Abstract
This research proposes a new approach to improve information retrieval systems based on a multinomial naive Bayes classifier (MNBC), Bayesian networks (BNs), and a multi-terminology which includes MeSH thesaurus (Medical Subject Headings) and SNOMED CT (Systematized Nomenclature of Medicine of Clinical Terms). Our approach, which is entitled improving semantic information retrieval (IMSIR), extracts and disambiguates concepts and retrieves documents. Relevant concepts of ambiguous terms were selected using probability measures and biomedical terminologies. Concepts are also extracted using an MNBC. The UMLS (Unified Medical Language System) thesaurus was then used to filter and rank concepts. Finally, we exploited a Bayesian network to match documents and queries using a conceptual representation. Our main contribution in this paper is to combine a supervised method (MNBC) and an unsupervised method (BN) to extract concepts from documents and queries. We also propose filtering the extracted concepts in order to keep relevant ones. Experiments of IMSIR using the two corpora, the OHSUMED corpus and the Clinical Trial (CT) corpus, were interesting because their results outperformed those of the baseline: the P@50 improvement rate was +36.5% over the baseline when the CT corpus was used.
1. Introduction
The amount of data and information on the web is permanently increasing. Indeed, the web represents the most important source of knowledge and information that is quick and easy to access. Several information retrieval systems (IRSs) are available to users. An IRS’s task is to identify the information most relevant to a user’s query. This information can be a document, an image, a video, etc. In this paper, we focus especially on retrieving documents. A query is a set of words that represents a user’s need for information. The two main tasks that characterize an IRS are the indexing task (documents and the query) and the matching task between the index documents and the index of the query. Indexing consists of extracting the most representative terms of the document (or of a query) that allows for an IRS to select a set of documents to respond to the users’ queries. Term disambiguation is an essential step to improve the performance of an IRS, given the use of multi-terminologies for indexing. In multi-terminologies, a term may be related to more than one concept, so it is ambiguous. For example, “implantation procedure” and “implantation in uterus” are two different SNOMED-CT concepts that have the same term: “implantation, nos”. This study proposes a new approach to improve information retrieval (IR) called “improving semantic information retrieval” (IMSIR). Our main contribution is to combine an unsupervised method (BN) and a supervised method (MNBC) to extract concepts and then filter the results using semantic information provided by the Unified Medical Language System (UMLS). The role of the filtering step is to retain the relevant concepts. We also exploited multi-terminologies instead of one terminology. The use of multi-terminologies has had good results in indexing biomedical documents [1], allowing IRSs to extract more concepts that are relevant to the user’s query. Our approach exploits the structure of biomedical terminologies and the semantic information that these terminologies provide. In addition, IMSIR is based on the mechanism of inference which characterizes a Bayesian network (BN) to disambiguate terms and extract concepts and to match documents and queries. A BN is a graph that exploits a robust inference process for reasoning under uncertainty. The BN exploited by IMSIR performs a partial match that allows it to extract concepts that occur in the documents as well as concepts that partially occur in the documents. Moreover, IMSIR uses a multinomial naive Bayes classifier (MNBC) to extract concepts. The MNBC allows the IMSIR to enrich the index with new concepts whose terms do not occur in the documents [2]. For example, the concept “Bronchodilator Agents” belongs to the index of the document having the PMID = 11115306, although this concept does not occur in the document. In fact, experts can judge that concepts are relevant even when they do not occur in the document or in the query because they correspond to the context of the document or the query. Due to the fact that MNBC exploits features, these concepts can be extracted using MNBC. Machine learning is an efficient method for classification and its exploitation has led to good results [3], especially naive Bayes, which has been exploited in different works [4,5,6,7].
