Noisy ECG Signal Analysis for Automatic Peak Detection
Abstract
1. Introduction
2. Artifacts/Noises Affecting the ECG
- Baseline wander.
- Power line interference.
- Motion artifacts.
- Muscle noise.
- Other interference.
2.1. Baseline Wander
2.2. Power Line Interference
2.3. Motion Artifacts
2.4. Muscle Noise
2.5. Other Interferences
3. Methods
3.1. Adopted Techniques
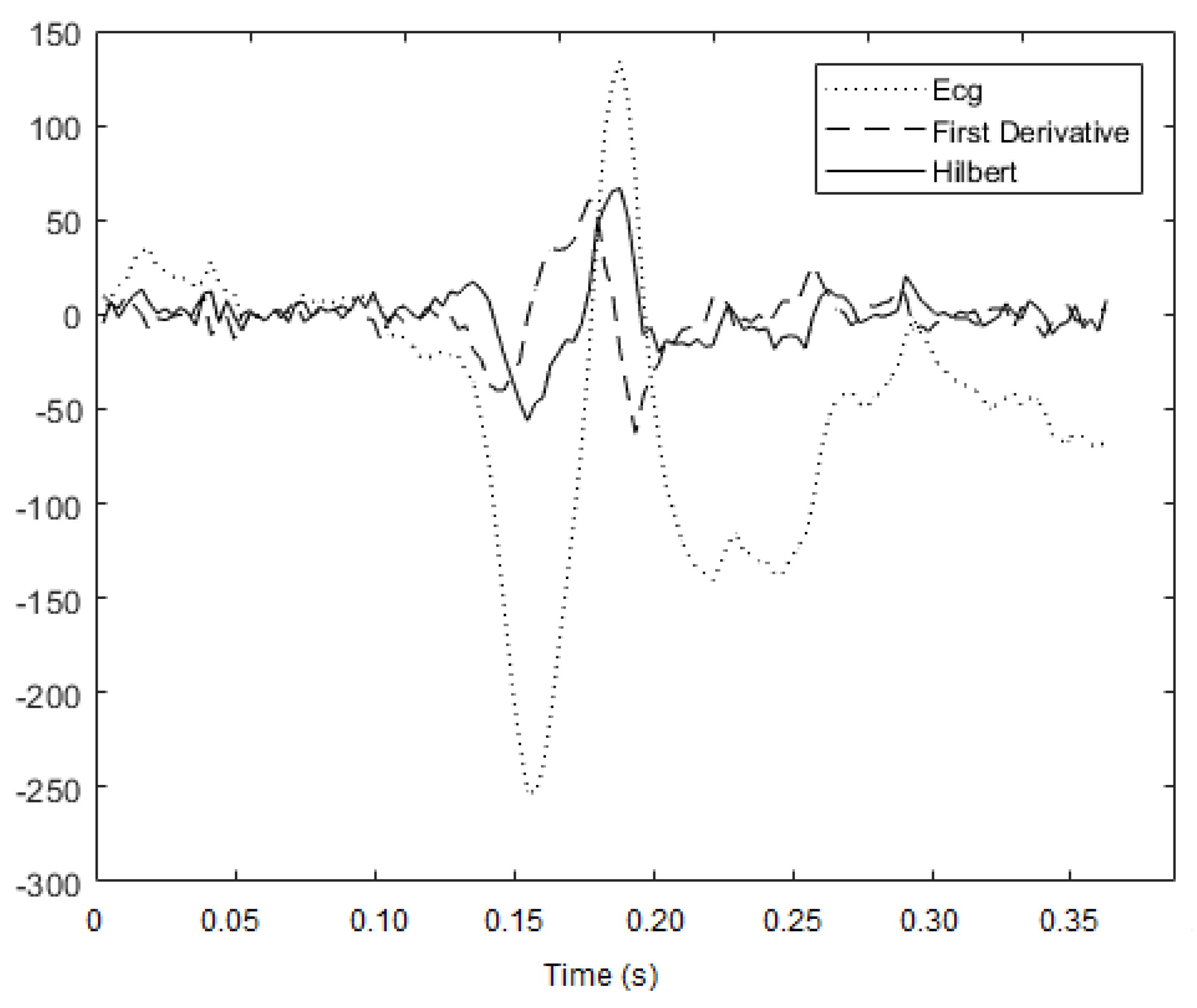
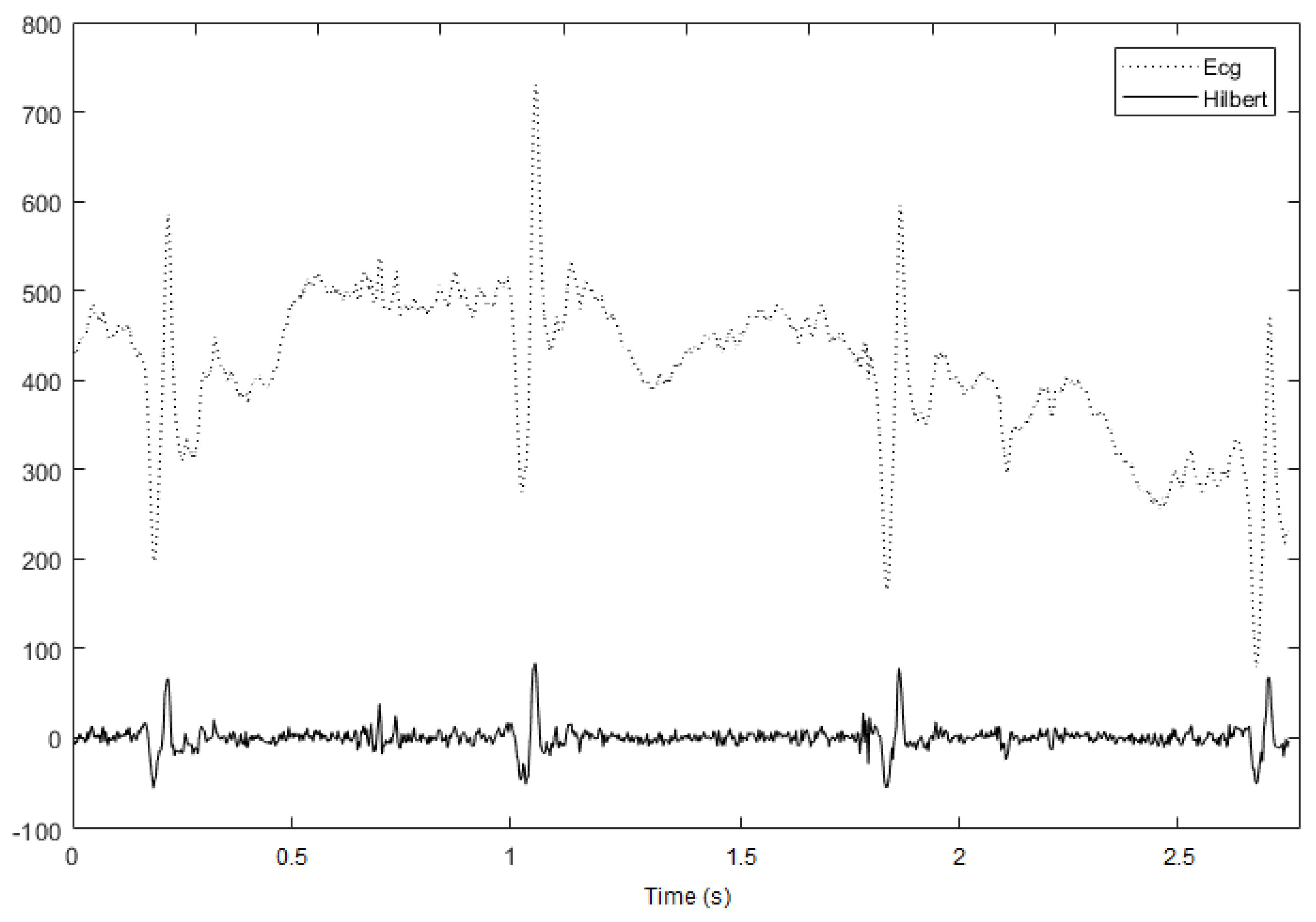
3.1.1. Hilbert Transform
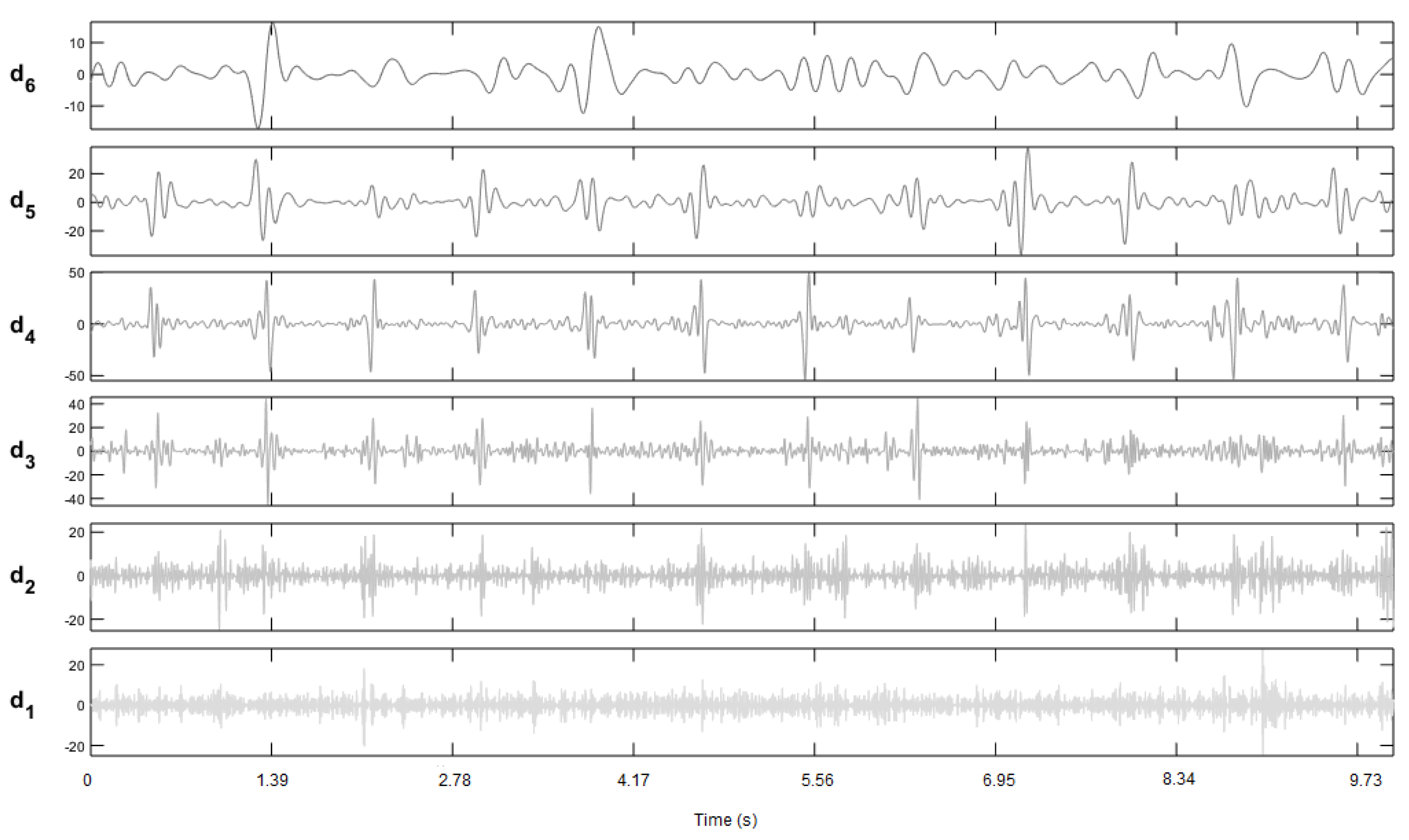
3.1.2. Wavelet Transform
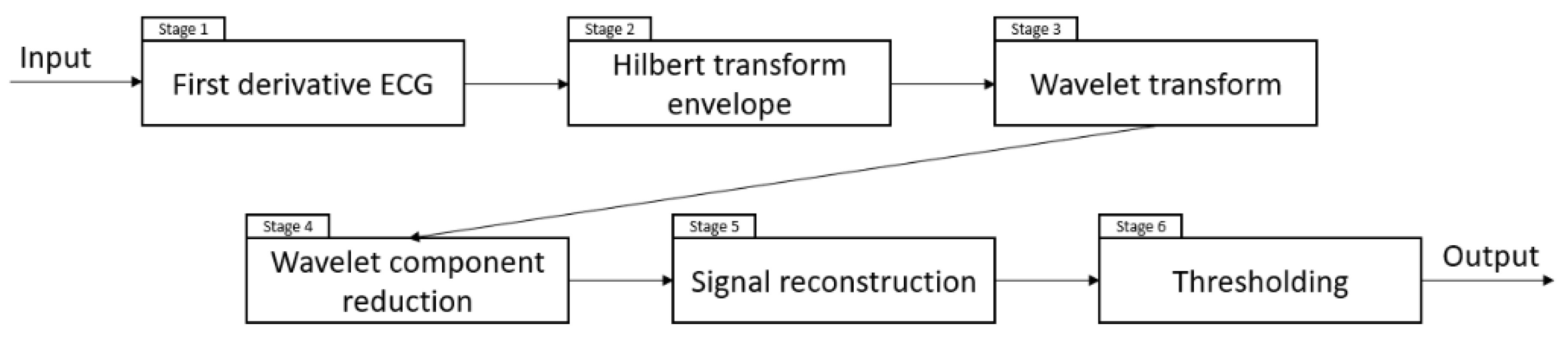
3.2. Implemented Method
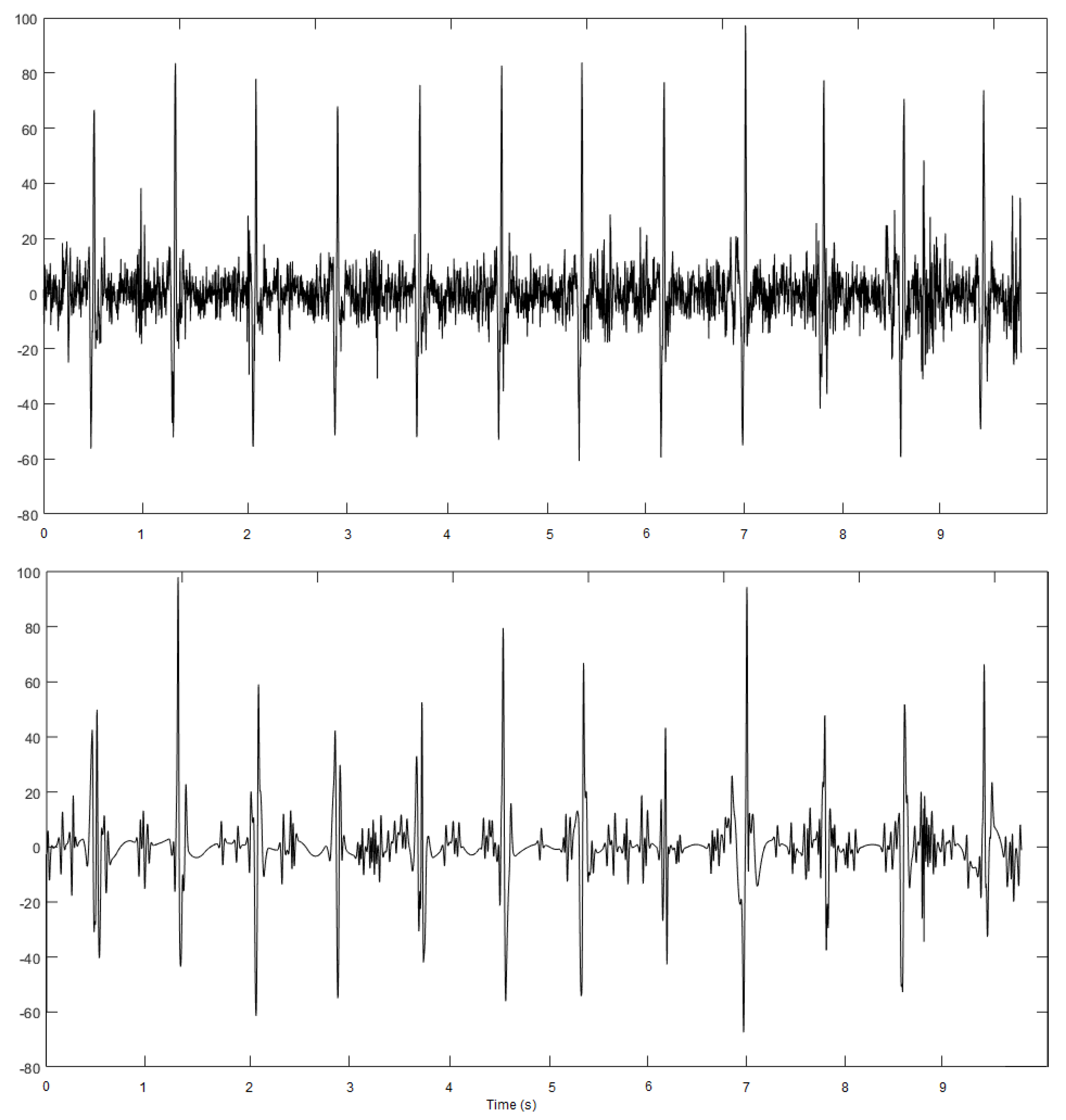
4. Results and Discussion
4.1. ECG Database Used as Test Bench
