Chronic kidney disease is a recognized public health problem. Chronic kidney disease is classified into stages according to the level of glomerular filtration rate, and stage-specific action plans facilitate the evaluation and the management of chronic kidney disease. Glomerular filtration rate can be estimated by means of empirical formulas that incorporate blood serum creatinine concentration, blood serum cystatin-C concentration as well as demographic and clinical variables such as age, gender, race, and body size. Glomerular filtration rate estimating formulas provide a more accurate assessment of the level of kidney function than bio-markers concentrations alone.
Measuring the glomerular filtration rate is crucial for determining appropriate drug dosing, monitoring the effects of therapeutic interventions, and for overseeing the progression of chronic kidney disease. For instance, in pediatric autologous hematopoietic stem cell transplantation treatment protocols, chemotherapy dosing is commonly based on renal function, as patients with a reduced GFR levels receive reduced dosages, which can affect toxicity profiles and therapeutic benefit [
26].
3.2. Experimental Results on Statistical Numerical Modeling of Pedriatic Patients Data
Existing multivariate formulas for GFR estimation have been compared and validated in [
38] over a dataset of 87 Chinese children and adolescents aged 1 through 18. The authors of the research have included their dataset with the publication. For each patient, the available data comprise age, gender, physical parameters (such as height and weight), GFR (measured using double-sample plasma clearance [
39]), two values for serum creatinine concentration as well as cystatine-C and blood urea nitrogen concentration. The two values of serum creatinine concentration correspond to two different measurement techniques, namely, the Jaffe method and the IDMS enzymatic method.
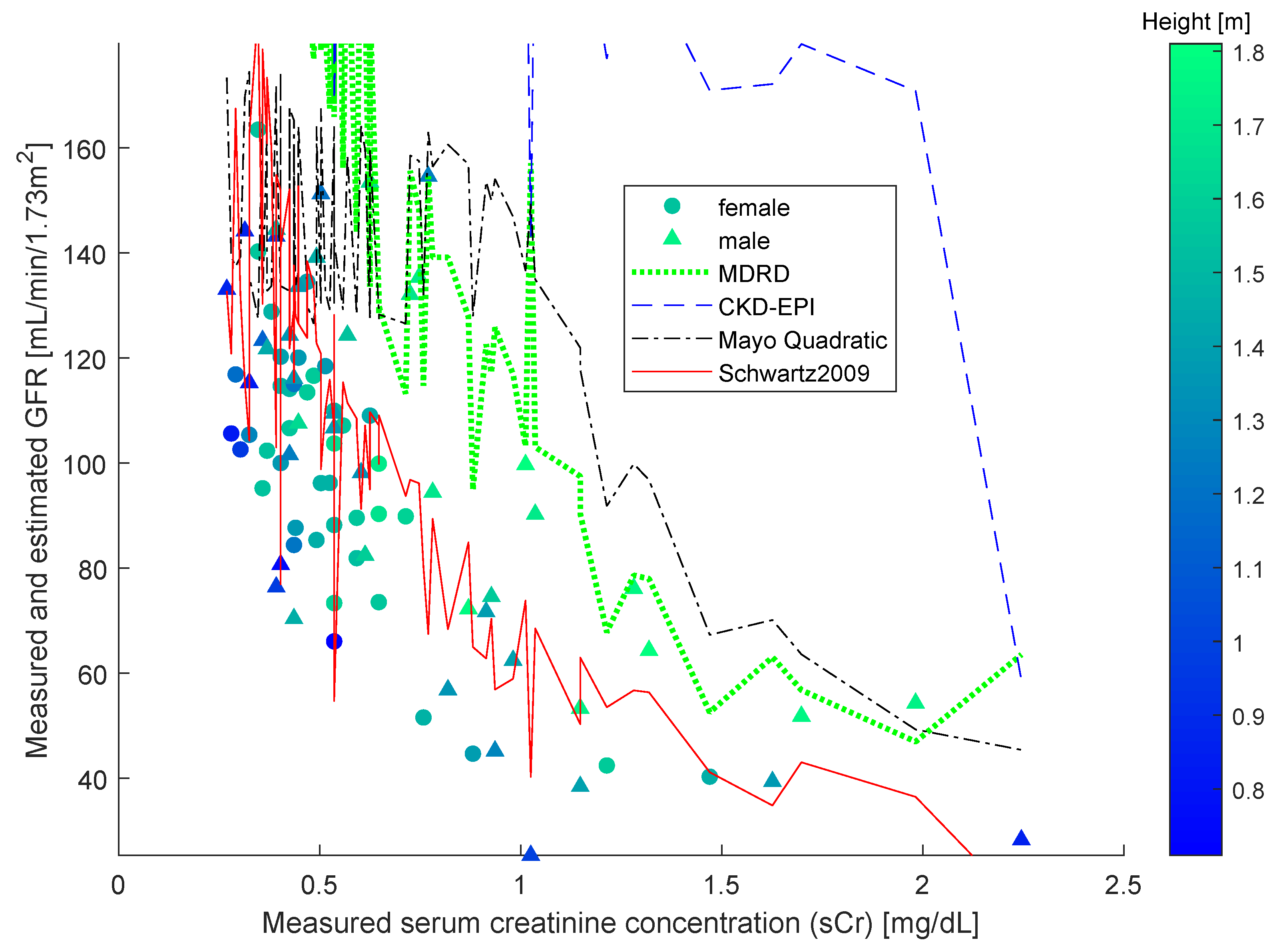
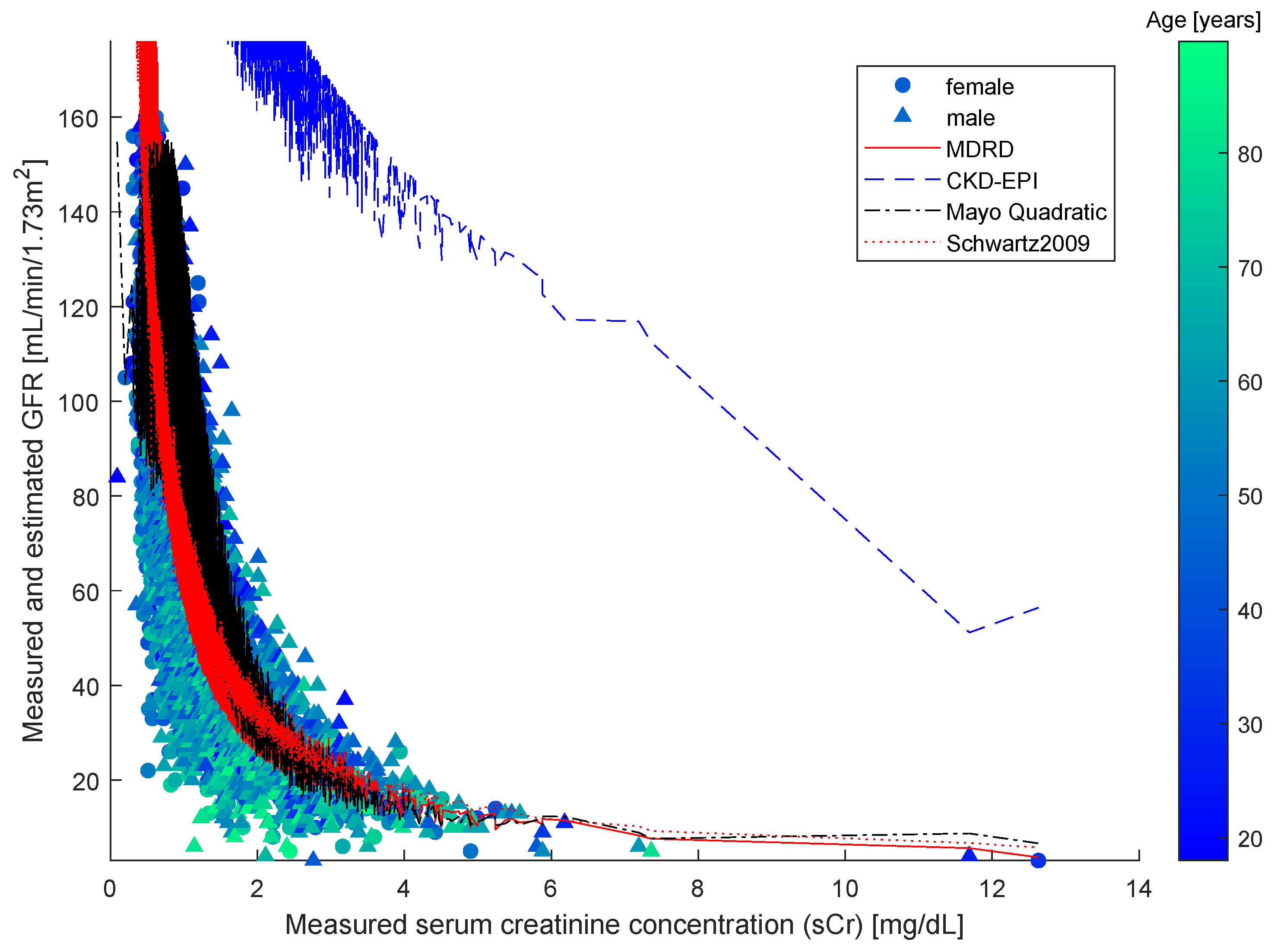
The study [
38] compared four different formulas, namely, the original Schwartz formula, the updated Schwartz formula, the Filler formula and the CKiD formula. The study found the most effective estimation formula to be the updated Schwartz one. Over said data, we also computed estimations using the other three widely employed formulas, namely MDRD, CKD-EPI and Mayo Quadratic, and compared them with the results of the updated Schwartz estimation formula. From the
Figure 2, it is clearly confirmed that the updated Schwartz formula outperforms all of the other functions.
