Abstract
Now, data collection and analysis are becoming more and more important in a variety of application domains, as long as novel technologies advance. At the same time, we are experiencing a growing need for human–machine interaction with expert systems, pushing research toward new knowledge representation models and interaction paradigms. In particular, in the last few years, eHealth—which usually indicates all the healthcare practices supported by electronic elaboration and remote communications—calls for the availability of a smart environment and big computational resources able to offer more and more advanced analytics and new human–computer interaction paradigms. The aim of this paper is to introduce the HOLMeS (health online medical suggestions) system: A particular big data platform aiming at supporting several eHealth applications. As its main novelty/functionality, HOLMeS exploits a machine learning algorithm, deployed on a cluster-computing environment, in order to provide medical suggestions via both chat-bot and web-app modules, especially for prevention aims. The chat-bot, opportunely trained by leveraging a deep learning approach, helps to overcome the limitations of a cold interaction between users and software, exhibiting a more human-like behavior. The obtained results demonstrate the effectiveness of the machine learning algorithms, showing an area under ROC (receiver operating characteristic) curve (AUC) of 74.65% when some first-level features are used to assess the occurrence of different chronic diseases within specific prevention pathways. When disease-specific features are added, HOLMeS shows an AUC of 86.78%, achieving a greater effectiveness in supporting clinical decisions.
Keywords:
eHealth; big data; deep learning; Watson; Spark; decision support system; prevention pathways 1. Introduction
Now, data collection and analysis are becoming more and more important in a large variety of application domains, as long as novel technologies advance. At the same time, we are experiencing a growing need for human–machine interaction with expert systems, pushing research toward new knowledge representation models and interaction paradigms [1].
This phenomenon can be observed in many fields, from e-commerce to medical diagnostics. For example, the world-famous retail business company Wal-Mart Stores Inc. produces more than one million customer transactions per hour by their point-of-sale terminals in their 6000 stores. A traditional data warehouse able to contain such an amount of information should be more than three petabytes. This implies that suitable data models and cluster-computing facilities are mandatory in order to use machine learning algorithms tailored to this kind and volume of data. As is well known, such approaches aim to extract patterns indicating the effectiveness of the business strategies, improving the inventory and supply chain management [2].
Similar problems could evidently arise in medical image processing applications that require elaborating a huge amount of data burdened by strong temporal constraints, computationally-heavy tasks, and privacy issues. For example, a clinical center of a typical size equipped with magnetic resonance imaging (MRI) appliances can provide up to 20–30 DCE-MRI (Dynamic Contrast-Enhanced Magnetic Resonance Imaging) scans per day, producing about 2–3 gigabytes of raw data. This amount can easily become tenfold when machine learning and pattern recognition are applied, growing up to 30 GB per day, requiring a suitable storage system, a dedicated computing architecture, and specific store-and-retrieve procedures [3,4].
As for medical imaging, electronic health records (EHRs) should be treated with the same carefulness. A clinical center may want to store all its health records within a database in order to have historical information, thus strongly improving further diagnosis. A reliable digital knowledge-base should include all the details about personal data (age, height, and weight), anamnesis (family history, patient lifestyle, and personal health status), clinical investigations (examination results and diagnostic images) [5], the applied treatments, and the resulting diagnosis [6]. It is worth noting that it is very important to store both positive and negative responses in order to have a reliable and complete knowledge-base. Moreover, each single case may have a specific data structure that may strongly differ from that of other ones. It follows that traditional relational databases may not be able to handle such a variety of data properly. Therefore, a proper data storage mechanism should be used to avoid data boundaries or constraints and to guarantee the modularity and upgradability of the system. In addition, the ubiquitous modern mobile and wearable medical devices (e.g., smartwatches, smartphones, smart sensors, and so on) continuously provide further information about the lifestyle (e.g., pedometer, sleep monitoring, dietary habits) and current clinical situation (e.g., level of glycemia in the blood, heartbeat rate) of patients, offering new ways and opportunities to track and analyze user behavior.
As evidence of this, we analyzed data growth models for a healthcare structure, showing how clinical data can easily increase in terms of both velocity and volume.
In more detail, Figure 1 shows real data collected from a medical examination center, used in this work to validate the proposed system (see Section 4.7). During the observation period from 2011–2017, the collection rate of clinical data showed a rapidly increasing rate. At the end of the observation period, there was a production of about 2000 patient records per quarter (about 600 GByte per quarter). An estimation of the growth factor indicates a production of about 5200 records per quarter (about 900 GByte per quarter) at the end of 2018 (collecting up to about 7500 records in late 2019, about 1.3 TByte per quarter). These numbers give an indication of how the data grow quickly, promising to reach large volumes of data in a few years.
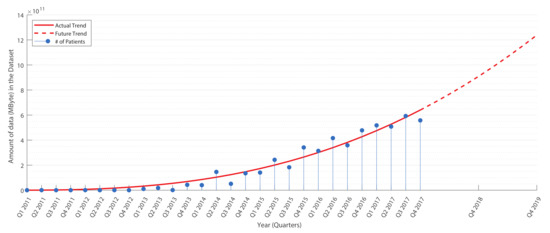
Figure 1.
Data production in a medium-sized medical center (see Section 4.7). The tendency curve (in red) shows the estimation for future years.
The medical context includes hundreds of thousands of known human diseases; however, a physician is able to recall only a small fraction of these diseases at the diagnosis moment. Even if the medical diagnoses are driven by clinical trials and patient anamnesis, disease misclassification in the diagnosis phase is frequent. According to a study performed in 2012 in an intensive care unit (ICU) of the U.S., up to 40,500 patients die annually due to misdiagnosis [7], while a 25 year-long study [8] showed that between 80,000 and 160,000 patients suffered from preventable injuries or deaths related to misdiagnosis, with more than $38 billion estimated to have been awarded to victims in settlements and verdicts.
Thus, we have to deal with collections of data that present high volume, variety, and velocity characteristics [9] that are essential to improve healthcare processes. In addition, for some of such data, we have to consider stream management issues that are typical of Internet of Thing (IoT) scenarios and to provide more and more advanced descriptive, predictive, and prescriptive analytics. Actually, Big Data technologies constitute the most natural and effective manner to face the cited problems [1].
The need for such smart environments and big computational resources thus pushes to search for proper and new solutions also in the eHealth domain.