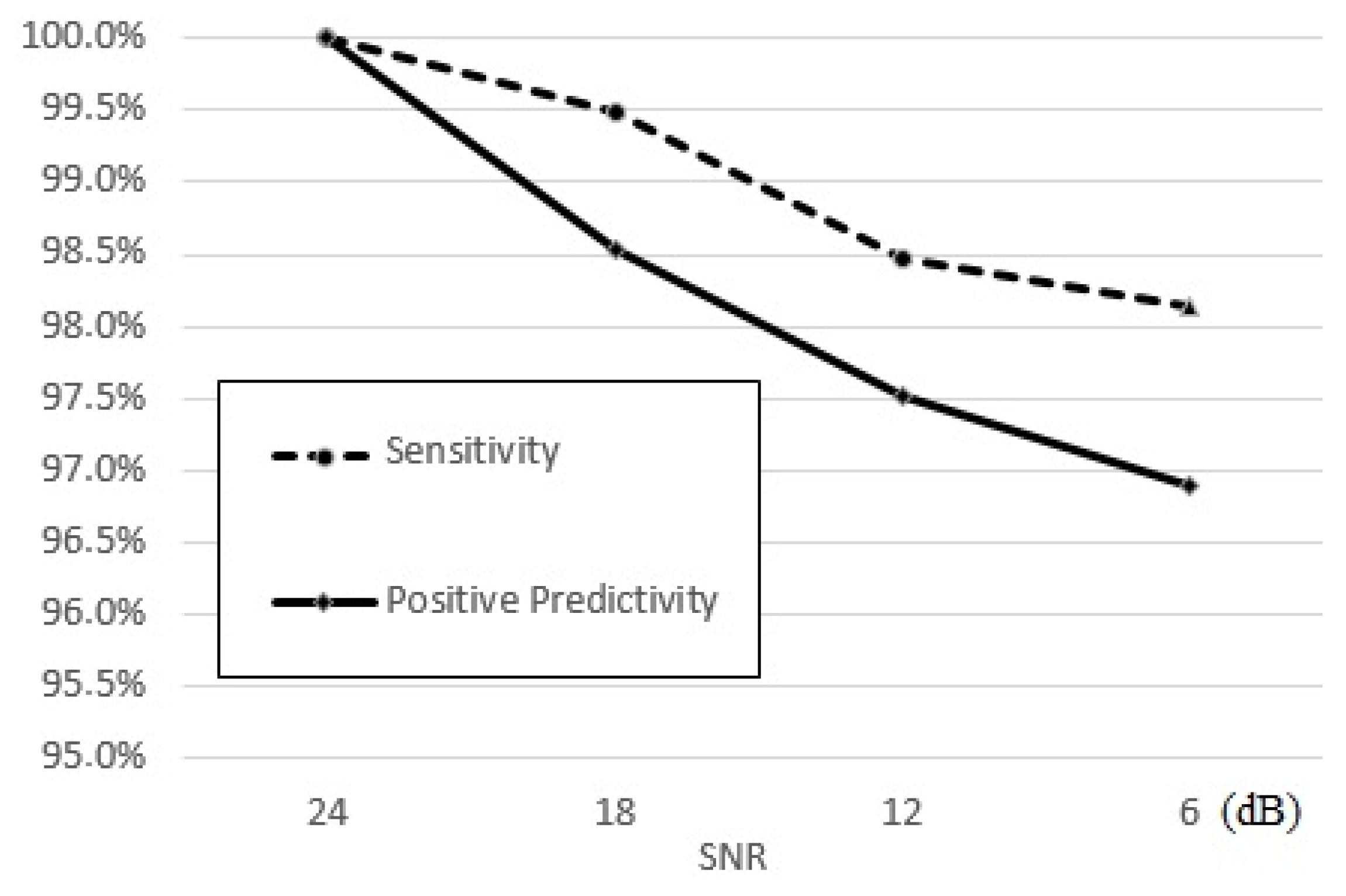
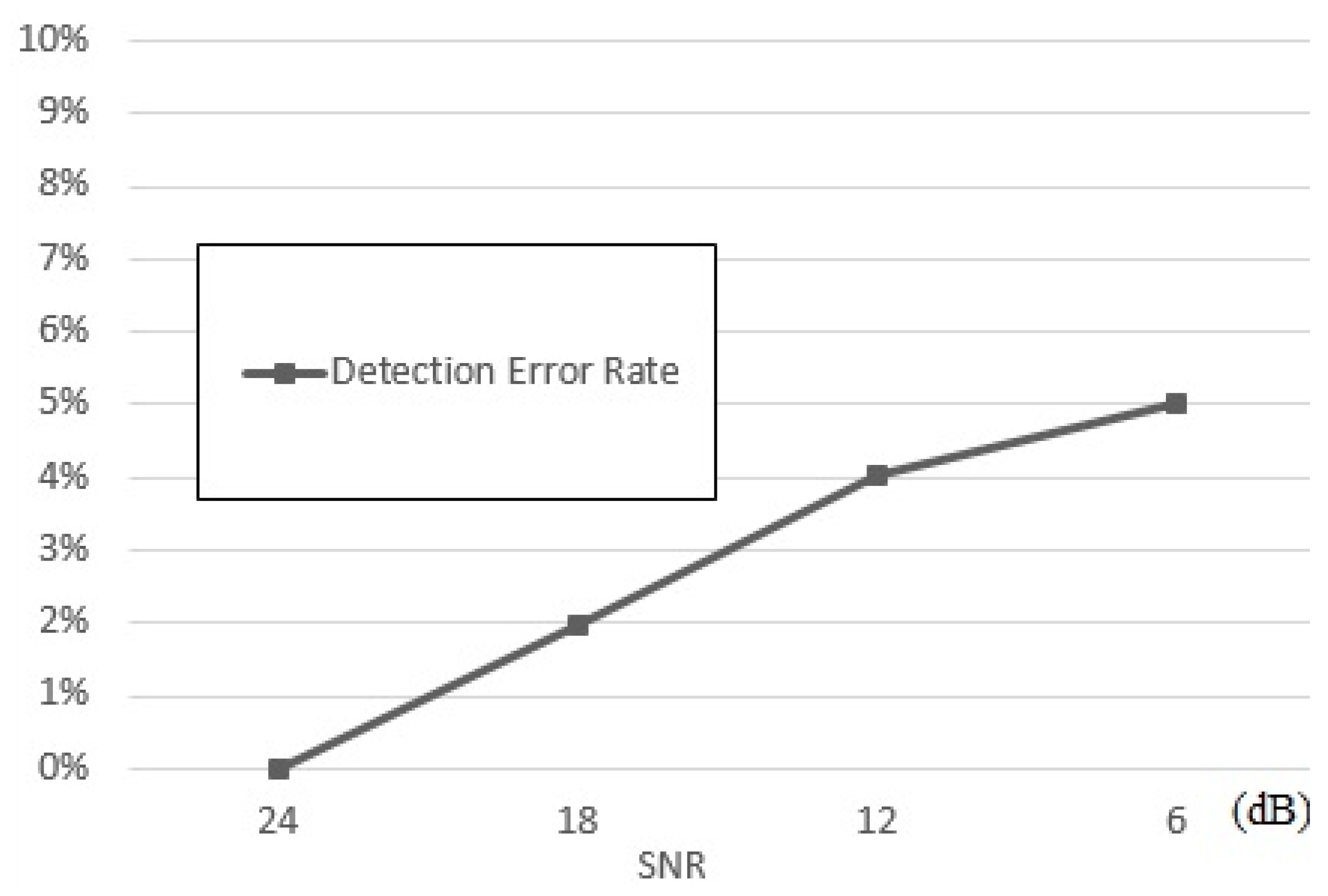
4.2. Performance Evaluation and Results
4.3. Performance Assessment and Comparisons
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pentel, T. ANS and heart rate variability in normal sleep and sleep disorders. Sleep Med. 2007, 8, 44–45. [Google Scholar]
- Agrawal, A.; Gawali, D.H. FPGA-based peak detection ECG signal using histogram approach. In Proceedings of the International Conference on Recent Innovations in Signal Processing and Embedded Systems (RISE 2017), Bhopal Madhya Pradesh, India, 27–29 October 2017. [Google Scholar]
