Identifying the Early Signs of Preterm Birth from U.S. Birth Records Using Machine Learning Techniques
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Data Elements
3. Model Development
3.1. Handling Missing Values and Categorical Variables
3.2. Interpretation Techniques
4. Results
4.1. Study Design
4.2. Results of the Machine Learning Algorithms
4.3. Comparison with Other Studies
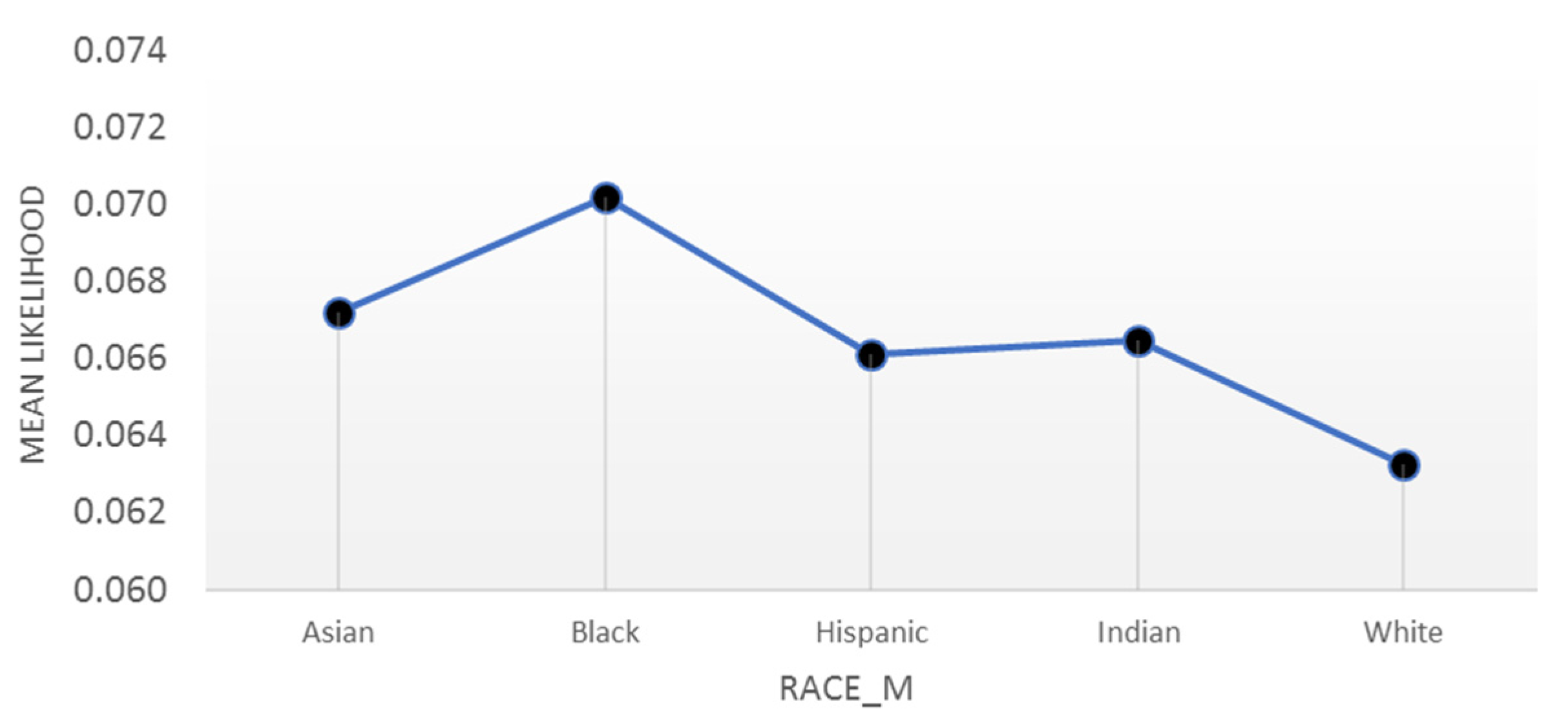
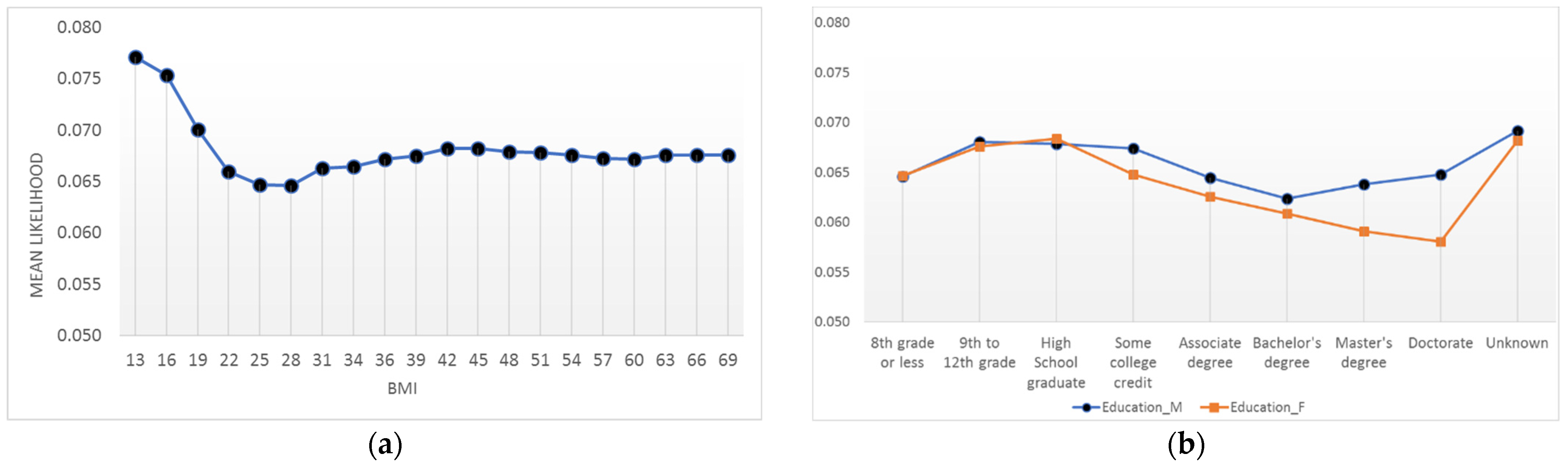
4.4. Interpretations
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blencowe, H.; Cousens, S.; Chou, D.; Oestergaard, M.; Say, L.; Moller, A.-B.; Kinney, M.; Lawn, J.; the Born Too Soon Preterm Birth Action Group. Born Too Soon: The global epidemiology of 15 million preterm births. Reprod. Health 2013, 10 (Suppl. S1), S2. [Google Scholar] [CrossRef] [PubMed]
- Mathews, T.J.; MacDorman, M.F.; Menacker, F. Infant Mortality Statistics from the 2013 Period: Linked Birth/Infant Death Data Set. Natl. Vital Stat. 2015, 64, 1–30. [Google Scholar]
- Ebrahimvandi, A.; Hosseinichimeh, N.; Iams, J. Understanding State-Level Variations in the US Infant Mortality: 2000 to 2015. Am. J. Perinatol. 2018, 36, 1271–1277. [Google Scholar] [CrossRef]
- Butler, A.S.; Behrman, R.E. Preterm Birth: Causes, Consequences, and Prevention; National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Saigal, S.; Doyle, L.W. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet 2008, 371, 261–269. [Google Scholar] [CrossRef]
- Iams, J.D. Identification of candidates for progesterone: Why, who, how, and when? Obstet. Gynecol. 2014, 123, 1317–1326. [Google Scholar] [CrossRef]
- Katz, K.S.; Blake, S.M.; Milligan, R.A.; Sharps, P.W.; White, D.B.; Rodan, M.F.; Rossi, M.; Murray, K.B. The design, implementation and acceptability of an integrated intervention to address multiple behavioral and psychosocial risk factors among pregnant African American women. BMC Pregnancy Childbirth 2008, 8, 22. [Google Scholar] [CrossRef]
- Goldenberg, R.L.; Culhane, J.F.; Iams, J.D.; Romero, R. Epidemiology and causes of preterm birth. Lancet 2008, 371, 75–84. [Google Scholar] [CrossRef]
- Singh, N.; Bonney, E.; McElrath, T.; Lamont, R.F.; Shennan, A.; Gibbons, D.; Guderman, J.; Stener, J.; Helmer, H.; Rajl, H. Prevention of preterm birth: Proactive and reactive clinical practice-are we on the right track? Placenta 2020, 98, 6–12. [Google Scholar] [CrossRef]