The term “eHealth” (also written “e-health”) indicates all the healthcare practice supported by electronic elaboration and remote communications [10]. Several different definitions of eHealth have been proposed [11]: Some authors consider the term as an extension of health informatics or digital processing of health data [12]; another point of view considers all the healthcare procedures delivered via the Internet as falling under the umbrella of eHealth [13,14]; the term can also refer to healthcare services delivered via mobile devices (mHealth) [15]; and, finally, in a more general meaning, all the services or systems laying on the edge of healthcare and information technology can also be considered under the eHealth term.
In the eHealth context, some additional challenges must be taken into account:
- Data sensitivity of medical records (privacy must be guaranteed) [3,4];
- Operational times comparable with clinical environment time [16,17];
- Patient trustiness, meaning that the system should represent a clinician (therefore, human-like interaction models may be provided with the aim of minimizing any kind of bias);
- Massive data handling, since a diagnostic by images produces huge amounts of data (not only structured as 3D volumes) [18];
- Context scalability, in order to take into account some sort of distributed computing [3,4];
- Legal responsibility for mistakes in acting or non-acting in an optimal way should be addressed by any clinical systems.
In the social network era, communication over the Internet has a strong influence from the chat-like conversation model. Indeed, this new communication approach influenced social interactions, business services, and the customer care philosophy.
eHealth should surely benefit from this new communication paradigm performing a human-like interaction that can remove any bias against a computer-based information system, giving the patient the feeling of a natural conversation and exploiting the possibility of a schema-free interaction.
Artificial intelligence, currently even more reliable thanks to deep learning approaches, provides automatic and adaptive human-like conversation behavior via chat-bot applications, enabling user–system interactions that are able to simulate human behavior.
A chat-bot (also known as a talk-bot, chatterbot, Bot, chatterbox, artificial conversational entity) is a software that holds a conversation via message exchange. The final goal of such programs is to convincingly simulate human behaviors to pass the Turing test.
Today, chat-bots are used in dialogue-based systems for several practical purposes, including customer services or data collection. The most performing bots use sophisticated natural language processing (NLP) techniques and are trained with deep neural networks to better emulate human behaviors. Many simpler systems scan for keywords within the input, then pull a reply with the most matching keywords, or the most similar wording patterns within a database.
Moreover, many application service providers deploy chat-bot services to be trained on a specific task via example databases and able to interact with the most spread social networks and instant messaging applications.
The main goal of the paper is twofold:
- On one hand, we introduce a general big data architecture which is useful to several eHealth applications;
- on the other hand, we present a particular instance/implementation of such an architecture—the HOLMeS (health online medical suggestions) system—for supporting the prevention of chronic diseases within the CMO (centro medico oplonti) diagnostic center (we note that this activity was performed in the context of a joint research project between the Data Life Research Consortium and the Department of Electrical Engineering and Information Technology of University of Naples), whose main novelty lies on:
- -
- the adoption of a new human–machine interaction paradigm where a machine learning algorithm, deployed on a big data computing cluster, provides online medical suggestions through a chat-bot module;
- -
- the training of the chat-bot in the medical domain using a deep learning approach, thus overcoming limitations of a biased interaction between users and software.
The cluster computing facility is provided by Databricks (https://databricks.com/), where a cluster of nodes allows us data processing capabilities on top of the Apache Spark technological stack. In turn, chat-bot implementation is obtained using the Watson conversation service, designed and trained via the Bluemix platform.
The paper is organized as follows. In Section 2, we briefly describe the evolution of decision support systems for eHealth applications, while in Section 3, we show the tools used to design and realize the HOLMeS system. In Section 4, we present the proposed system, explaining each module and providing an overview of the complete architecture. Finally, the obtained results are presented in Section 5 and discussed in Section 6, where we also draw some conclusions and suggest future work.
2. Related Work
Actually, eHealth does not represent a new approach in supporting physicians in the hard task of giving the right diagnosis. For many years, in such a context, clinical decision support systems [19] (CDSSs) had a fundamental role in assisting physicians and other health professionals with decision-making tasks, such as determining diagnosis on the basis of patient clinical data.
Early CDSSs were designed as a two-stage interaction system, where the physician inputs the patient data, the symptoms, and a few outcomes from clinical tests, receiving the diagnosis. More advanced CDSSs are able to interact with the physicians and guide them in a progressive process of successive refinement to the final diagnosis.
There are two main types of CDSS:
- Knowledge-based CDSSs consist of three parts: A mechanism to communicate (GUI), which allows the system to show the results to the user as well as submit the input to the system; a knowledge-base (KB), which contains a set of rules and associations; the inference engine (IE), which combines the rules from the knowledge-base with the patients’ data.
- Nonknowledge-Based CDSSs, based on a machine learning system which allows learning from past experiences (ground truth) and/or finds patterns in clinical data; among the most common types of nonknowledge-based systems are supervised machine learning algorithms (as for developing computer-aided diagnosis systems [20,21]) and unsupervised machine learning algorithms (such as clustering or genetic algorithms).
Perhaps one of the first CDSSs (in early 1975) was MYCIN [22], an expert system that operated using a fairly simple inference engine and a knowledge-base of about 600 rules to identify bacteria causing severe infections, such as bacteremia and meningitis, and to recommend antibiotics, with the dosage adjusted for the patient’s body weight. During the same time (1970–1986), CADUCEUS was developed; it worked using an inference engine similar to MYCIN’s, but it made a number of changes (like incorporating abductive reasoning) to deal with problems such as the additional complexity of internal diseases, a number of simultaneous diseases, and generally flawed and scarce data. With DXplain (1984–1986), CDSSs began to be web-oriented, providing access through the Internet; in the 1990s (precisely with RODIA in 1997), they began to be used in medical imaging, diagnostics, orthopaedic, and other, more complex medical disciplines. RODIA provides two major functionality areas: Image (x-ray and ultrasonography) quantitative evaluation and fracture healing monitoring [23].
A modern CDSS is DiagnosisPro [24], developed in the 2000s, which is also a web-oriented medical expert system that provides exhaustive diagnostic possibilities for 11,000 diseases and 30,000 findings, providing the most appropriate differential diagnosis. The actual web-oriented trend is still growing, both in order to facilitate the diffusion and to exploit the greater computing capabilities of servers not always available on conventional workstations. Moreover, this allowed to greatly increase the number of diseases and to engage in more complex tasks, such as the analysis of biomedical images. More recently, it has been claimed that decision support will begin to replace clinicians in common tasks in the future [25].
Among the major services currently provided under the eHealth term, there are electronic health record management systems, ePrescribing, Telemedicine, and many others that aim at assisting physicians, patients or both.
In the last decade, several proposals to introduce chat-bot and social functionalities in eHealth have been carried out.