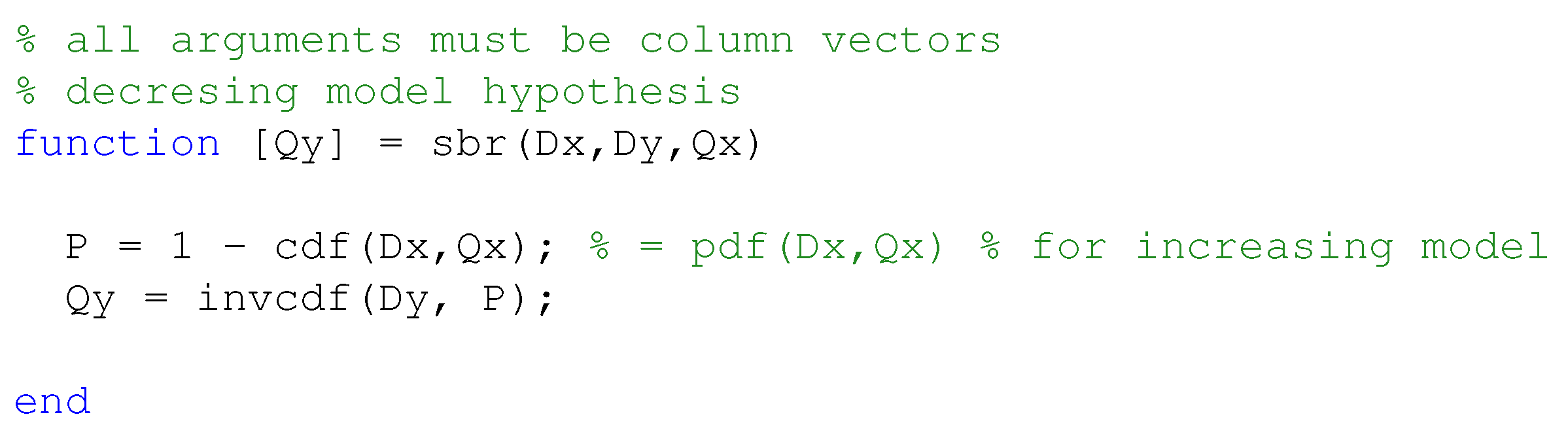
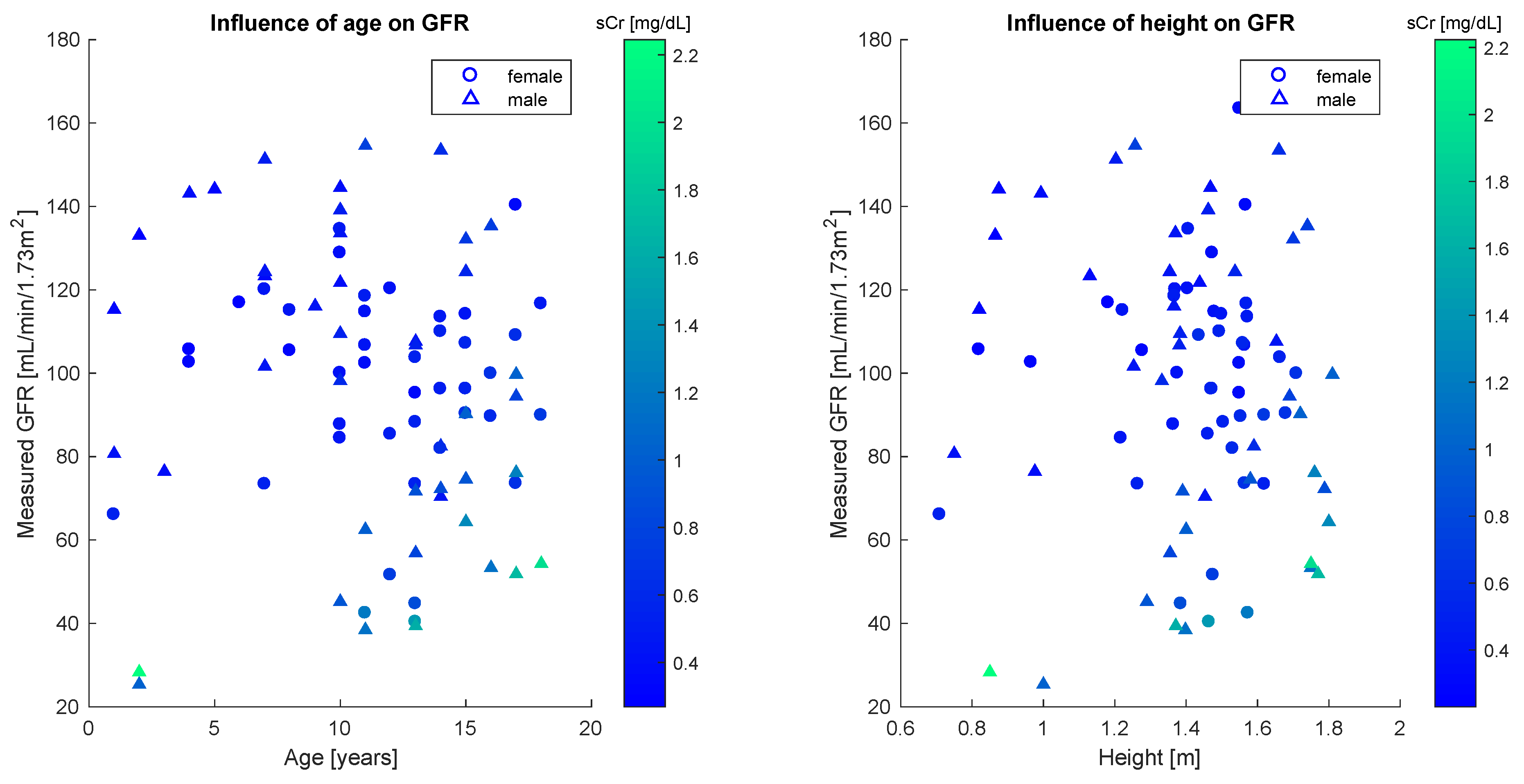
In order to apply the statistical bivariate numerical modeling algorithm developed in the
Section 2, we first assessed the existence of a single dominant independent variable. This was clearly found to be the serum creatinine concentration. Other variables used to estimate the glomerular filtration rate are age and height, whose effect however is marginal, as the scatter plots in
Figure 3 reveal no strong statistical features.
Quantitatively, this analysis is confirmed by the sample correlation coefficient [
40].
Table 2 shows correlation coefficients between the GFR and each of age, height and sCr for the whole population and the gender-defined subsets. The results illustrated in the table confirm the weak statistical correlation between glomerular filtration rate and age, as well as between glomerular filtration rate and height, especially for female patients.