2. Related Work
In this section, we highlight the main approaches that have been proposed an IRS. The proposed approaches for IR that we cite are divided into unsupervised approaches and supervised approaches. We can cite some unsupervised approaches. In the work of Salton et al. [8], a similarity is computed using a vector space model (VSM) between the indexing terms of the query and the indexing terms of the documents. Based on a mathematical model, the authors of [9] proposed a probabilistic model that computes the likelihood of a document’s relevance for a query [10]. These two IR models [8,9] do not use semantic resources, which leads to less precision. The possibility and necessity measures are used to map the query to the documents [2]. For document ranking, Ref. [11] suggested a generalized ensemble model (gEnM) that linearly merges numerous rankers. The authors in [12] proposed the matching of concepts and queries with a possibilistic network (PN) that is also used to match concepts and documents and to retrieve and rank documents. To retrieve documents, Ensan and Bagheri [13] presented a cross-language information retrieval approach using a language different from the one used by the user when writing the query. The work reported in [14] performed a new approach that exploited the proximity and co-occurrence of query terms in the document. Moreover, VSM is used to retrieve documents. The work in [15] proposed an unsupervised neural vector space model (NVSM) that defined representations of documents. NVSM learns document and word representations and rank documents based on their similarity to query representations. To improve IRS, the authors in [16] propose enriching the query by combining domain-specific and global ontologies. The authors computed weights for both semantic relationships and the occurrence of each concept. To evaluate the query expansion process, this was integrated into current search engines. The results showed an improvement of 10% in terms of precision. A user’s profile and the context of their web history were exploited in [17] to improve IR.
We can site also some proposed supervised approaches for IR. The work in [18] defined relevance between a keyword style query and a document using a new deep learning model. The next and final step was a deep convolution stage, where, in order to compute the relevance, a deep feed-forward network is defined. The major limitation of this work is the small amount of training data used. The work described in [19] proposed the use of multinomial naive Bayes to improve IRS. The authors enriched the user’s query using the following process: after retrieving documents for a user’s query, the multinomial naive Bayes is exploited to extract relevant terms from retrieved documents. The document corpus is then processed and indexed. A limitation of this approach is that it depends on text and does not use semantic knowledge, which leads to low accuracy. The authors in [20] presented a neural semi-supervised framework to improve information retrieval. The framework is composed of two neural networks: an unsupervised network, which is a self-attention convolutional encoder–decoder network, and a supervised and sentence-level attention scientific literature retrieval network. The aim of combining the two networks is to detect the semantic information and learn the semantic representations in scientific literature datasets. Experiments using two datasets have shown encouraging results. The work of Prasath, Sarkar, and O’Reilly [21] proposed a supervised method to improve users’ queries and ranking candidates’ terms for indexing the query. The proposed framework is composed of two steps: the training stage and the testing stage. Pseudo-relevance feedback is used to have a set of candidates’ terms. These are illustrated as a feature vector. These vectors contain the extracted context-based feature and the extracted resource-based features. A supervised method is exploited to refine and rank terms.
According to their theoretical methods and also when analyzing the index of documents and queries, we can conclude that the proposed unsupervised methods for IR ignore relevant concepts that do not occur in the documents [2]. In fact, these approaches extract only concepts that occur or partially occur in the document. The missed concepts can be extracted using supervised methods. The proposed supervised methods for IR suffer from low performance in indexing biomedical documents due to the lack of efficient features and a training corpus. To deal with the limitations of both supervised and unsupervised methods, we propose combining both using a BN that shows a good performance in indexing biomedical documents [22] and a MNBC that allows the extraction of new relevant concepts. The results are then filtered using UMLS [23].
To further improve an IRS, especially when using multi-terminologies, it is essential to include a word sense disambiguation (WSD) step. We can classify the WSD approaches as either supervised, external resource-based approaches [24] or free-knowledge and unsupervised approaches. We now describe some knowledge-based approaches. The work reported in [25] proposed implementing a supervised WSD using two deep learning-based models. The first model is dependent on a bi-directional long short-term memory (BiLSTM) network. The second is a neural network model with an appropriate top-layer structure. The authors in [26] developed an approach called deepBioWSD. It takes advantage of current deep learning and UMLS breakthroughs to build a model that exploits one single BiLSTM network. The proposed model produces a logical prediction for any ambiguous phrase. These embeddings were used to initialize a network to be trained. According to the experiments, WSD approaches based on supervised methods outperform other approaches. However, developing a distinct classifier for each ambiguous phrase necessitates a large amount of training data, which may not be available. The work described in [27] builds concept embeddings using recent approaches in neural word embeddings. Cosine similarity combined with the embeddings and an external-based method is exploited to find the correct meaning of a word, leading to high accuracy. The probability measure used by the naive Bayes was exploited in work [28], which evaluated the context of an ambiguous word. The relevant concept with the highest score was kept to represent the sense of the polysemic word. A similarity was computed in [29] between the description of the candidates’ concepts and the context of the ambiguous word. [30] maps the documents to WordNet synsets. Definitions of UMLS [1] concepts were combined with word representations created on large corpora [31] to create a conceptual representation. The description of ambiguous terms’ context was compared to the conceptual representation. However, a large training set is needed to test the method. Machine learning is an efficient approach exploited for classification in different fields
3. Materials and Method
The process of our information retrieval system IMSIR is composed of the following steps, as illustrated in Figure 1:
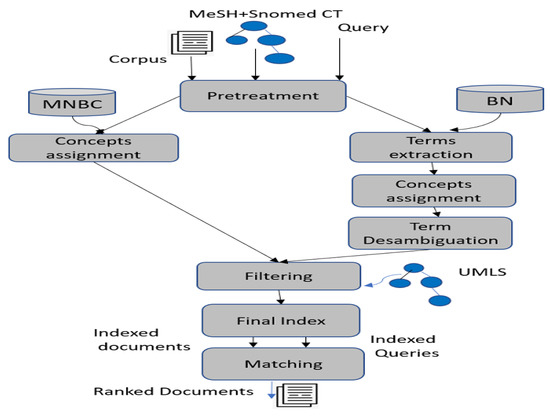
Figure 1.