- GYANT (www.gyant.com): Using artificial intelligence, GYANT is able to analyze symptoms and provide recommendations tied to the user answer, with full medical explanations. The result is not a real diagnosis but provided in terms of a follow-up to be held.
- Babylon Health (www.babylonhealth.com): It is a health service provider able to connect patients and doctors using mobile messaging and a video functionality allowing users to obtain drug prescriptions, consult health specialists, and book exams with nearby facilities. It is provided with a nonfree subscription paradigm with a web-oriented and mobile-oriented GUI. Its novelty is in the communication protocol, but it does not enable any machine learning or artificial intelligence algorithms. In the last update (2016), an AI-like feature was released, providing prescreening feedback on the user health condition.
- Brook (www.brook.org.uk): Logging all health-related data may be a long and boring procedure. Book makes this procedure as simple as messaging and uses mobile phone notifications for quick and easy updates. Brook was borne as a disease-specific application allowing to control sugar-safe zones in patients with diabetes. Today, it represents a general-level application able to keep a logbook of our health data with a specific focus on food habits and lifestyle.
- Forksy (www.getforksy.com): It is defined as a robot-coach that supports the users in improving their eating habits. The chat-bot starts talking about some personal information to estimate a proper calorie limit. During the day, Forksy sends a message asking what users ate or drank and keeps it all in a food diary. Finally, Forksy provides a personal nutritional assessment to improve: Digestion, sleep, brain and cognitive performance, and productive hours per day.
Most of the proposals miss a machine learning core providing simple advice to the final user. In some cases, the application does not represent a real clinical center and, therefore, has limited functionalities (no physicians on the back-end, limited clinical data, and no possibility of booking a de-visu consult).
To the best of our knowledge, our proposal, HOLMeS (health online medical suggestions) is a novelty in the eHealth field, since it offers most of the services required by eHealth in an innovative way.
3. Big Data Tools Supporting eHealth Applications
In recent years, the growing amount of data and the need to extrapolate useful information from them motivated many big players to develop their own deep learning and big data application frameworks. The most common service-oriented architectures are deployed using different service models, such as: Software as a service (SaaS), platform as a service (PaaS), and infrastructure as a service (IaaS) [26]. In order to understand the techniques, methods, and approaches proposed in this work, it is necessary to describe some of these frameworks.
3.1. Apache Spark Cluster
Apache Spark is the general-purpose system for cluster computing developed by the Apache foundation [27]. It is designed to provide its services through high-level APIs (application programming interfaces) available for Java, Scala, Python, and R, also supporting different higher-level tools, among which it is worth mentioning SQL support, structured data processing, and MLlib (a machine learning library) [28]. Spark is able to run both by itself or over three different cluster managers:
- Standalone, using the included simple cluster manager;
- Apache Mesos [29], a more general cluster manager able to run Hadoop MapReduce and service applications;
- Hadoop YARN [30], the standard resource manager for Hadoop 2.
3.1.1. Hadoop Distributed File System
The Hadoop distributed file system (HDFS) is a distributed file system designed to run on commodity hardware [31]. It inherits some characteristics from the other distributed file systems, but, by design, it is high fault-tolerant and well-suited to work on commodity hardware. It also provides high throughput access to applications that use a large dataset, even if it is not fully compliant with the POSIX standard in order to provide data streaming. HDFS was originally intended to meet some specific goals:
- Hardware failure, since it has to run over hundreds or thousands of commodity server machines;
- Streaming data access, since applications that use HDFS usually need batch processing and, thus, hight throughput is preferred over low access latency;
- Large data, supporting tens of millions of files, each of a size in gigabytes to terabytes;
- Simple coherency model, in order to simplify application access policies, since many applications usually need a write-once-read-many access model for files;
- Portability both across heterogeneous hardware and software, in order to sustain its usage and diffusion.
3.1.2. MapReduce
MapReduce is a software framework originally introduced by Google to support computing on big data sets with a parallel, distributed algorithm on a cluster [32]. A typical MapReduce application is made of two steps:
- The map step, which filters and sorts data (for example sorting patient by age, arranging a different queue for each possible age value);
- The reduce step, which summarizes the data (for example, counting the number of patients in each queue, producing the age frequencies).
The core application manages all operations, primarily by distributing data over different servers, executing all tasks in parallel, and managing communication and fault tolerance schemas. One of the most popular open-source implementations of MapReduce is Apache Hadoop [33].
3.1.3. Spark.ML and ML Pipelines
Spark.ml, introduced starting from Spark 1.2, is a package that collects the inheritance of the old Spark MLlib, standardizing the Spark API for machine learning applications. ML Pipelines is a set of high-level API designed to provide a uniform development strategy to help users to create or combine multiple machine learning algorithms into a single pipeline (also called a workflow) [34]. A pipeline is a set of subsequent stages, each of which can be either a transformer (an abstraction including both feature transformers and trained models) or an estimator (an abstraction of any king of learning concept or algorithm that trains on data). Stages within a pipeline are executed in order, where the output of a given stage represents the input for the subsequent one. Apache Spark ML supports several high-level programming languages, including java, python. and scala.
3.2. Databricks
Databricks refers both to the company founded by the creators of Apache Spark and to the relative cluster infrastructure [35]. It aims to assist users in developing cloud-based big data processing applications using Spark by selling both a hosted cloud product (built on Spark) and relative training and courses. Databricks provides, in an IaaS model, a virtual analytic platform based on a web-based platform for working with Spark, which provides automated cluster management and IPython-style notebooks.
3.3. Watson
Watson is an advanced question-answering infrastructure, part of the Bluemix platform developed by IBM, able to interact in natural language processing. Its development started in 2006, but it was only in 2011 that it became world-famous when it was demonstrated to able to answer questions from the quiz show ‘Jeopardy!’ [36], winning the final prize. To achieve such a task, the Watson inference engine was able to process structured and unstructured contents, producing and elaborating up to four TeraBytes of data. In 2013, IBM devoted Watson to commercial applications involving the supercomputer in a decision support task for lung cancer treatment at Memorial Sloan Kettering Cancer Center, in New York City. From that application, several winning strategies led IBM to successful customer service applications.
In health-care, the natural language skills, the inference engine, and the evidence-based learning capabilities have been applied to contribute to clinical decision support systems for use by several medical professionals. Moreover, Watson cognitive services are able to draw from 600,000 medical evidence reports, 1.5 million patient records and clinical trials, and two million pages of text from medical journals to help doctors to develop treatment plans tailored to patients’ individual symptoms, genetics, and histories.