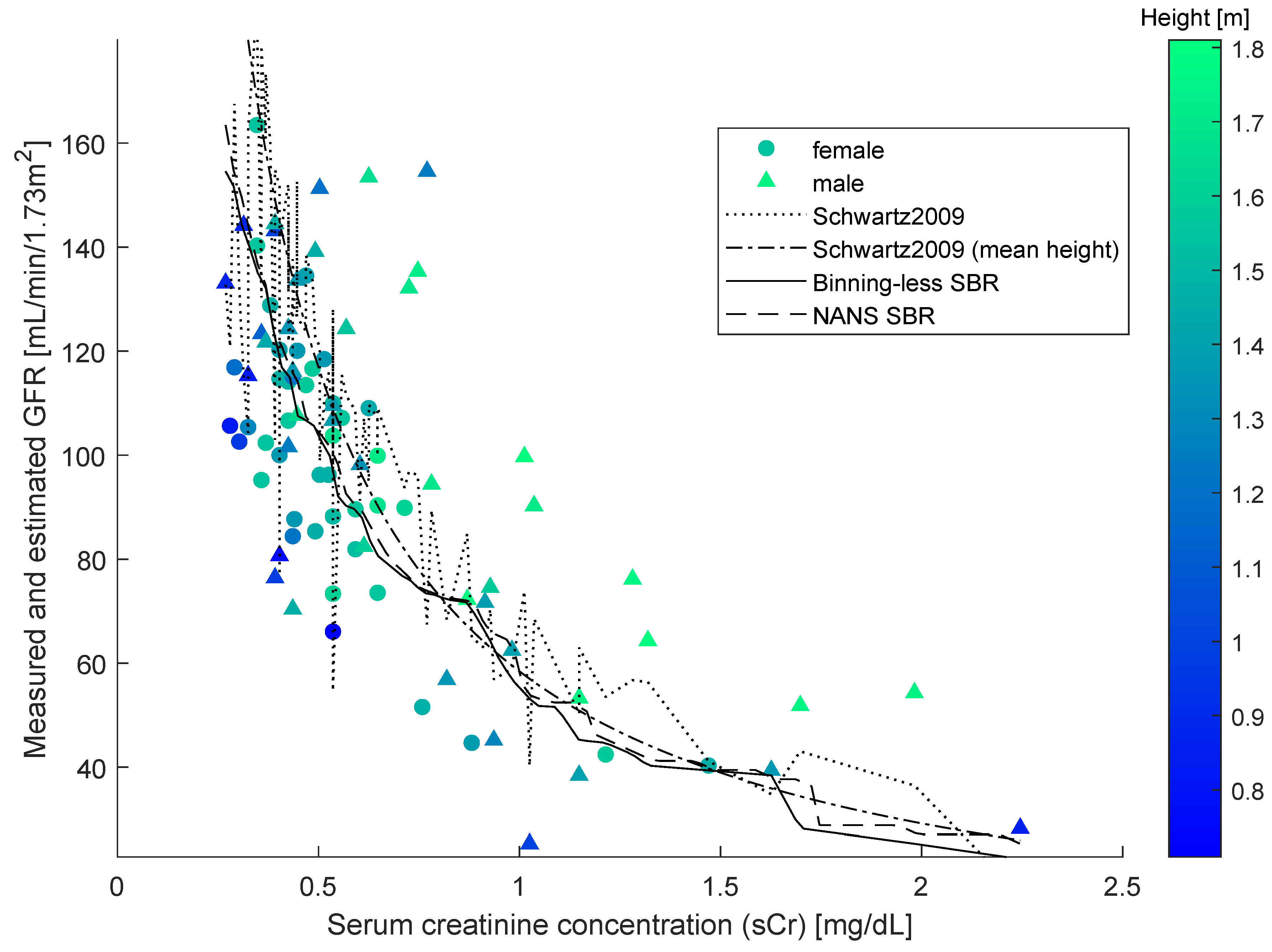
From the
Figure 2, it is readily appreciated that the sCr–GFR relationship presents a monotonically decreasing trend, which enables us to apply the SBR numerical modeling algorithm presented in the
Section 2. According to the observations drawn about the performances of the closed-form models, we did not compare the SBR numerical modeling algorithm with the MDRD, the CKD-EPI and the Mayo Quadratic formulas.