- Pan, J.; Tompking, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, P.S.; Tompkins, W.J. Quantitative investigation of QRS detection rules using MIT/BIH arrhythmia database. IEEE Trans. Biomed. Eng. 1986, 12, 1157–1165. [Google Scholar] [CrossRef]
- He, R.; Wang, K.; Li, Q.; Yuan, Y.; Zhao, N.; Liu, Y.; Zhang, H. A novel method for the detection of R-peaks in ECG based on K-Nearest Neighbors and Particle Swarm Optimization. EURASIP J. Adv. Signal Process. 2017, 2017. [Google Scholar] [CrossRef]
- Benitez, D.; Gaydecki, P.A.; Zaidi, A.; Fitzpatrick, A.P. The use of the Hilbert transform in ECG signal analysis. Comput. Biol. Med. 2001, 31, 399–406. [Google Scholar] [CrossRef]
- Qin, Q.; Li, J.; Yue, Y.; Liu, C. An Adaptive and Time-Efficient ECG R-Peak Detection Algorithm. J. Healthc. Eng. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Pang, D.; Igasaki, T. A Combined Syntactical and Statistical Approach for R Peak Detection in Real-Time Long-Term Heart Rate Variability Analysis. Algorithms 2018, 11, 83. [Google Scholar] [CrossRef]
- Kathirvel, P.; Sabarimai Manikandan, M.; Prasanna, S.R.M.; Soman, K.P. An efficient R-peak detection based on new nonlinear transformation and first-order Gaussian differentiator. Cardiovasc. Eng. Technol. 2011, 2, 408–425. [Google Scholar] [CrossRef]
- Chanwimalueang, T.; von Rosenberg, W.; Mandic, D.P. Enabling R-peak Detection in Wearable ECG: Combining Matched Filtering and Hilbert Transform. In Proceedings of the 2015 IEEE International Conference on Digital Signal Processing (DSP), Singapore, 21–24 July 2015. [Google Scholar] [CrossRef]