To provide advanced services deployed over the Watson supercomputer, IBM released Bluemix, a cloud platform as a service (PaaS) supporting several programming languages and cognitive services. Among the newer services provided via the Bluemix platform, there is the Watson conversation service. The IBM Watson conversation service can create, via API interfaces, an application that understands natural-language inputs and uses machine learning to respond to customers in a way that simulates a conversation between humans.
4. System Description
4.1. System Overview
In this work, first of all, we have designed a big data infrastructure to support different eHealth analytics applications as required for the last generation CDSSs. Figure 2 describes, at a glance, a functional overview of the proposed system that presents a layered architecture typical of a big data platform.
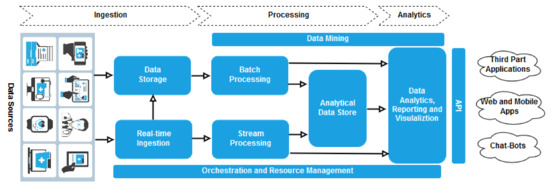
Figure 2.
System architecture.
During the ingestion stage, information coming from different data sources (e.g., EHR repositories, medical sensors and mobile devices) is gathered by the system. The architecture includes facilities to store data in a distributed file system that can hold high volumes of large files in various formats (data storage) and to capture and store real-time messages for stream processing (real time ingestion).
The system supports two different kinds of processing on ingested information:
- Data are processed using long-running batch jobs to filter, aggregate, and otherwise prepare the data themselves for analysis (batch processing);
- After capturing real-time messages, the architecture has to process them by filtering, aggregating, and otherwise preparing the data for analysis (strema processing).
Our architecture prepares data for analysis and then serves the processed data in a structured format that can be queried using analytical tools (analytical data store). Analytics is to provide insights into the data through analysis, reporting, and visualization (data analytic, reporting and visualization). To empower users to analyze the data, the architecture may include a data modeling layer. Analysis and reporting can also take the form of interactive data exploration by data scientists or data analysts. Eventually, data mining facilities can be exploited during processing and analytics tasks to support advanced advanced descriptive, predictive, and prescriptive analysis.
The resource management layer coordinates and optimizes the use of the different resources needed for implementing the whole system; in turn, orchestration allows to execute some of the workflow for data processing.
Finally, our system provides a set of API to applications (mobile and web apps, chat-bots, third-part, etc.) for invoking the analytical services.
4.2. Implementation Details
A particular instance/implementation of the described architecture is provided by the HOLMeS (health online medical suggestions) system.
It is composed of different modules that collaborate with each other to provide an advanced eHealth service through an intuitive chat application (Figure 3).
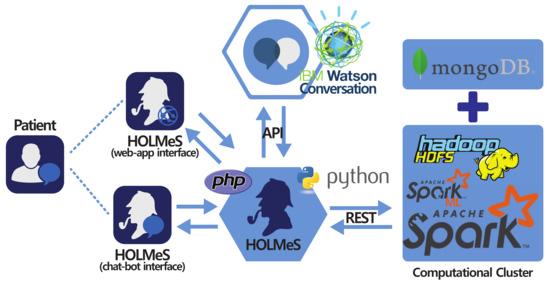
Figure 3.
HOLMeS (health online medical suggestions) system main modules with interaction paradigm: At the bottom-centre, the HOLMeS application core; On the left, the HOLMeS chat-bot (bottom) and the patient (top) interacting with HOLMeS; At the top-centre the, IBM Watson conversation logic adopted by HOLMeS through its API; On the right, the computational cluster providing storage, computing, and machine learning services used by HOLMeS.
The HOLMeS system general architecture is composed as follows:
- The HOLMeS Application is the HOLMeS system core. Written in Python, it implements the main logic and orchestrates module communications and functions. In particular, it communicates with the user through the chat-bot, interpreting their requests using the functionalities provided by the Watson conversation API, managing requests and responses between the patient and application modules in order to provide to the user with whom it is interacting (through the chat-bot or through the web-app) with the result of the machine learning algorithms with respect to the possibility of having a disease.
- HOLMeS chat-bot is the module dedicated to interacting with the user in order to let them feel more conformable. The bot is designed to understand different kinds of chat interactions, from formal writing to more handy ones. It is one of the HOLMeS system entry points and interacts with the user to let them choose the required service. It is also intended to kindly ask the user for required information (such as age, height, weight, smoking status, and so on), just as a human physician would.
- HOLMeS web-app is the interface dedicated to performing the medical interview via dynamic web forms. The forms change the question fields according to the previous answers, minimizing the interaction while maximizing the quality of the information. The results are provided by bar plots and gauges. Although the web-form interfaces are not as comfortable as a chat-bot, they offer a suitable means for Internet browsers and mobile devices (via mobile-ready interfaces).
- IBM Watson provides the service needed to establish a written conversation, simulating human interactions, through its conversation APIs. Main features include natural language processing and text mining through machine learning approaches (in particular, convolutional neural networks and support vector machines).
- Computational cluster implements the decision making logic on the basis of the described big data architecture. It uses the Apache Spark cluster executed over the Databricks infrastructures, in order to be fast and scalable enough to be effectively used in a very big clinical scenario, where many requests from different patients come together, ensuring a response time comparable to that of a human physician. It uses machine learning algorithms from Spark ML library, previously trained on many clinical features, in order to predict the expected occurrence probability for different diseases. Finally, the storage service is delegated to Hadoop HDFS and to Mongo DB for storing patient clinical records.
HOLMeS is able to handle four possible use-case scenarios:
- Providing general information about itself or the affiliated medical centre;
- Collecting general patient information in order to provide general prevention pathways indications among different disease (the result is provided in terms of probabilities, per each prevention pathway, of a positive response to each of the screening procedures);
- Collecting detailed patient information (clinical, examination results and so on) in order to evaluate the needs of a specific disease prevention partway (the result is provided in terms of probability of a positive response to the screening procedure);
- Booking a de-visu examination with the affiliated medical centre.
As described, HOLMeS was designed to be modular, where each concern is associated with a given module. This allows to design and develop each piece separately, in order to better fit specific requirements, such as scalability, speed, availability, and so on. In the following, we describe the HOLMeS main functionalities. Finally, the used dataset is introduced, laying the foundation for results shown in Section 5.