The process of IMSIR.
- (1)
- Document, query and term pretreatment [12,32]
- (2)
- Concept extraction using a multinomial naive Bayes classifier (MNBC)
- (3)
- Term and concept extraction and disambiguation using a Bayesian network
- (4)
- Filtering concepts
- (5)
- Final indexes
- (6)
- Matching queries and documents
Let us consider a document denoted , a concept denoted , and a term denoted . is a document that belongs to the corpus of documents that will be indexed, a ∈, and U is the number of documents in the corpus. A ∈ with M as the number of concepts in UMLS that correspond to MeSH descriptors and SNOMED-CT concepts. A concept is composed of a set of terms, for example, “Abortion, induced” is a concept and its terms are, respectively, “Abortion, induced”, “Abortion, Rivanol”, “Fertility Control, Post conception”, “Abortion Failure”, and “Adverse effects” [2]. A ∈ with P is the number of terms that belong to all the concepts. A term can be composed of one or more than one word. A word ∈, L is the number of words that belong to terms and documents. A query is denoted and ∈, A being the number of queries. First of all, documents, queries, and terms are pretreated. The pretreatment step consists of removing punctuation, pruning stop words, stemming the text, and dividing phrases into words. Then, the concepts are extracted using MNBC. The outputs of this step are concepts (classes) mapped to documents. In the next step, terms are extracted using BN, and the concepts are assigned and disambiguated. The output of this step is the indexes of the concepts. The two indexes of each document are merged and filtered. Thus, we obtain a final index for each query and document. Finally, documents are retrieved for each query by matching a query to each index of document and documents are ranked according to the score Equation (17).
3.1. Concept Extraction Using a Multinomial Naïve Bayes Classifier
In this step, we use the MNBC to map concepts with documents with the aim of obtaining an index of concepts that represent the document. An MNBC is exploited for document classification [33], which consists of mapping classes and documents, using the statistical analysis of their contents. The classification is performed based on the documents that have already been classified. MNBC assumes the independence of variables and exploits probabilistic measures. It is characterized by the fact that the occurrence of one feature does not affect the probability of the occurrence of the other feature that characterized that category. A main advantage of MNBC is that it considers the Goss frequency, which is the frequency of the word and not the binary occurrence (whether the word occurs or not). The process of concept extraction using the MNBC is composed of the following steps (Figure 2): the training step and the classification step. The inputs of the training step are the already indexed documents and a set of classes C = that corresponds to the set of MeSH and SNOMED CT concepts. The documents of the training set (v is the number of documents) were indexed manually by experts with concepts that represent the classes of the document. The outputs of the training step are the probabilities . The probabilities, a test corpus, and the set of classes are the inputs of the classification step. A set of classified documents is the output of the last step (Equation (3)). The concepts (classes) are assigned to documents by computing the probabilities of documents knowing concepts , which is based on the probability that a word belongs to a given class (concept), also called likelihood. is calculated as follows (Equations (1) and (2)) [19]:
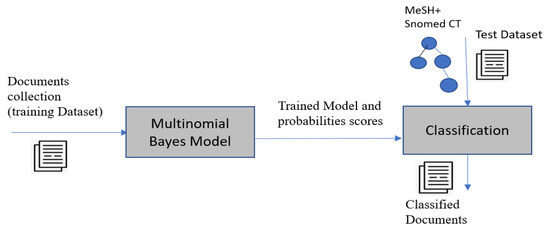
Figure 2.