- Hooft, J.V.; Duffy, J.M.N.; Daly, M.; Williamson, P.R.; Meher, S.; Thom, E.; Saade, G.R.; Alfirevic, Z.; Mol, B.W.J.; Khan, K.S. A Core Outcome Set for Evaluation of Interventions to Prevent Preterm Birth. Obstet. Gynecol. 2016, 127, 49–58. [Google Scholar] [CrossRef]
- Hosseinichimeh, N.; Kim, H.; Ebrahimvandi, A.; Iams, J.; Andersen, D.J.S.R.; Science, B. Using a Stakeholder Analysis to Improve Systems Modelling of Health Issues: The Impact of Progesterone Therapy on Infant Mortality in Ohio. Syst. Res. Behav. Sci. 2019, 36, 476–493. [Google Scholar] [CrossRef]
- Darabi, N.; Ebrahimvandi, A.; Hosseinichimeh, N.; Triantis, K. A DEA evaluation of US States’ healthcare systems in terms of their birth outcomes. Expert Syst. Appl. 2021, 182, 115278. [Google Scholar] [CrossRef]
- Boots, A.B.; Sanchez-Ramos, L.; Bowers, D.M.; Kaunitz, A.M.; Zamora, J.; Schlattmann, P. The short-term prediction of preterm birth: A systematic review and diagnostic metaanalysis. Am. J. Obstet. Gynecol. 2014, 210, 54.e1–54.e10. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.-A.; Watson, L.; Rayner, J.A.; Rowlands, S. Risk-scoring systems for predicting preterm birth with the aim of reducing associated adverse outcomes. Cochrane Database Syst. Rev. 2015, 2015, CD004902. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Raja, E.A.; Mirazo, E.R.; Campbell, D.M.; Lee, A.J.; Norman, J.E.; Bhattacharya, S. Inherited Predisposition to Spontaneous Preterm Delivery. Obstet. Gynecol. 2010, 115, 1125–1133. [Google Scholar] [CrossRef] [PubMed]
- Laughon, S.K.; Albert, P.S.; Leishear, K.; Mendola, P. The NICHD Consecutive Pregnancies Study: Recurrent preterm delivery by subtype. Am. J. Obstet. Gynecol. 2013, 210, 131.e1–131.e8. [Google Scholar] [CrossRef]
- Webb, D.A.; Mathew, L.; Culhane, J.F. Lessons learned from the Philadelphia Collaborative Preterm Prevention Project: The prevalence of risk factors and program participation rates among women in the intervention group. BMC Pregnancy Childbirth 2014, 14, 368. [Google Scholar] [CrossRef]
- Belaghi, R.A.; Beyene, J.; McDonald, S.D. Clinical risk models for preterm birth less than 28 weeks and less than 32 weeks of gestation using a large retrospective cohort. J. Perinatol. 2021, 41, 2173–2181. [Google Scholar] [CrossRef]