4.3. Data Storage
The clinical service supporting the screening procedures generates a huge amount of data. The bigger amount of data is represented by the images from CT (computed tomography), MRI, and RX (X-Rays) and their storage is managed by a clinical PACS (picture archiving and communication system). The remaining of the produced data consist of clinical records, medical annotations, and diagnoses (directly derived from the clinical images stored on the PACS). Since the ML algorithms work on the clinical records and on the diagnosis, the storing of the latter is crucial and it is delegated to MongoDB.
MongoDB is a cross-platform document-oriented database classified as a NoSQL-like database. It uses JSON-like (Javascript object notation) documents with not-fixed schemata organized for our goals as in Figure 4:
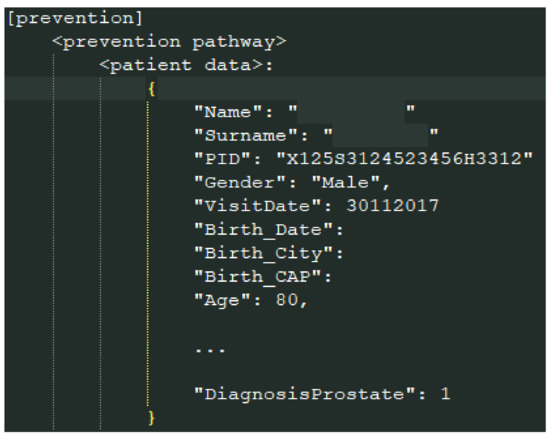
Figure 4.
The MongoDB data format.
The main container is the document prevention; for each prevention pathway, a subdocument, named according to the prevention name, contains the patients data involved in such a pathway. In Figure 4, part of the data of a patient is shown.
4.4. Chat-Bot Interaction Skills
The HOLMeS chat-bot module mimics human behavior in order to let the user feel more conformable, overcoming the bias of a ‘machine interaction’. It relies on the IBM Watson conversation APIs that are able to hold a complete automatic chat with the user. Applying natural language processing and machine learning algorithms, the Watson service identifies the user’s intents and the concerning entities.
Watson provides a high-level dialogue flow design allowing to write down the entire conversation example-by-example. These examples and the interaction model, composed of intents and entities, is used to fine-tune a pretrained deep neural network.
In order to fulfill the four possible use-cases (described above), the Watson conversation service has been designed to recognize the following intents:
- #greetings: To handle the initial conversation preamble;
- #book: To describe actions as reserving a de-visu examination;
- #get: To ask to receive something, such as prevention pathway indications or information about the centre;
- #put: To catch the intent of giving the required information to the system.
Moreover, several entities useful to contextualize the above intents have been described. The entities are formalized by synonyms. The more equivalent formulations of an entity are provided, the more precise the subject recognition will be. Some of the used entities are in the following list:
- @HOLMeS: “holmes”, ”you”, ”system”;
- @HOLMeS_functionalities: ”functionalities”, ”skills”, capabilites”;
- @clinical_centre: ”CMO”, ”centre”, ”clinical”;
- @address, ”address”, ”location”, ”position”;
- @de_visu_examination: ”visit”, ”appointment”, ”medical consult”;
- @specific_pathway_examination: ”specific visit”, ”specific examination”, ”specific pathway”;
- @general_pathway_examination: ”general visit”, ”general examination”, ”general pathway”;
- @age: ”age”, ”years”, ”years old”;
- @sex: ”gender”, ”sex”, ”sexuality”;
- @birthday: ”birthday”, ”BD”;
- @height: ”height”, ”highness”;
Intents and entities are then combined to achieve a fully automated conversation flow using the ‘dialog design toolbox’ of the Bluemix platform. For example, to achieve the initial greeting preamble, the application can catch the #greeting intent and, thus, will answer with random greeting sentences. Only after the greeting step is passed can the user ask for more intents, such as #get, referring to a specific entity, for example, the @HOLMeS_functionalities. In this case, HOLMeS will answer with the list of all the available functionalities the user can ask for. An example of the functionalities described above is depicted in Figure 5.
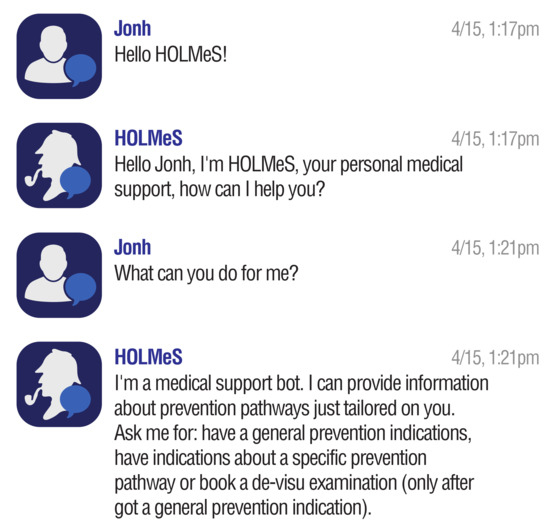
Figure 5.
Conversing with HOLMeS: Greating preamble.
4.5. The Machine Learning Algorithm
Even if it is able to understand natural language, the chat-bot alone is not able to provide any kind of medical advice. IBM Watson conversation APIs offer a broad range of features to design a fully operating conversation chat-bot but miss a trainable machine learning module for custom data. To overcome that limitation, HOLMeS contains a machine learning core, directly connected with the orchestrator and the Watson APIs, enabling it to act as a clinical decision support system. The machine learning core (a) is able to derive inferences based on uses clinical data, (b) autonomously learn the best representation of diseases after a proper training stage, and, thus, (c) automatically generate the knowledge-base if new diseases have to be introduced to the system. In particular, to enhance HOLMeS upgradeability, we proposed to implement a different classifier for each disease the system has to be able to predict. Finally, in order to combine all classifier results and to return simple and clear information to the user, a suitable combiner is required. Since results consist of a set of risk probabilities associated to each disease, the simpler way to combine them in a single-shot graph is to use a histogram.
The main advantages of the multiclassifier architecture are:
- Improved scalability, since classifiers can be easily distributed over a cluster to accommodate a growing amount of user requests;
- Reduced computational time, since the schema is highly parallelizable because different classifier predictions can be evaluated in parallel;
- High maintainability, since any single classifier can be adapted, corrected or improved without any impact on any other system components;
- High upgradability, since a new classifier can be easily added to make HOLMeS able to deal with any new desired diseases.