- Sabherwal, P.; Agrawal, M.; Singh, L. Automatic detection of the R peaks in single-lead ECG signal. Circuits Syst. Signal Process. 2017, 36, 4637–4652. [Google Scholar] [CrossRef]
- Khriji, L.; Al-Busaidi, A.M. New Adaptive Thresholding-based ECG R-peak Detection Technique. In Proceedings of the 2018 IEEE 4th Middle East Conference on Biomedical Engineering (MECBME), Gammarth, Tunisia, 28–30 March 2018; pp. 147–152. [Google Scholar]
- Falconi, J.A.; Osman, H.A.; El Saddik, A. R-Peak Detection Algorithm Based on Differentiation. In Proceedings of the 2015 IEEE 9th International Symposium on Intelligent Signal Processing (WISP), Siena, Italy, 15–17 May 2015. [Google Scholar] [CrossRef]
- Kaur, H.; Rajni, R. Electrocardiogram signal analysis for R-peak detection and denoising with hybrid linearization and principal component analysis. Turk. J. Electr. Eng. Comput. Sci. 2017, 25, 2163–2175. [Google Scholar] [CrossRef]
- Dev Sharma, L.; Sunkaria, R.K. A robust QRS detection using novel pre-processing techniques and kurtosis based enhanced efficiency. Measurement 2016, 87, 194–204. [Google Scholar] [CrossRef]
- Rakshit, M.; Panigrahy, D.; Sahu, P.K. An improved method for R-peak detection by using Shannon energy envelope. Sadhana 2016, 41, 469–477. [Google Scholar] [CrossRef]
- Clifford, G.D.; Azuaje, F.; Mesharry, P. ECG statistics, noise artifacts and missing data. In Advanced Methods and Tools for ECG Data Analysis; Artech. House Inc.: London, UK, 2006. [Google Scholar]
- Gaceck, A.; Pedryez, W. (Eds.) ECG Signal Processing, Classification and Interpretation: A Comprehensive Framework of Computational Intelligence; Springer: Berlin, Germany, 2011. [Google Scholar]
- Gupta, P.; Sharma, K.K.; Joshi, D. Baseline wander removal of electrocardiogram signals using multivariate empirical mode decomposition. Healthc. Technol. Lett. 2015, 2, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Kschischang, F.R. The Hilbert Transform. Available online: http://www.comm.utoronto.ca/frank/notes/hilbert.pdf (accessed on 19 January 2019).
