellite Markers for the Green Toad, Bufotes viridis, to Assess Population Structure at Its Northwestern Range Boundary in Germany. Salamandra 2019, 55, 191–198. [Google Scholar] [CrossRef]
- Giacoma, C.; Zugolaro, C.; Beani, L. The Advertisement Calls of the Green Toad (Bufo viridis): Variability and Role in Mate Choice. Herpetologica 1997, 53, 454–464. [Google Scholar]
- Lörcher, K.; Schneider, H. Vergleichende Bio-Akustische Untersuchungen an der Kreuzkröte, Bufo calamita (Laur.), und der Wechselkröte, Bufo v. Viridis (Laur.). Z. Tierpsychol. 1973, 32, 506–521. [Google Scholar] [CrossRef] [PubMed]
- Vargová, V.; Čerepová, V.; Balogová, M.; Uhrin, M. Calling Activity of Urban and Rural Populations of Green Toads Bufotes viridis Is Affected by Environmental Factors. N.-West. J. Zool. 2023, 19, 46–50. [Google Scholar]
- Ooms, J. Working with Audio and Video in R; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Ligges, U.; Krey, S.; Mersmann, O.; Schnackenberg, S. tuneR: Analysis of Music and Speech; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Sueur, J.; Aubin, T.; Simonis, C. Seewave: A Free Modular Tool for Sound Analysis and Synthesis. Bioacoustics 2008, 18, 213–226. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Mehdizadeh, R.; Eghbali, H.; Sharifi, M. Vocalization Development in Geoffroy’s Bat, Myotis emarginatus (Chiroptera: Vespertilionidae). Zool. Stud. 2021, 60, 20. [Google Scholar] [CrossRef]
- Castellano, S.; Rosso, A.; Doglio, S.; Giacoma, C. Body Size and Calling Variation in the Green Toad (Bufo viridis). J. Zool. 1999, 248, 83–90. [Google Scholar] [CrossRef]
- Chou, W.-H.; Lin, J.-Y. Geographical Variations of Rana seuterl (Anura: Ranidae) in Taiwan. Zool. Stud. 1997, 36, 201–221. [Google Scholar]
- Rowley, J.J.; Callaghan, C.T.; Cutajar, T.; Portway, C.; Potter, K.; Mahony, S.; Trembath, D.F.; Flemons, P.; Woods, A. FrogID: Citizen Scientists Provide Validated Biodiversity Data on Frogs of Australia. Herpetol. Conserv. Biol. 2019, 14, 155–170. [Google Scholar]
- Zilli, D. Smartphone-Powered Citizen Science for Bioacoustic Monitoring. Ph.D. Thesis, University of Southampton, Southampton, UK, 2015. [Google Scholar]
- Pérez-Granados, C.; Feldman, M.J.; Mazerolle, M.J. Combining Two User-Friendly Machine Learning Tools Increases Species Detection from Acoustic Recordings. Can. J. Zool. 2024, 102, 403–409. [Google Scholar] [CrossRef]
- Shonfield, J.; Heemskerk, S.; Bayne, E.M. Utility of Automated Species Recognition for Acoustic Monitoring of Owls. J. Raptor Res. 2018, 52, 42. [Google Scholar] [CrossRef]
- Dorcas, M.E.; Price, S.J.; Walls, S.C.; Barichivich, W.J. Auditory Monitoring of Anuran Populations. In Amphibian Ecology and Conservation; Dodd, C.K., Ed.; Oxford University Press: Oxford, UK, 2009; pp. 281–298. ISBN 978-0-19-954118-8. [Google Scholar]
- Rubbens, P.; Brodie, S.; Cordier, T.; Destro Barcellos, D.; Devos, P.; Fernandes-Salvador, J.A.; Fincham, J.I.; Gomes, A.; Handegard, N.O.; Howell, K.; et al. Machine Learning in Marine Ecology: An Overview of Techniques and Applications. ICES J. Mar. Sci. 2023, 80, 1829–1853. [Google Scholar] [CrossRef]
- Arch, V.S.; Narins, P.M. Sexual Hearing: The Influence of Sex Hormones on Acoustic Communication in Frogs. Hear. Res. 2009, 252, 15–20. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).