To deploy our machine learning algorithms, we chose to use Spark as cluster-computing framework deployed over the Databricks infrastructures. Spark provides data storage, implicit data parallelism, and fault-tolerance features. The Spark.ML library provides all the data-preparation functionalities and the machine learning algorithms to train the random forest models using the collected training data. The same Spark.ML library is, then, used to achieve the final classification for each disease prevention pathway. Moreover, via the ‘ML pipeline’ paradigm, Spark allows to easily deploy and generate the knowledge-base (represented by the trained model) every time a retraining is required (such as when new diseases or new patients are stored in the database).
To be more specific, the HOLMeS machine learning core provides two different working modalities:
- General-level prevention evaluation, in which the user can query the HOLMeS system, through the chat bot, in order to have the first medical prevention advice, submitting some simple clinical features, such as age, height, weight, living place, smoking status, some disease familiarity, and so on.
- Specific disease prevention evaluation, in which a user can query the HOLMeS System, through the chat bot, in order to have more detailed prevention advice about a specific disease by submitting more specific features, such as results examination, blood pressure, respiratory and heart rate, oxygen saturation, body temperature, and so on.
The output of the machine learning algorithm, deployed in Spark, is different according to the required working modality. When a general-level evaluation is required, the algorithm will produce a histogram graph containing the disease occurrence probability per each of the disease available in the dataset. This result will be stored in the database and can be retrieved by the physician when a de-visu examination will occur or can be used by the patient to choose the specific disease to be evaluated in the second working modality. In this last working modality, the user will receive a probability of being affected by the requested disease.
4.6. General Functionalities
The core of the system is in charge of orchestrating the several data flows. Data from and to the patient, through the chat-bot (Figure 6) or web-app (Figure 7) interactions, have to be routed to the computational cluster, where the trained machine learning models produce the diagnosis. Such a diagnosis is directly inferred, using the trained models, from the data collected in the chat-bot session or from the web-app form, and then shown to the patient. Only the chat-bot skills require the Watson services using a conversation API. The interaction between the user and the application occurs without any further elaboration from the HOLMeS Application that observes the data flow, memorizing the information of interest.
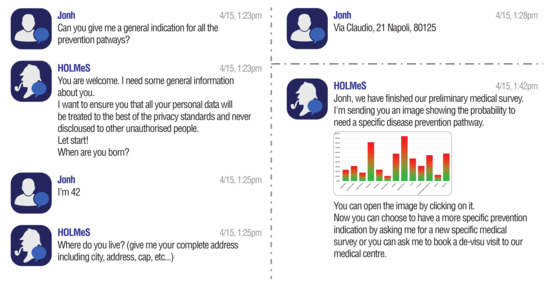
Figure 6.
Conversing with HOLMeS via the chat-bot interface: Asking for general indications about the available prevention pathways.
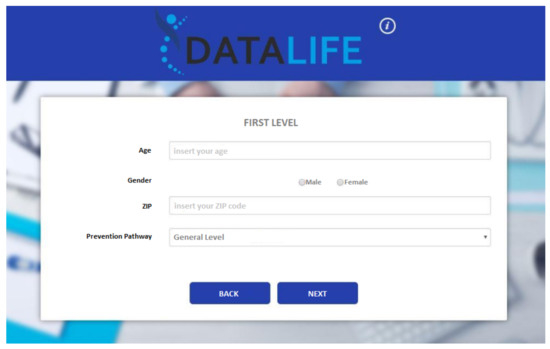
Figure 7.
Layout of the deployed web application: Interface asking for the preliminary information.
When interaction requires the elaboration of machine learning algorithms or access to the data storage, the core system contacts the cluster to accomplish the specific task.
In order to fulfill the four possible use-cases (as described above), the core application needs to catch the following intents:
- The user wants information about the affiliated medical centre;
- The user asks for general information about the system and its functionalities;
- The patient desires to obtain general-level evaluation about the available prevention pathways;
- The patient desires to obtain detailed indications about a specific disease prevention pathway (only after a general survey has been carried out);
- The patient wants a de-visu examination (only after a general survey has been carried out).
For example, to meet the 2nd use-case scenario, after the greeting preamble, HOLMeS recognizes the #get user intent combined with the @general_patways_examination entity. Such interaction yields to the data collecting chat flows with the aim of achieving a general-level prevention pathway evaluation.
The results of a general-level evaluation conversation are similar to those reported by the graph in Figure 8. The idea is that a histogram can easily highlight the patient’s occurrence risk associated to each disease on the basis of the collected data, guiding him (i) to take a de-visu examination with the proper physician or (ii) to proceed to the specific-level evaluation for the most risky pathologies, in order to have more reliable results (since based on the answers to more specific questions).
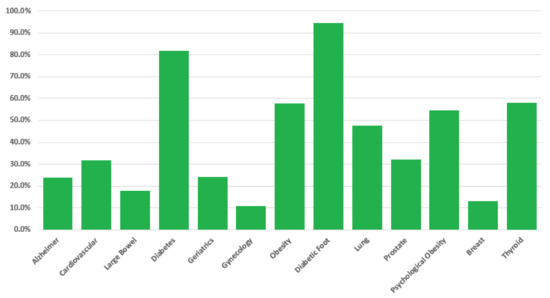
Figure 8.
Histogram produced in response to the general-level evaluation (both for chat-bot and web-app interface). For each disease (x axes), the bar reports the associated occurrence risk (y axes), evaluated on the basis of the answers provided by the user.
Once the patient obtains an overview of the risk associated to each analyzed disease, they can proceed to a second-level interview, to get more specific insights about a given pathology (one per time). It is worth noting that in this case, questions are more specific and often concern the outcomes of other investigations, like blood analysis, ultra-sounds, etc. The reason is that while the general-level investigation aims at giving an early picture of the most likely issues, the specific-level investigation aims at helping the patient to find the most suitable physician expert.
4.7. Dataset
In this paper, we focus on the preventive healthcare for 13 different prevention pathways. A total of 16,733 patients’ prevention records have been collected from the clinical teams of each prevention pathway, led by 13 expert physicians, one per each specific medical screening field. Figure 1 shows the timeline of the data acquisition. Each patient contains a positive or negative ground truth, indicating whether its prevention pathway leads to a positive diagnosis (the presence of the diseased subject of the screening program). For each pathway, Table 1 reports the number of involved patients and their average age.

Table 1.
Dataset size and stats per each prevention pathway.