The process of concept extraction using MNBC.
is the probability of a word that occurs in a class in the training documents.
is the number of occurrences of in the class . is the total number of words in the class .
is the length of the vocabulary,
The concept that will index a query or a document is selected using the maximizing function (Equation (3)):
3.2. Concept Extraction Using BN
To extract concepts, we employ a three-layer Bayesian network [22] (Figure 3). The network represents the following nodes: (i) the document to be indexed (ii) a word of the document and of the term , (iii) the term and (iv) the dependency relationships that exist between the nodes. A document belongs to the set of documents that will be indexed using our approach (U is the number of documents that will be indexed). A term belongs to the set of terms of MeSH and Snomed CT (P is the number of terms).
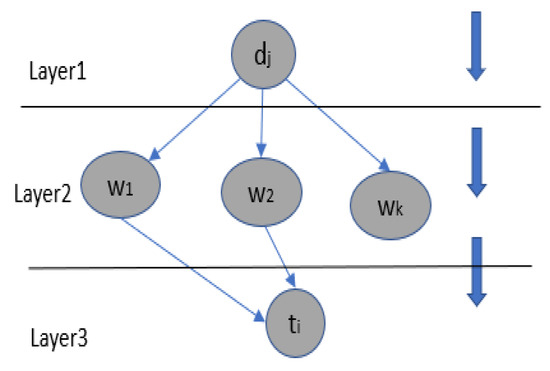
Figure 3.
The BN for term extraction.
3.2.1. Evaluation of a Term
A term is evaluated through the propagation of information given by the indexing term in the network once it is instantiated. Edges are activated by instantiating the term to the document. For each node, the conditional and marginal posterior probability are calculated given the conditional and marginal prior probability calculated according to Equations (4) and (5). According to the topology of the graph [22], we have:
represents the set of possible configurations of the parents of the instantiated term . is a possible configuration in .
are the possible configurations of the words (the parents of t).
a is a coefficient whose values are included in the interval [0, 1] with a < 1 if the words of a term are not in the same sentence. In the case where the words of a term are not in the same sentence, the coefficient a was tuned. : is the set of words of the document D. : is the set of words of the term T.
3.2.2. Computing the Weight of the Arc
To weigh the arc that links the nodes words to the document that will be indexed, we used the word frequency-inverse document frequency () measure. Thus, (Equations (6)–(8)):
Nu is the number of documents in the corpus test. is the number of documents in which the word k appears. In addition, m denotes the total number of words in the document. Finally, is the number of times the word k appears in the document . p is the number of words in the document that will be indexed. is the frequency of the word k in the document .
3.2.3. Aggregation of Words of Terms
In our model, we adopted the five canonical forms proposed by Turtle in their Bayesian network Information Retrieval (IR) model for each type of search [34]. In fact, we replaced the query by an indexing term. Thus, an indexing term can be aggregated by a probabilistic sum or a Boolean operator (OR, AND, NOT) or one of its variations, the weighted sum. The aggregations are defined in Equations (9)–(13) to evaluate the conditional probabilities ( is all the set of parents of T) of a node T having n parents (n words) and
The weight of the term and the word are denoted by , , respectively. A partial match between documents and terms is performed using our method. As a result, we used the disjunction to solve . If we consider a term as a disjunctive Boolean query, candidate terms are those that have at least one word in the document . However, is the formula for a phrase with p words.
3.2.4. Concept Assignment and Terms Disambiguation
To assign concepts to the terms, we compute the following Equation (Equation (14))
With as a set of terms of a concept .
The score of the sense of an ambiguous term is computed as follows (Equation (15))
3.3. Filtering Based on UMLS
We merge the two indexes that are composed of concepts from both methods (BN and MNBC), putting the concepts with the highest scores in the first ranks, and we delete the duplicated concepts. Then, the UMLS is exploited to filter the concepts extracted in the previous step while keeping the relevant ones. Both the MNBC and BN methods can produce irrelevant concepts that contain a part of the words of their terms or all the words of their terms (in the case of using MNBC) and do not occur in the document. To deal with this limitation, we divide the set of concepts into two indexes: the secondary index (SI) and main index (MI). The MI is a set of concepts that have at least one term that has all of its words occurring in the document. The SI is a set of concepts where the words of all of their terms do not occur in the document.