- Rizzi, M.; D’Aloia, M.; Castagnolo, B. Computer aided detection of microcalcifications in digital mammograms adopting a wavelet decomposition. Integr. Comput.-Aided Eng. 2009, 16, 91–103. [Google Scholar] [CrossRef]
- Wilson, J.D.; Govindan, R.B.; Hatton, J.O.; Lowery, C.L.; Preissl, H. Integrated approach for fetal QRS detection. IEEE Trans. Biomed. Eng. 2008, 55, 2190–2197. [Google Scholar] [CrossRef] [PubMed]
- Shensa, M.J. The discrete wavelet transform: Wedding the a’ trous and Mallat algorithms. IEEE Trans. Signal Process. 1992, 40, 2464–2482. [Google Scholar] [CrossRef]
- MIT-BIH Noise Stress Test Database. Available online: https://physionet.org/physiobank/database/nstdb/ (accessed on 19 January 2019).
- Vollmer, M. Noise resistance of several top-scored heart beat detectors. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017. [Google Scholar]
- Pangerc, U.; Jager, F. Robust detection of heart beats in multimodal records using slope and peak sensitive band-pass filters. Physiolog. Meas. 2015, 36. [Google Scholar] [CrossRef] [PubMed]
- Antink, C.H.; Brüser, C.; Leonhardt, S. Detection of heart beats in multimodal data: A robust beat-to-beat interval estimation approach. Physiolog. Meas. 2015, 36. [Google Scholar] [CrossRef]
- De Cooman, T.; Goovaerts, G.; Varon, C.; Widjaja, D.; Willemen, T.; Van Huffel, S. Heart beat detection in multimodal data using automatic relevant signal detection. Physiolog. Meas. 2015, 36. [Google Scholar] [CrossRef]
- Vollmer, M. Robust detection of heart beats using dynamic thresholds and moving windows. Comput. Cardiol. 2014, 41, 569–572. Available online: http://cinc.org/archives/2014/pdf/0569.pdf (accessed on 19 January 2019).
- Moody, G.; Moody, B.; Silva, I. Robust detection of heart beats in multimodal data: The PhysioNet/Computing in Cardiology Challenge 2014. Comput. Cardiol. 2014, 41, 549–552. [Google Scholar]
| SNR = 6 dB | SNR = 0 dB | |||
|---|---|---|---|---|
| Method | Se | +P | Se | +P |
| Pangerc U. et al. | 99.91 | 95.91 | 83.97 | 68.92 |
| Antink C.H. et al. | 84.89 | 76.40 | 72.20 | 66.37 |
| De Cooman T. et al. | 99.47 | 73.30 | 96.51 | 59.36 |
| Vollmer M. | 98.50 | 96.73 | 77.10 | 74.91 |
| This study | 98.13 | 96.91 | 78.98 | 75.25 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Aloia, M.; Longo, A.; Rizzi, M. Noisy ECG Signal Analysis for Automatic Peak Detection. Information 2019, 10, 35. https://doi.org/10.3390/info10020035
D’Aloia M, Longo A, Rizzi M. Noisy ECG Signal Analysis for Automatic Peak Detection. Information. 2019; 10(2):35. https://doi.org/10.3390/info10020035
Chicago/Turabian StyleD’Aloia, Matteo, Annalisa Longo, and Maria Rizzi. 2019. "Noisy ECG Signal Analysis for Automatic Peak Detection" Information 10, no. 2: 35. https://doi.org/10.3390/info10020035
APA StyleD’Aloia, M., Longo, A., & Rizzi, M. (2019). Noisy ECG Signal Analysis for Automatic Peak Detection. Information, 10(2), 35. https://doi.org/10.3390/info10020035