5. Results
In this section, we report the results obtained by applying the HOLMeS system to the use-case scenario introduced in the previous section. Starting from the very beginning, our idea was to develop a system able to be effectively used in a real clinical scenario, with the aim of assisting both physicians and patients. However, given the nature of the problem under consideration, to fulfill this goal, it is crucial to validate the reliability of the obtained disease risk factor outcomes. To this aim, we performed a 10-fold cross validation for each disease prevention pathway, separately, for both working modalities. It is worth noting that since the system operates in a clinical environment, not only does it have to correctly identify all risky patients, but it is crucial to minimize the false negative rate over the false positive rate: It is more important to not miss a diseased patient. Therefore, we chose to compare classifier results in term of the area under the ROC curve (AUC–ROC) that both considers the accuracy and the error-rates. For both the working modalities, Table 2 reports the mean values of the 10-fold cross validation evaluated for each disease by means of random forest classifiers made of 200 decision trees and without any limitation on the depth of the trees; the table also reports the 95% confidence interval and the median value for both working modalities, evaluated over all diseases.

Table 2.
Classification performance of the general and specific pathway evaluation (first level) per each disease in the dataset. The performances are in terms of area under the ROC (receiver operating characteristic) curve (AUC–ROC) obtained from a 10-fold cross validation. 95% confidence intervals and median value are reported.
6. Conclusions
The aim of this paper was to propose HOLMeS (health online medical suggestions) system.
The proposed approach aims at supporting eHealth applications using a trained machine learning algorithm, deployed on a cluster-computing framework, which provides medical suggestion via a chat-bot module. The chat-bot, trained with a deep learning approach, helps in overcoming the limitation of a cold interaction between users and the software simulating human behavior.
The results presented in Section 5 validate the machine learning algorithms both for the general-level prevention indications and for the disease-specific prevention evaluation. It is worth noting that the second-level evaluation leads to better results (improving the AUC by about 12%) because it exploits more specific features provided by physicians or obtained via further examination.
We want to note that using machine learning in a clinical scenario requires taking into account the explainability of the choices made by the model, in order to make it possible to understand the processes that lead the classifier to make its predictions and ensure that no critical (illegal or discriminatory) actions were performed. Among all the available white-box algorithms, we chose the random forest classifier because it turned out to be the most reliable (the one with the greater generalization ability) in some preliminary results we performed on the same data (with a lower temporal resolution) in a previous activity.
We are currently working on a GUI that allows to use the knowledge-base and the machine-learning algorithms deployed in the cluster infrastructure without the chat-bot. That will turn HOLMeS into a classical clinical decision support system giving to a physician the possibility of continuing the disease prevention evaluation starting from the second level, when the patient ask for a de-visu examination.
Future work will also focus on improving the trustiness of the chat-bot, providing more human-like behaviors and improving the so-far implemented use-case scenarios. To this aim, we are currently working on (i) training the chat-bot on a set of real conversations collected during the patient examinations and (ii) voice recognition and text-to-speech modules, in order to not only provide a simpler usage modality, but to also allow HOLMeS to be used by disabled patients (for example, those affected by blindness or those with reduced arms/hands ability).
Moreover, additional prevention pathways should be added and more patients will be recruited in order to have complete prevention and a more reliable result.
Further future work will concern interoperability issues. We are planning to open up the system to third party research, considering, as an example, the PMML (predictive model markup language) standard for ML model description and HL7 and the related extension to support communication with other healthcare information systems.
Finally, Watson provides support for different natural languages (such as Brazilian Portuguese, English, French, Italian, Spanish, German, Traditional Chinese, Simplified Chinese, Dutch, and Arabic). This multilanguage feature could be added to our system, pushing the spread of HOLMeS to different countries and helping in sharing medical knowledge.
Author Contributions
Conceptualization, G.P. and S.M.; Methodology, G.P. and S.M.; Software, F.A. and A.P.; Validation, A.P. and C.S.; Formal analysis, F.A.; Investigation, G.P. and S.M.; Resources, C.S. and V.M.; Data curation, G.P., S.M. and F.A.; Writing—original draft preparation, G.P. and S.M.; Writing—review and editing, C.S., V.M. and A.P.; Supervision, C.S., V.M. and A.P.; Project administration, V.M.; All authors read and approved the final manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors would like to thank CMO—one of the partners of the DATALIFE Research Consortium—for the availability of the dataset that allowed us to perform the experimentation of this paper. The authors would also like to thank all anonymous reviewers and editors for their helpful suggestions for the improvement of this paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Buyya, R.; Yeo, C.S.; Venugopal, S. Market-oriented cloud computing: Vision, hype, and reality for delivering it services as computing utilities. In Proceedings of the 10th IEEE International Conference on High Performance Computing and Communications, 2008. HPCC’08, Dalian, China, 25–27 September 2008; pp. 5–13. [Google Scholar]
- Bryant, R.; Katz, R.H.; Lazowska, E.D. Big-Data Computing: Creating Revolutionary Breakthroughs in Commerce, Science and Society, 2008. Available online: https://www.immagic.com/eLibrary/ARCHIVES/GENERAL/CRA_US/C081222B.pdf (accessed on 21 January 2019).