MI = , is a main concept. v is the number of MC. SI = , is a secondary concept. K is the number of SC.
The SCs are then ranked according to the score computed in Equation (16). We hypothesize that if an SC is co-occurring and has semantic links (according to the UMLS) with the MI’s L-initial MCs, it is more likely to be relevant. Finally, the n concepts with the highest scores are kept for indexing documents (n is tuned).
For example, the MeSH concepts “imaging, Three-Dimensional” and “coronary artery disease” are linked with the semantic relation “diagnoses”, and the MeSH concept “Endocarditis, Bacterial” co-occurs 100 times with the MeSH concept “Penicillins” in MEDLINE.
The number of semantic relations is expressed by NR, and the frequency of co-occurrence is CF. z is the total number of co-occurrences between all MC and all SC. s is the total number of semantic relations between all MC and all SC.
3.4. Computing a Similarity between Queries and Documents
We computed the similarity between a query and a document using a Bayesian network.
Thus, we computed using Equation (4) by replacing a term with a query. To compute we used , which is computed using and the Bayes rule as follows:
4. Results
Two corpora were used to evaluate our IR approach:
- (1)
- OHSUMED (https://trec.nist.gov/ (Hersh et al., 1994) accessed on 23 April 2023), is a document collection that was used for the TREC-9 filtering track. This corpus is the same as that used in [12]. Details on this corpus are presented in [12].
- (2)
- The Clinical Trial corpus 2021, which is composed of topics (descriptions of the user needs), clinical documents, and relevance judgments evaluated by experts. The topics correspond to the queries. This is the link to the corpus: http://www.trec-cds.org accessed on 12 May 2022.
We chose these two corpora because the first one is characterized by short queries and the second is characterized by long queries, which allowed us to test the performance of IMSIR using the two types of queries. Below is an example of a topic (query) in Clinical Trial corpus:
<topics task=“2021 TREC Clinical Trials”>
<topic number=“-1”>
A 2-year-old boy is brought to the emergency department by
their parents for 5 days of high fever
and irritability…
topic>
topics>
To test our approach, we indexed queries and documents using IMSIR and we computed the score (Equation (16)) between each query and document. The documents were then retrieved and ranked according to the score (Equation (16)) as a response to the query.
To evaluate our proposed information retrieval approach, we opted for the mean precision (MAP) (Equation (18)). We also computed the precision at ranks 5, 20, and 50. We compared the performance of our approach that exploits MNBC with the performance of our approach using a support vector machine (SVM) or a random forest classifier (RFC) instead of MNBC (Table 1). In addition, we computed the improvement rate () (19)), which highlights the added value of our contributions compared to a baseline, which is the work of [35] (Table 2 and Table 3). This is a recent approach that exploits supervised methods and terminologies to improve the IRS. We also compared our work that exploits BN to match queries and documents with our work that exploits BM25 or VSM (vector space model) instead of BN (Table 2 and Table 3) and with the approach of Mingying et al. [20], which is a recent approach that exploits a semi-supervised method. We also tested CIRM [12] (Table 2 and Table 3). Moreover, we computed Students’ t-tests between the ranks (P@10, P@20, P@50, and MAP) obtained by each method tested and the baseline.

Table 1.
Evaluation of IMSIR using different supervised methods when the corpus OHSUMED is exploited.

Table 2.
Evaluation of IMSIR when the corpus OHSUMED is exploited.

Table 3.
Evaluation of IMSIR when the Clinical Trial corpus is exploited.
The total number of documents is n. The number of relevant documents is N. In addition, P@i indicates the accuracy of document retrieval. Finally, if the document is not relevant, then is equal to 0 and if it is relevant, then is equal to 1.