- Martin, J.A.; Hamilton, B.E.; Osterman, M.J.; Driscoll, A.K.; Drake, P. Births: Final Data for 2016; National Vital Statistics Reports: Hyattsville, MD, USA, 2018. [Google Scholar]
- Fuchs, F.; Monet, B.; Ducruet, T.; Chaillet, N.; Audibert, F. Effect of maternal age on the risk of preterm birth: A large cohort study. PLoS ONE 2018, 13, e0191002. [Google Scholar] [CrossRef]
- Newman, R.; Goldenberg, R.; Iams, J.; Meis, P.; Mercer, B.; Moawad, A.; Thom, E.; Miodovnik, M.; Caritis, S.; Dombrowski, M. Preterm prediction study: Comparison of the cervical score and Bishop score for prediction of spontaneous preterm delivery. Obstet. Gynecol. 2008, 112, 508. [Google Scholar] [CrossRef]
- Magee, L.; Von Dadelszen, P.; Chan, S.; Gafni, A.; Gruslin, A.; Helewa, M.; Hewson, S.; Kavuma, E.; Lee, S.; Logan, A. The control of hypertension in pregnancy study pilot trial. BJOG Int. J. Obstet. Gynaecol. 2007, 114, 770-e20. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Alleman, B.W.; Smith, A.R.; Byers, H.M.; Bedell, B.; Ryckman, K.K.; Murray, J.C.; Borowski, K.S. A proposed method to predict preterm birth using clinical data, standard maternal serum screening, and cholesterol. Am. J. Obstet. Gynecol. 2013, 208, 472.e1–472.e11. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Darmstadt, G.L.; Gruber, S.; Foeller, M.E.; Carmichael, S.L.; Stevenson, D.K.; Shaw, G.M. Application of machine-learning to predict early spontaneous preterm birth among nulliparous non-Hispanic black and white women. Ann. Epidemiol. 2018, 28, 783–789.e1. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Osmundson, S.; Edwards, D.R.V.; Jackson, G.P.; Malin, B.A.; Chen, Y. Deep learning predicts extreme preterm birth from electronic health records. J. Biomed. Inform. 2019, 100, 103334. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, L.; VanDyne, M.; Lin, S.; Talbert, S. Data mining issues and opportunities for building nursing knowledge. J. Biomed. Inform. 2003, 36, 379–388. [Google Scholar] [CrossRef]
- Woolery, L.K.; Grzymala-Busse, J. Machine Learning for an Expert System to Predict Preterm Birth Risk. J. Am. Med. Inform. Assoc. 1994, 1, 439–446. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Chuang, C.-H.; Yang, Y.-J.; Wu, T.-P. Exploring the risk factors of preterm birth using data mining. Expert Syst. Appl. 2011, 38, 5384–5387. [Google Scholar] [CrossRef]