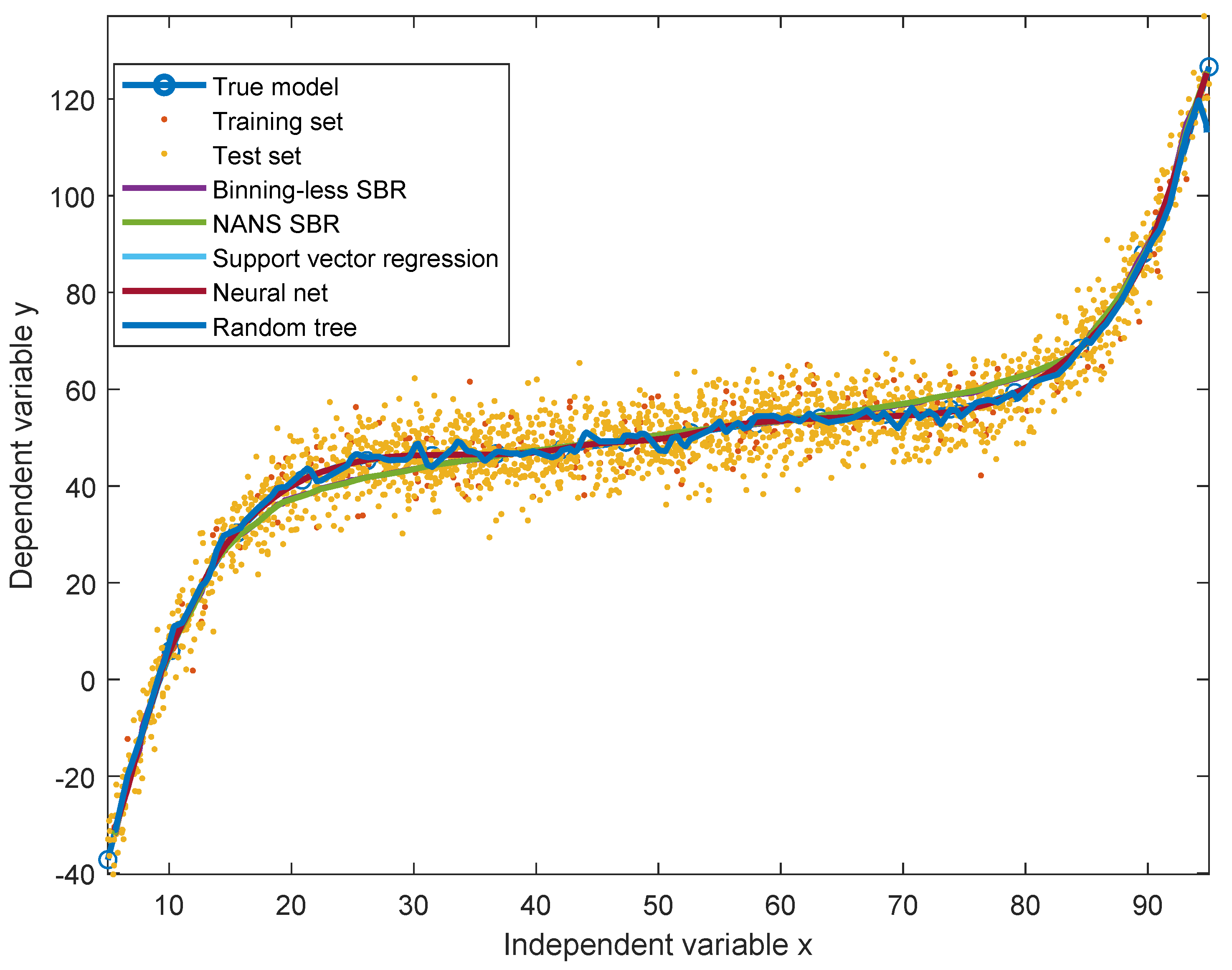
As SBR generates a bivariate numerical model, for the sake of the comparison, a simplified version of the updated Schwartz formula was introduced to be independent of height. This was done by replacing the variable height with a constant equal to the mean height of all individuals in the dataset. This model is illustrated in the
Figure 4, along with the datapoints, the numerical model obtained by SBR using the numerical-algebraic neural system NANS method explained in [
21], the original updated Schwartz formula and the numerical model obtained using the binning-less method described in
Section 2. Notice that the GFR estimations pertaining to the updated Schwartz formula were not calculated as they were provided within the dataset.
The input-output nature of bivariate numerical modeling grants the use of functional notation (i.e., given a value for the independent variable x, the prediction made for the value of the dependent variable y can be expressed as ). The notation is commonly used in reference to closed-form models and will be adopted in this paper to also indicate predictions made using the Algorithm 3 and by interpolating the curve obtained using the NANS method.
The two closed-form models and the two numerical modeling algorithms displayed in the
Figure 4 were compared on prediction performance using four indexes: mean squared error (MSE), mean absolute error (MAE), variance-accounted-for (VAF) and coefficient of correlation (
), as defined in the
Section 2.2. In the present context, each
represents an instance of serum creatinine concentration sCr, each
represents an instance of glomerular filtration rate GFR, and
.