As shown in Table 1, Table 2 and Table 3, the performance of our information retrieval system (IMSIR) is better than the baseline and the approach of [20] in terms of MAP and precision in different ranks of documents. Moreover, our proposed approach shows comparable results with CIRM. Furthermore, compared to the baseline, IMSIR is statistically significant. These results highlight the interest in the similarities proposed in IMSIR, which exploits a statistical and semantic weight for ranking concepts and proves that the structure of RB and the information propagation mechanism are adequate for controlled indexing. In addition, MNBC brings new relevant concepts, especially those whose terms do not occur in the document or in the query. The combination of BNs, a BNC, and the use of UMLS for filtering contributes to the retaining of relevant concepts and improvement of extraction and ranking of concepts. Table 4 shows the interest in using the filtering step in the process of IMSIR. In fact, the performance of IMSIR becomes greater when applying the filtering step. Using the co-occurrences and semantic relations provided by UMLS allows for the deletion of irrelevant concepts, especially those where a part of their words do not occur in the document or all of their words do not occur in the document. It is also clear also that IMSIR performs better when using the Clinical Trial corpus (CTC) than when using the OHSUMED corpus (Table 2). These results are explained by the fact that IMSIR exploits statistic measures that demonstrate good results when using long queries. Moreover, according to Table 1, our approach returns better results when using the supervised method MNBC than when using SVM or RFC. Table 2 and Table 3 also highlight the use of BN to match queries, as the performance of IMSIR-BN outperforms those of IMSIR-VSM and IMSIR-BM25. Moreover, IMSIR-BN achieved better performance when NC = 5 (Table 5 and Figure 4).

Table 4.
Evaluation of IMSIR with and without the step of filtering.

Table 5.
The performance of IMSIR-BN using different values of NC.
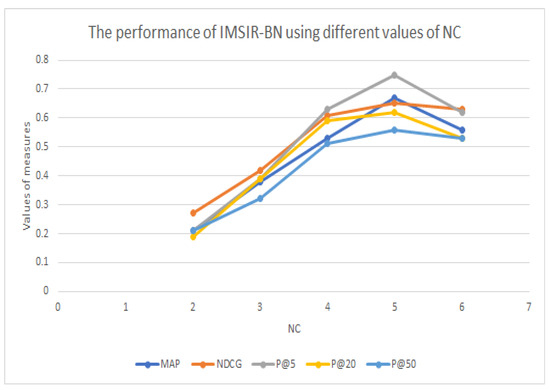
Figure 4.
Tuning the value of NC. C is a concept, is a term, is a word belonging to a document or to a term, and is a document.
5. Conclusions
This study developed a novel IRS called IMSIR that allows the improvement of the process of indexing documents and queries by adding new relevant concepts to the indexes. In fact, our approach combines a BN with three layers, MNBC, and terminologies to extract, disambiguate, and rank concepts. The BN allows extraction of concepts that occur and partially occur in the documents, and MNBC allows for enrichment of the index with relevant concepts that do not occur in the document. A semantic method is also exploited in IMSIR by using the terminologies; in fact, concepts are extracted when their terms occur in the documents or queries. An added value of our approach is the filtering step after the extraction of concepts using the supervised and the unsupervised methods. These methods do not perform an exact match; thus, irrelevant concepts may be extracted, and a filtering step is required in order to keep relevant concepts. This step exploits the properties of UMLS which are semantic relations and co-occurrences. In addition, IMSIR aims to enhance the ranking of retrieved unstructured documents in an IRS by using an efficient score to rank documents. Moreover, the experiments with IMSIR using the Clinical Trial corpus highlighted the added value of combining the inference mechanism of BN, MNBC, and the biomedical terminologies’ structure and their semantics to extract, disambiguate, and rank concepts and documents. Furthermore, the experiments allowed us to determine how many concepts were used to index the queries. In the future, we aim to use the same methods described in this paper to enhance IRS through query expansion. In addition, we intend to employ more terminologies, as it will obtain a performance increase over the use of one terminology alone. Moreover, we aim to improve the ranking of concepts step after filtering.