- Van Dyne, M.; Woolery, L.; Gryzmala-Busse, J.; Tsatsoulis, C. Using machine learning and expert systems to predict preterm delivery in pregnant women. In Proceedings of the Tenth Conference on Artificial Intelligence for Applications, San Antonia, TX, USA, 1–4 March 1994; pp. 344–350. [Google Scholar]
- Sun, Q.; Zou, X.; Yan, Y.; Zhang, H.; Wang, S.; Gao, Y.; Liu, H.; Liu, S.; Lu, J.; Yang, Y.; et al. Machine Learning-Based Prediction Model of Preterm Birth Using Electronic Health Record. J. Health Eng. 2022, 2022, 9635526. [Google Scholar] [CrossRef]
- Kim, J.-I.; Lee, J.Y. Systematic Review of Prediction Models for Preterm Birth Using CHARMS. Biol. Res. Nurs. 2021, 23, 708–722. [Google Scholar] [CrossRef]
- Pereira, G.; Regan, A.K.; Wong, K.; Tessema, G.A. Gestational age as a predictor for subsequent preterm birth in New South Wales, Australia. BMC Pregnancy Childbirth 2021, 21, 607. [Google Scholar] [CrossRef]
- Bertini, A.; Salas, R.; Chabert, S.; Sobrevia, L.; Pardo, F. Using Machine Learning to Predict Complications in Pregnancy: A Systematic Review. Front. Bioeng. Biotechnol. 2022, 9, 780389. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.B.; McIntire, D.D.; McDonald, J.; Gard, J.; Turrichi, P.; Leveno, K.J. 17-alpha Hydroxyprogesterone caproate did not reduce the rate of recurrent preterm birth in a prospective cohort study. Am. J. Obstet. Gynecol. 2017, 216, 600.e1–600.e9. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.N.; Norwitz, E. Preterm Birth: Risk Factors, Interventions for Risk Reduction, and Maternal Prognosis. Available online: https://www.uptodate.com/contents/preterm-birth-risk-factors-interventions-for-risk-reduction-and-maternal-prognosis (accessed on 2 March 2019).
- Iams, J.D. Prevention of preterm parturition. N. Engl. J. Med. 2014, 370, 254–261. [Google Scholar] [CrossRef] [PubMed]
- He, J.-R.; Ramakrishnan, R.; Lai, Y.-M.; Li, W.-D.; Zhao, X.; Hu, Y.; Chen, N.-N.; Hu, F.; Lu, J.-H.; Wei, X.-L.; et al. Predictions of Preterm Birth from Early Pregnancy Characteristics: Born in Guangzhou Cohort Study. J. Clin. Med. 2018, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Linked Birth/Infant Death Records 2007–2019. Available online: https://wonder.cdc.gov/lbd-current.html (accessed on 2 March 2019).