Comparisons were also made to evaluate the generalization ability of the closed-form model (simplified Schwartz) as well as of the considered numerical numerical modeling algorithms (Binning-less SBR and NANS SBR). This was achieved by measuring the “roughness” of the the numerical models [
41] through the index
G defined by
on the basis of the second-order differences of a sequence
. By definition, the index
increases with sharp changes in slope. The reference value for a well-performing algorithm is
as close as possible to zero. To be useful, the
values have to be sorted in some significant manner: in the present context, for each model
to be evaluated,
assumes the predictions at
equally spaced, increasing, values of serum creatinine concentration, namely:
where
and
are respectively the smallest and largest measured creatinine concentration levels. The same index cannot be applied to multivariate functions, therefore the updated Schwartz equation was not tested with this criterion. An index similar to
was discussed in [
42] to prevent overfitting of a neural-network model. The value of
G is expected to be large for irregular curves and indeed it is close to zero for the simplified Schwartz model (independent of height), which is essentially a hyperbola, graph of a smooth function.
The results of comparison are summarized in the
Table 3. Notice that, in this experiment, the number
N of model-points exceeds the number
n of data-points. Among the four methods considered, the binning-less statistical bivariate numerical modeling algorithm exhibits the lowest
and
values and highest
value, that shows how SBR is very effective at fitting data, as well as the lowest computation time (the
value pertaining to the simplified Schwartz formula is negative and hence non-meaningful; the values pertaining to the updated Schwartz formula were provided in the dataset, hence no computation time is available for this method). Taking the
G-value of the numerical model provided by the simplified Schwartz formula as a baseline, the binning-less algorithm exhibits a closer value to such baseline than the NANS SBR: this result shows that the novel SBR algorithm returns a smoother numerical model compared to the NANS SBR one.
3.3. Experimental Results on Statistical Numerical Modeling of Adult Patients Data
In this section, we illustrate and discuss experimental results about statistical numerical modeling of sCr–GFR dependency in adult patients data. The accessed data-set is a large database drawn from [
43] that contains 10,610 records of mixed children, adolescents an adults. The study summarized in [
43] used a dataset of subjects 3 to 90 years old, referred between July 2003 and July 2014 to a single university hospital to undergo GFR measurement for suspected or established renal dysfunction, renal risk, or before kidney donation. The exclusion criteria were being treated with dialysis at the time of the study or taking cimetidine, trimethoprim or intravenous injections of albumin or diuretics before GFR measurement. From this dataset, we excluded all those records corresponding to patients aged 18 and below, so as to isolate 9530 adults. Given the large number of data-pairs available, the numerical models were build on a training set of
of the records and tested on a test set made of the