Author Contributions
Conceptualization, W.C.; methodology, W.C.; software, W.C. and M.W.; validation, W.C., M.W., R.A., M.A. and O.E.; formal analysis and investigation, W.C. and M.W.; resources, W.C. and M.W.; data curation, W.C.; writing—original draft preparation, W.C.; writing—review and editing, R.A., M.A. and O.E.; visualization, all the authors; supervision, all the authors; project administration, all the authors; funding acquisition, all the authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data exploited in our experiments are available at the following links: https://trec.nist.gov/, accessed on 1 August 2022 and http://www.cs.cmu.edu/~rafa/ir/ohsumed.html, accessed on 12 September 2022.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chebil, W.; Soualmia, L.F.; Omri, M.N.; Darmoni, S.J. Indexing biomedical documents with a possibilistic network. J. Assoc. Inf. Sci. Technol. 2016, 67, 928–941. [Google Scholar] [CrossRef]
- Chebil, W.; Soualmia, L.F.; Dahamna, B.; Darmoni, S.J. Indexation automatique de documents en santé: Évaluation et analyse de sources d’erreurs. IRBM 2012, 33, 316–329. [Google Scholar] [CrossRef]
- Alazab, M. Automated malware detection in mobile app stores based on robust feature generation. Electronics 2020, 9, 435. [Google Scholar] [CrossRef]
- De Stefano, C.; Fontanella, F.; Marrocco, C.; di Freca, A.S.A. Hybrid Evolutionary Algorithm for Bayesian Networks Learning: An Application to Classifier Combination. In Applications of Evolutionary Computation. EvoApplications 2010; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6024. [Google Scholar]
- Shen, Y.; Zhang, L.; Zhang, J.; Yang, M.; Tang, B.; Li, Y.; Lei, K. CBN: Constructing a Clinical Bayesian Network based on Data from the Electronic Medical Record. J. Biomed. Inform. 2018, 88, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Malviya, S.; Tiwary, U.S. Knowledge-Based Summarization and Document Generation using Bayesian Network. Procedia Comput. Sci. 2018, 89, 333–340. [Google Scholar] [CrossRef]
- de Campos, C.P.; Zeng, Z.; Ji, Q. Structure Learning of Bayesian Networks Using Constraints. In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; Association for Computing Machinery: New York, NY, USA. [Google Scholar]
- Salton, G.; Wong, A.; Yang, C. A vector space model for automatic indexing. Commun. ACM 1975, 18, 438–446, 613–620. [Google Scholar] [CrossRef]
- Robertson, S.; Jones, K.S. Relevance weighting of search terms. J. Am. Soc. Inf. Sci. 1976, 27, 129–146. [Google Scholar] [CrossRef]
- Salton, G.; Fox, E.A.; Wu, H. Extended boolean information retrieval. Commun. ACM 1983, 26, 1022–1036. [Google Scholar] [CrossRef]
- Wang, Y.; Choi, I.; Liu, H. Generalized ensemble model for document ranking in information retrieval. arXiv 2015, arXiv:1507.08586. [Google Scholar] [CrossRef]
- Chebil, W.; Soualmia, L.F.; Omri, M.; Darmoni, S.J. Possibilistic Information Retrieval Model Based on a Multi-Terminology. In Proceedings of the ADMA Advanced Data Mining and Applications, Nanjing, China, 18 November 2018. [Google Scholar]
- Ensan, F.; Bagheri, E. Retrieval model through semantic linking. In Proceedings of the 10th ACM International Conference on Websearch and Data Mining, Cambridge, UK, 6–10 February 2017; pp. 181–190. [Google Scholar]
- Sneiders, E. Text retrieval by term cooccurrences in a query based vector space. In Proceedings of the COLING 2016 the 26th International Conference on Computational Linguistics, Osaka, Japan, 11–16 December 2016; Technical Papers. pp. 2356–2365. [Google Scholar]
- Kenter, T.; Borisov, A.; Van Gysel, C.; Dehghani, M.; de Rijke, M.; Mitra, B. Neural networks for information retrieval. In Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval, Tokyo, Japan, 7–11 August 2017; pp. 1403–1406. [Google Scholar]
- Jaina, S.; Seejab, K.; Jindal, R. A fuzzy ontology framework in information retrieval using semantic query expansion. J. Inf. Manag. Data Insights 2021, 1, 300–307. [Google Scholar] [CrossRef]
- Chebil, W.; Wedyan, M.O.; Lu, H.; Elshaweesh, O.G. Context-Aware Personalized Web Search Using Navigation History. Int. J. Semant. Web Inf. Syst. 2020, 16, 91–107. [Google Scholar] [CrossRef]