- Bengio, Y. Deep learning of representations: Looking forward. In Proceedings of the International Conference on Statistical Language and Speech Processing, Tarragona, Spain, 29–31 July 2013; pp. 1–37. [Google Scholar]
- Goldstein, M.; Uchida, S. A Comparative Evaluation of Unsupervised Anomaly Detection Algorithms for Multivariate Data. PLoS ONE 2016, 11, e0152173. [Google Scholar] [CrossRef] [PubMed]
- Leng, Q.; Qi, H.; Miao, J.; Zhu, W.; Su, G. One-Class Classification with Extreme Learning Machine. Math. Probl. Eng. 2015, 2015, 412957. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. The Elements of Statistical Learning; Series in Statistics; Springer: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Schapire, R.E.; Freund, Y. Boosting: Foundations and algorithms. Kybernetes 2013, 42, 164–166. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 3146–3154. [Google Scholar]
- Goodwin, L.K.; Iannacchione, M.A.; Hammond, W.E.; Crockett, P.; Maher, S.; Schlitz, K. Data Mining Methods Find Demographic Predictors of Preterm Birth. Nurs. Res. 2001, 50, 340–345. [Google Scholar] [CrossRef]
- Vovsha, I.; Rajan, A.; Salleb-Aouissi, A.; Raja, A.; Radeva, A.; Diab, H.; Tomar, A.; Wapner, R. Predicting preterm birth is not elusive: Machine learning paves the way to individual wellness. In Proceedings of the 2014 AAAI Spring Symposium Series, Palo Alto, CA, USA, 24–26 March 2014. [Google Scholar]
- Meis, P.J.; Goldenberg, R.L.; Mercer, B.M.; Iams, J.D.; Moawad, A.H.; Miodovnik, M.; Menard, M.; Caritis, S.; Thurnau, G.R.; Bottoms, S.F.; et al. The preterm prediction study: Risk factors for indicated preterm births. Am. J. Obstet. Gynecol. 1998, 178, 562–567. [Google Scholar] [CrossRef]
- Manuck, T.A. Racial and ethnic differences in preterm birth: A complex, multifactorial problem. Semin. Perinatol. 2017, 41, 511–518. [Google Scholar] [CrossRef]
- Lu, M.C.; Kotelchuck, M.; Hogan, V.; Jones, L.; Wright, K.; Halfon, N. Closing the Black-White gap in birth outcomes: A life-course approach. Ethn. Dis. 2010, 20 (Suppl. S2), 62–76. [Google Scholar]
- Wadhwa, P.D.; Entringer, S.; Buss, C.; Lu, M.C. The Contribution of Maternal Stress to Preterm Birth: Issues and Considerations. Clin. Perinatol. 2011, 38, 351–384. [Google Scholar] [CrossRef] [PubMed]