- Piantadosi, G.; Marrone, S.; Sansone, M.; Sansone, C. A Secure OsiriX Plug-In for Detecting Suspicious Lesions in Breast DCE-MRI. In Algorithms and Architectures for Parallel Processing; Springer International Publishing: Berlin, Germany, 2013; pp. 217–224. [Google Scholar]
- Piantadosi, G.; Marrone, S.; Sansone, M.; Sansone, C. A secure, scalable and versatile multi-layer client–server architecture for remote intelligent data processing. J. Reliab. Intell. Environ. 2015, 1, 173–187. [Google Scholar] [CrossRef]
- Pérez, A.; Gojenola, K.; Casillas, A.; Oronoz, M.; de Ilarraza, A.D. Computer aided classification of diagnostic terms in spanish. Expert Syst. Appl. 2015, 42, 2949–2958. [Google Scholar] [CrossRef]
- Arsene, O.; Dumitrache, I.; Mihu, I. Expert system for medicine diagnosis using software agents. Expert Syst. Appl. 2015, 42, 1825–1834. [Google Scholar] [CrossRef]
- Winters, B.; Custer, J.; Galvagno, S.M.; Colantuoni, E.; Kapoor, S.G.; Lee, H.; Goode, V.; Robinson, K.; Nakhasi, A.; Pronovost, P.; et al. Diagnostic errors in the intensive care unit: A systematic review of autopsy studies. BMJ Qual. Saf. 2012, 21, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Tehrani, A.; Lee, H.; Mathews, S.; Shore, A.; Makary, M.; Pronovost, P. Diagnostic Errors More Common, Costly and Harmful Than Treatment Mistakes; John Hopkins Medicine: Baltimore, Maryland, 2013. [Google Scholar]
- Ongenae, F.; Claeys, M.; Dupont, T.; Kerckhove, W.; Verhoeve, P.; Dhaene, T.; De Turck, F. A probabilistic ontology-based platform for self-learning context-aware healthcare applications. Expert Syst. Appl. 2013, 40, 7629–7646. [Google Scholar] [CrossRef]
- Della Mea, V. What is e-Health: The death of telemedicine? J. Med. Internet Res. 2001, 3, e22. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Rizo, C.; Enkin, M.; Jadad, A. What is eHealth? A systematic review of published definitions. J. Med. Internet Res. 2005, 7, e1. [Google Scholar] [CrossRef] [PubMed]
- Healy, J. Implementing e-Health in developing countries: Guidance and principles. In ICT Applications and Cyber Security Division (CYB). Policies and Strategies Department. [Monografía en Internet]; International Telecommunication Union: Geneva, Switzerland, 2008. [Google Scholar]
- Ball, M.J.; Lillis, J. E-health: Transforming the physician/patient relationship. Int. J. Med. Inform. 2001, 61, 1–10. [Google Scholar] [CrossRef]
- Eysenbach, G.; Diepgen, T.L. The role of e-health and consumer health informatics for evidence-based patient choice in the 21st century. Clin. Dermatol. 2001, 19, 11–17. [Google Scholar] [CrossRef]
- O’donoghue, J.; Herbert, J. Data management within mHealth environments: Patient sensors, mobile devices, and databases. J. Data Inf. Qual. 2012, 4, 5. [Google Scholar] [CrossRef]
- Marrone, S.; Piantadosi, G.; Fusco, R.; Petrillo, A.; Sansone, M.; Sansone, C. A Novel Model-Based Measure for Quality Evaluation of Image Registration Techniques in DCE-MRI. In Proceedings of the 2014 IEEE 27th International Symposium on Computer-Based Medical Systems, New York, NY, USA, 27–29 May 2014; pp. 209–214. [Google Scholar]
- Piantadosi, G.; Marrone, S.; Fusco, R.; Petrillo, A.; Sansone, M.; Sansone, C. Data-driven selection of motion correction techniques in breast DCE-MRI. In Proceedings of the 2015 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Torino, Italy, 15–18 May 2015; pp. 273–278. [Google Scholar]
- Marrone, S.; Piantadosi, G.; Fusco, R.; Petrillo, A.; Sansone, M.; Sansone, C. Breast segmentation using Fuzzy C-Means and anatomical priors in DCE-MRI. In Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 1472–1477. [Google Scholar]
- Berner, E.S. Clinical Decision Support Systems; Springer: Berlin, Germany, 2007. [Google Scholar]
- Marrone, S.; Piantadosi, G.; Fusco, R.; Petrillo, A.; Sansone, M.; Sansone, C. Automatic Lesion Detection in Breast DCE-MRI. In Image Analysis and Processing (ICIAP); Springer: Berlin/Heidelberg, Germany, 2013; pp. 359–368. [Google Scholar]
- Piantadosi, G.; Fusco, R.; Petrillo, A.; Sansone, M.; Sansone, C. LBP-TOP for Volume Lesion Classification in Breast DCE-MRI. In Lecture Notes in Computer Science (including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2015; Volume 9279, pp. 647–657. [Google Scholar]
- Buchanan, B. Rule based expert systems. In The MYCIN Experiments of the Stanford Heuristic Programming Project; Wiley Online Library: Hoboken, NJ, USA, 1984. [Google Scholar]
- Musen, M.A.; Middleton, B.; Greenes, R.A. Clinical decision-support systems. In Biomedical Informatics; Springer: Cham, Switzerland, 2014; pp. 643–674. [Google Scholar]
- Aronson, A.R. DiagnosisPro: The ultimate differential diagnosis assistant. JAMA 1997, 277, 426–426. [Google Scholar] [CrossRef]
- Khosla, V. Technology Will Replace 80% of What Doctors Do. Available online: http://tech.fortune.cnn.com/2012/12/04/technology-doctors-khosla/ (accessed on 1 September 2018).
- Papazoglou, M.P.; Traverso, P.; Dustdar, S.; Leymann, F. Service-oriented computing: State of the art and research challenges. Computer 2007, 40. [Google Scholar] [CrossRef]
- Apache Spark. Apache Spark: Lightning-Fast Cluster Computing. Available online: http://spark.apache.org/ (accessed on 21 January 2019).
- Meng, X.; Bradley, J.; Yavuz, B.; Sparks, E.; Venkataraman, S.; Liu, D.; Freeman, J.; Tsai, D.; Amde, M.; Owen, S.; et al. Mllib: Machine learning in apache spark. J. Mach. Learn. Res. 2016, 17, 1–7. [Google Scholar]
- Kakadia, D. Apache Mesos Essentials; Packt Publishing Ltd.: Birmingham, UK, 2015. [Google Scholar]
- Vavilapalli, V.K.; Murthy, A.C.; Douglas, C.; Agarwal, S.; Konar, M.; Evans, R.; Graves, T.; Lowe, J.; Shah, H.; Seth, S.; et al. Apache hadoop yarn: Yet another resource negotiator. In Proceedings of the 4th Annual Symposium on Cloud Computing, Indianapolis, IN, USA, 10–11 June 2013; p. 5. [Google Scholar]
- Shvachko, K.; Kuang, H.; Radia, S.; Chansler, R. The hadoop distributed file system. In Proceedings of the 2010 IEEE 26th Symposium on Mass Storage Systems and Technologies (MSST), Lake Tahoe, NV, USA, 3–7 May 2010; pp. 1–10. [Google Scholar]
- Dean, J.; Ghemawat, S. MapReduce: Simplified data processing on large clusters. Commun. ACM 2008, 51, 107–113. [Google Scholar] [CrossRef]
- Apache hadoop. Available online: http://hadoop.apache.org/ (accessed on 21 January 2019).
- Meng, X.; Bradley, J.; Sparks, E.; Venkataraman, S. ML Pipelines: A New High-Level API for MLlib. Databricks Blog. Available online: https://databricks.com/blog/2015/01/07/ml-pipelines-a-new-high-level-api-for-mllib.html (accessed on 1 November 2018).
- Databricks.com. Databricks. Available online: https://databricks.com/ (accessed on 1 November 2018).
- Ferrucci, D.; Levas, A.; Bagchi, S.; Gondek, D.; Mueller, E.T. Watson: Beyond jeopardy! Artif. Intell. 2013, 199, 93–105. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).