remaining records, randomly selected. The number of model-points was again selected to be
which, in this case, is far less than the number of available data-points.
Over said data, we computed estimations using MDRD, CKD-EPI, Mayo Quadratic and updated Schwartz estimation formula. The results shown in the
Figure 5 confirm the widely taken assumption that the MDRD outperforms all other models when it comes to process adult patients data.
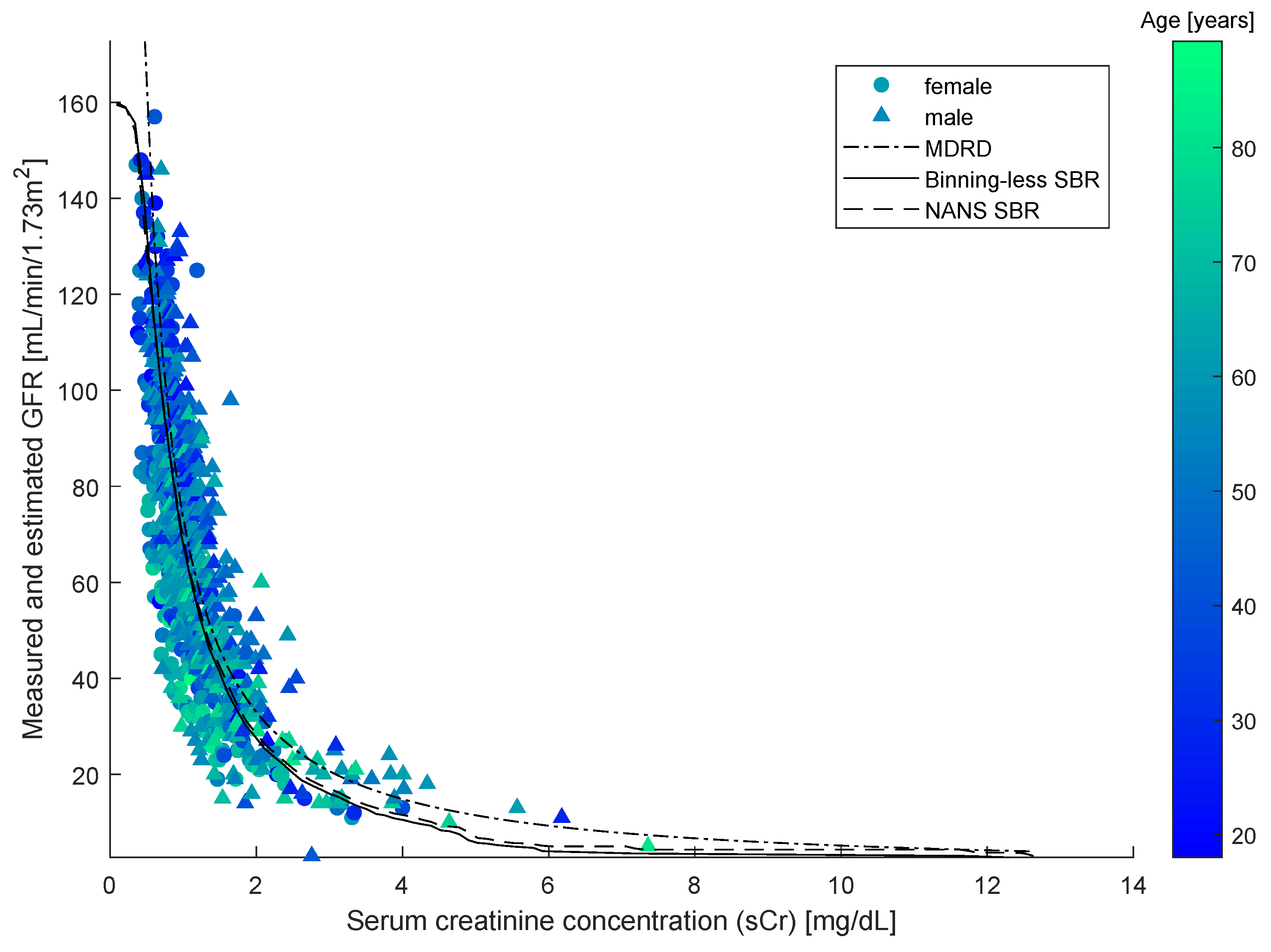
As SBR generates a bivariate numerical model, for the sake of the comparison, a simplified version of the MDRD formula was introduced to be independent of age and gender. This was done by averaging out the age and the term in the MDRD equation that depends on the gender over the population in the dataset. The resulting models are illustrated in the
Figure 6, along with testing-set datapoints.
The closed-form model and the two numerical modeling algorithms displayed in the
Figure 6 were compared on prediction performance using again four indexes: mean squared error (MSE), mean absolute error (MAE), variance-accounted-for (VAF), coefficient of correlation (
) and runtime (seconds). In the present context, each
represents an instance of serum creatinine concentration sCr, each
represents an instance of glomerular filtration rate GFR, and
(that is the size of the testing set).
The results of comparison are summarized in the
Table 4. Among the three methods considered, the statistical bivariate numerical modeling algorithms (NANS SBR and Binning-less SBR) exhibits the lowest
and
values and the highest
values, which shows how SBR methods are effective at fitting data. The extremely large value of the roughness
G exhibited by the Simplified MDRD method is due to the fact that, for low values creatinine (say, for
), the model is unreliable, as can be directly seen from the
Figure 6. The last column of the
Table 4 evidences how the Binning-less SBR algorithm is not only way more efficient than the older version (NANS SBR), but even of the Simplified MDRD formula. Since the performances exhibited by the old version of the modeling algorithm (NANS SBR) and the newest version proposed in this paper (Binning-less SBR) are very close to one another, but the computational complexity of the latter is lesser than the former, we drew the conclusion that the novel, simplified version is preferable to the older.
Example 2. In order to highlight the practical usage of the developed SBR modeling procedure, we discuss here an example of how a numerical model may be taken advantage of to infer the glomerular filtration rate from a serum creatinine assay. The Table 5 shows a portion of the SBR model inferred on the adult patients dataset. The whole model includes 100
sCR–GFR pairs and the table shows 10
pairs for sCr ranging in . As a numerical example, one may assume a reading of . The nearest values in the table are sCr and sCr , to which correspond GFR and GFR , respectively. By a linear interpolation, one gets the linear equationthat leads to the estimation . The actual value in the dataset is , hence the prediction by the model is off of about . Notice that the newly acquired value of serum creatinine may be merged to the sCr dataset in order to make the CDF estimation—and hence the model predictions—more accurate the next time.