- Krishna, U.; Bhalerao, S. Placental Insufficiency and Fetal Growth Restriction. J. Obstet. Gynecol. India 2011, 61, 505–511. [Google Scholar] [CrossRef]
- Fraser, A.M.; Brockert, J.E.; Ward, R. Association of Young Maternal Age with Adverse Reproductive Outcomes. N. Engl. J. Med. 1995, 332, 1113–1118. [Google Scholar] [CrossRef]
- Hendler, I.; Goldenberg, R.L.; Mercer, B.M.; Iams, J.D.; Meis, P.J.; Moawad, A.H.; MacPherson, C.A.; Caritis, S.N.; Miodovnik, M.; Menard, K.M.; et al. The Preterm Prediction study: Association between maternal body mass index and spontaneous and indicated preterm birth. Am. J. Obstet. Gynecol. 2005, 192, 882–886. [Google Scholar] [CrossRef]
- Honest, H.; Bachmann, L.M.; Ngai, C.; Gupta, J.K.; Kleijnen, J.; Khan, K.S. The accuracy of maternal anthropometry measurements as predictor for spontaneous preterm birth—A systematic review. Eur. J. Obstet. Gynecol. Reprod. Biol. 2005, 119, 11–20. [Google Scholar] [CrossRef]
- El-Sayed, A.M.; Galea, S. Temporal Changes in Socioeconomic Influences on Health: Maternal Education and Preterm Birth. Am. J. Public Health 2012, 102, 1715–1721. [Google Scholar] [CrossRef]
- Auger, N.; Abrahamowicz, M.; Park, A.L.; Wynant, W. Extreme maternal education and preterm birth: Time-to-event analysis of age and nativity-dependent risks. Ann. Epidemiol. 2013, 23, 1–6. [Google Scholar] [CrossRef]
- Luo, Z.-C.; Wilkins, R.; Kramer, M.S. Effect of neighbourhood income and maternal education on birth outcomes: A population-based study. Can. Med. Assoc. J. 2006, 174, 1415–1420. [Google Scholar] [CrossRef] [PubMed]
- Meertens, L.J.; Van Montfort, P.; Scheepers, H.C.; Van Kuijk, S.M.; Aardenburg, R.; Langenveld, J.; Van Dooren, I.M.; Zwaan, I.M.; Spaanderman, M.E.; Smits, L.J. Prediction models for the risk of spontaneous preterm birth based on maternal characteristics: A systematic review and independent external validation. Acta Obstet. Gynecol. Scand. 2018, 97, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Osterman, M.J.K.; Kirmeyer, S.E.; Gregory, E.C.W. Measuring Gestational Age in Vital Statistics Data: Transitioning to the Obstetric Estimate. Natl. Vital Stat. Rep. 2015, 64, 1–20. [Google Scholar] [PubMed]