- Mohan, S.; Fiorini, N.; Kim, S.; Lu, Z. A fast deep learning model for textual relevance in biomedical information retrieval. In Proceedings of the 2018 World Wide Web Conference, Lyon, France, 23–27 April 2018; pp. 77–86. [Google Scholar]
- Silva, S.; Seara, V.A.; Celard, P.; Iglesias, E.L.; Borrajo, L. A query expansion method using multinomial naive bayes. Appl. Sci. 2021, 11, 10284. [Google Scholar] [CrossRef]
- Xu, M.; Du, J.; Xue, Z.; Kou, F.; Xu, X. A semi-supervised semantic-enhanced framework for scientific literature retrieval. Neurocomputing 2021, 461, 450–461. [Google Scholar] [CrossRef]
- Prasath, R.; Sarkar, S.; OReilly, P. Improving cross language information retrieval using corpus based query suggestion approach. In Proceedings of the International Conference on Intelligent Text Processing and Computational Linguistics, Cairo, Egypt, 14–20 April 2015; pp. 448–457. [Google Scholar]
- Chebil, W.; Soualmia, L.F.; Omri, M.; Darmoni, S.J. Indexing biomedical documents with Bayesian networks and terminologies. In Proceedings of the 12th International Conference on Intelligent Systems and Knowledge Engineering (ISKE), Nanjing, China, 24–26 November 2017. [Google Scholar]
- Bodenreider, O. The unified medical language system umls integrating biomedical terminology. Nucleic Acids Res. 2004, 32, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Wedyan, M.; Alhadidi, B.; Alrabea, A. The effect of using a thesaurus in Arabic information retrieval system. Int. J. Comput. Sci. 2012, 9, 431–435. [Google Scholar]
- Zhang, C.; Bis, D.; Liu, X. Biomedical word sense disambiguation with bidirectional long short-term memory and attention-based neural networks. BMC Bioinform. 2019, 28, 159–182. [Google Scholar] [CrossRef] [PubMed]
- Pesaranghader, A.; Matwin, S.; Sokolova, M.; Pesaranghader, A. Deepbiowsd effective deep neural word sense disambiguation of biomedical text data. J. Am. Med. Inform. Assoc. 2020, 26, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Sabbir, A.; Jimeno-Yepes, A.; Kavuluru, R. Knowledge-Based Biomedical Word Sense Disambiguation with Neural Concept Embeddings. In Proceedings of the 2017 IEEE 17th International Conference on Bioinformatics and Bioengineering (BIBE), Washington, DC, USA, 23–25 October 2017. [Google Scholar] [CrossRef]
- Yepes, A.; Berlanga, R. Knowledge based word concept model estimation and refinement for biomedical text mining. J. Biomed. Inform. 2015, 53, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Lesk, M. Automatic sense disambiguation using machine readable dictionaries how to tell a pine cone from an ice cream cone. In SIGDOC ’86: Proceedings of the 5th Annual International Conference on Systems Documentation; Association for Computing Machinery: Washington, DC, USA, 1986; pp. 24–26. [Google Scholar]
- Voorhees, E.M. Using Wordnet to Disambiguate Word Senses for Text Retrieval. In SIGIR ’93: Proceedings of the ACM SIGIR, Conference on Research and Development in Information Retrieval; Association for Computing Machinery: Washington, DC, USA, 1993; pp. 171–180. [Google Scholar]
- Tulkens, S.; Suster, S.; Daelemans, W. Using distributed representations to disambiguate biomedical and clinical concepts. In Proceedings of the 15th Workshop on Biomedical Natural Language Processing, Berlin, Germany, 12 August 2016; Volume 53, pp. 77–82. [Google Scholar]
- Chebil, W.; Soualmia, L.F. Improving semantic information retrieval by combining possibilistic networks, vector space model and pseudo-relevance feedback. J. Inf. Sci. 2023. [Google Scholar] [CrossRef]
- Raschka, S. Naive Bayes and Text Classification I-Introduction and Theory. arXiv 2014. arXiv:1410-5329. [Google Scholar]
- Turtle, H.; Croft, W.B. Inference Networks for Document Retrieval. In Proceedings of the ACM SIGIR Conference on Research and Development in Information Retrieval, Association for Computing Machinery, Brussels Belgium, 5–7 September 1990; pp. 1–24. [Google Scholar]
- Xu, B.; Lin, H.; Yang, L.; Xu, K.; Zhang, Y.; Zhang, D.; Yang, Z.; Wang, J.; Lin, Y.; Yin, F. A supervised term ranking model for diversity enhanced biomedical information retrieval. BMC Bioinform. 2019, 20, 590. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).