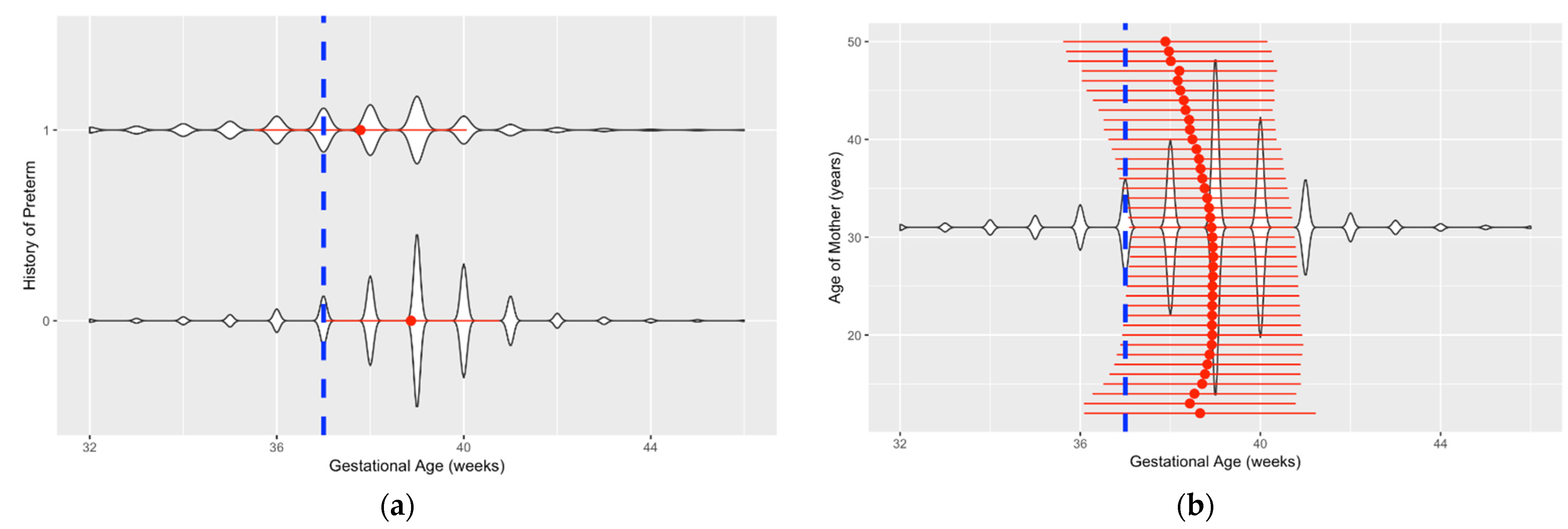
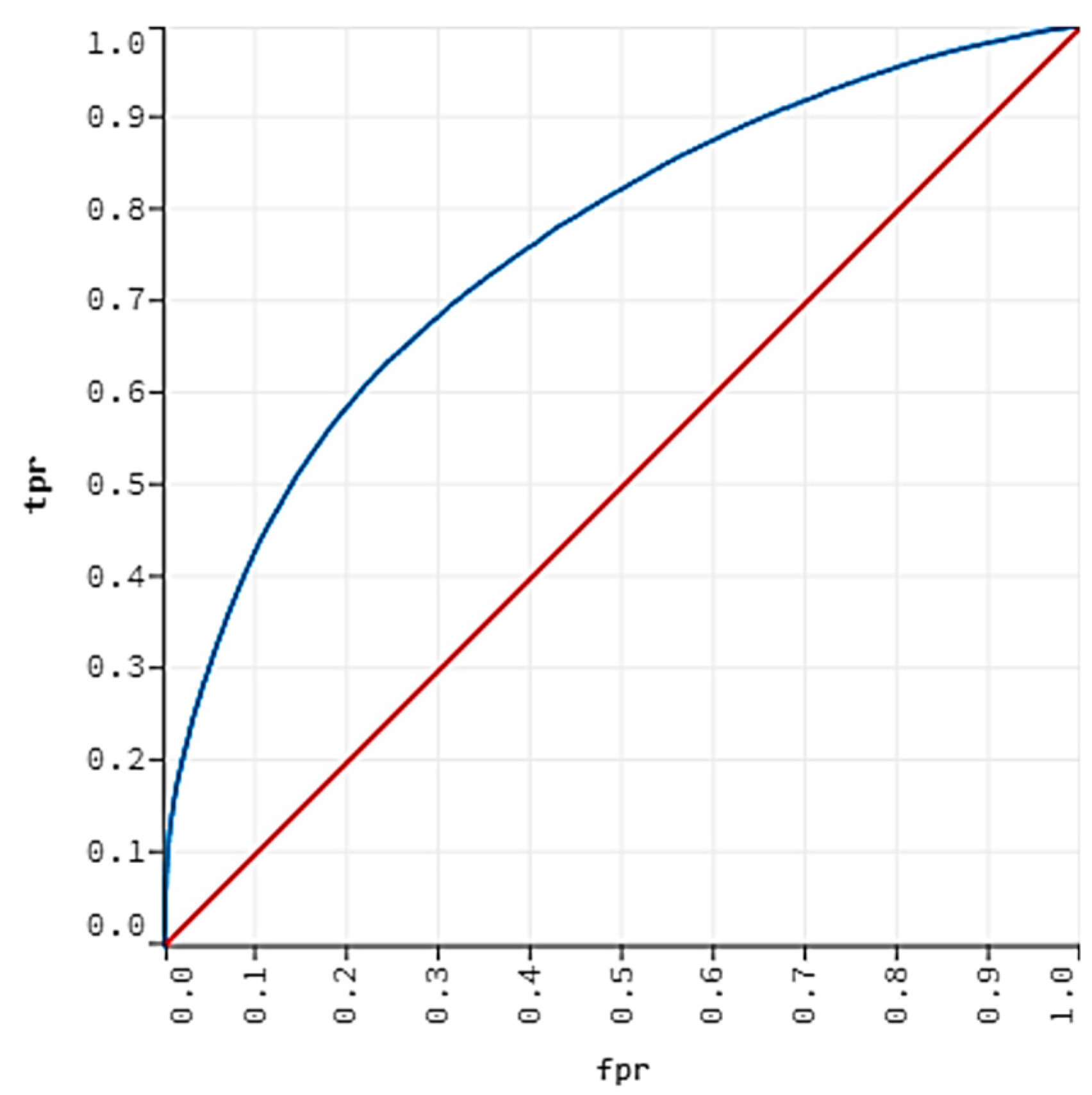

| Name | Number of Missing Values | Percent Missing |
|---|---|---|
| Father’s Age (Age_F) | 450,617 | 11.85 |
| Number of prior live births (priorlive) | 6608 | 0.17 |
| Number of prior dead births (priordead) | 13,552 | 0.36 |
| Number of daily cigarettes before pregnancy (cig_before) | 18,119 | 0.48 |
| Number of daily cigarettes during the first trimester of pregnancy (cig_1) | 18,481 | 0.49 |
| Number of daily cigarettes during the second trimester of pregnancy (cig_2) | 18,734 | 0.49 |
| Body mass index (BMI) | 102,932 | 2.71 |
| Sexually transmitted disease (STD) | 6592 | 0.17 |
| Nutrition Program for Women, Infants, and Children (WIC) | 46,744 | 1.23 |
| Access_to_Exercise | 3441 | 0.09 |
| Alchohol_Driving_Death | 536 | 0.01 |
| Prev_Hospital_Stays | 45,901 | 1.21 |
| Method | Parameter | Optimal Value | Range |
|---|---|---|---|
| RF | |||
| Number of trees | 220 | [20, 500] | |
| Max depth | 13 | [5, 20] | |
| Sample rate | 0.65 | [0.1, 1] | |
| GBM | |||
| Learning rate | 0.04 | [0.001, 0.05] | |
| Learning rate annealing | 0.99 | [0.9, 1] | |
| Number of trees | 800 | [50, 1000] | |
| Max depth | 13 | [5, 15] | |
| Sample rate | 0.55 | [0.1, 1] | |
| Min child row | 50 | [10, 100] | |
| Column sample rate | 0.8 | [0, 1] | |
| LightGBM | |||
| Learning rate | 0.08 | [0.01, 1] | |
| Number of trees | 280 | [50, 600] | |
| Max depth | 14 | [5, 15] | |
| Sample rate | 0.55 | [0.2, 0.8] | |
| Min child row | 20 | [10, 100] | |
| Column sample rate | 0.80 | [0.1, 1] |
| Method | Train AUC (%) | Test AUC (%) | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|
| LR-EN | 66.59 | 66.61 | 51.98 | 71.68 | 70.22 |
| RF | 70.24 | 70.78 | 57.36 | 73.01 | 71.78 |
| GBM | 77.94 | 75.58 | 64.82 | 73.01 | 72.37 |
| LightGBM | 78.34 | 75.91 | 62.24 | 73.93 | 72.99 |
| Model | Method | Sample Size (n) | Prevalence of Positive Class (%) | Test AUC (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| Goodwin, Iannacchione, Hammond, Crockett, Maher and Schlitz [49] | Neural nets, Stepwise LR | 19,970 | 22.20 | 72.00 | NR | NR |
| Vovsha, et al. [50] | SVM with Radial Basis kernel | 3002 | NR | NR | 57.60 | 62.10 |
| Alleman, Smith, Byers, Bedell, Ryckman, Murray and Borowski [24] | LR | 2509 | 7.50 | 69.50 ** | 31.20 | 90.60 |
| Weber, Darmstadt, Gruber, Foeller, Carmichael, Stevenson and Shaw [25] * | Super learner (Combination of RF, lasso, ridge) | 336,214 | 1.02 * | 67.00 | 62.00 | 65.00 |
| Best model in this study | GBM | 3,610,827 | 7.73 | 75.58 | 64.82 | 73.01 |
| Variable | Relative Importance | Scaled Importance | Scaled Percentage |
|---|---|---|---|
| hyper | 312,937.2813 | 1 | 0.1092 |
| interval | 171,301.625 | 0.5474 | 0.0598 |
| Previous_preterm | 161,489.9844 | 0.516 | 0.0563 |
| bmi | 154,535.0781 | 0.4938 | 0.0539 |
| Education_F | 121,954.1797 | 0.3897 | 0.0425 |
| Education_M | 104,375.5859 | 0.3335 | 0.0364 |
| Age_M | 96,351.9531 | 0.3079 | 0.0336 |
| Adequate | 84,977.4063 | 0.2715 | 0.0296 |
| Age_F | 72,292.6953 | 0.231 | 0.0252 |
| Race_F | 56,246.9297 | 0.1797 | 0.0196 |
| Race_M | 54,843.5078 | 0.1753 | 0.0191 |
| Healthcare_Cost | 49,813.7344 | 0.1592 | 0.0174 |
| priorlive | 43,385.2227 | 0.1386 | 0.0151 |
| Females_Under_65_no_Health_Ins | 42,884.2578 | 0.137 | 0.015 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebrahimvandi, A.; Hosseinichimeh, N.; Kong, Z.J. Identifying the Early Signs of Preterm Birth from U.S. Birth Records Using Machine Learning Techniques. Information 2022, 13, 310. https://doi.org/10.3390/info13070310
Ebrahimvandi A, Hosseinichimeh N, Kong ZJ. Identifying the Early Signs of Preterm Birth from U.S. Birth Records Using Machine Learning Techniques. Information. 2022; 13(7):310. https://doi.org/10.3390/info13070310
Chicago/Turabian StyleEbrahimvandi, Alireza, Niyousha Hosseinichimeh, and Zhenyu James Kong. 2022. "Identifying the Early Signs of Preterm Birth from U.S. Birth Records Using Machine Learning Techniques" Information 13, no. 7: 310. https://doi.org/10.3390/info13070310
APA StyleEbrahimvandi, A., Hosseinichimeh, N., & Kong, Z. J. (2022). Identifying the Early Signs of Preterm Birth from U.S. Birth Records Using Machine Learning Techniques. Information, 13(7), 310. https://doi.org/10.3390/info13070310






