Characterization of TGFβ Signaling Components in Large Yellow Croaker (Larimichthys crocea) and Their Role in Growth and Body Shape Regulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Fish
2.2. Sequence Analysis
2.3. mRNA Expression of TGFβ Signaling Pathway Genes
3. Results
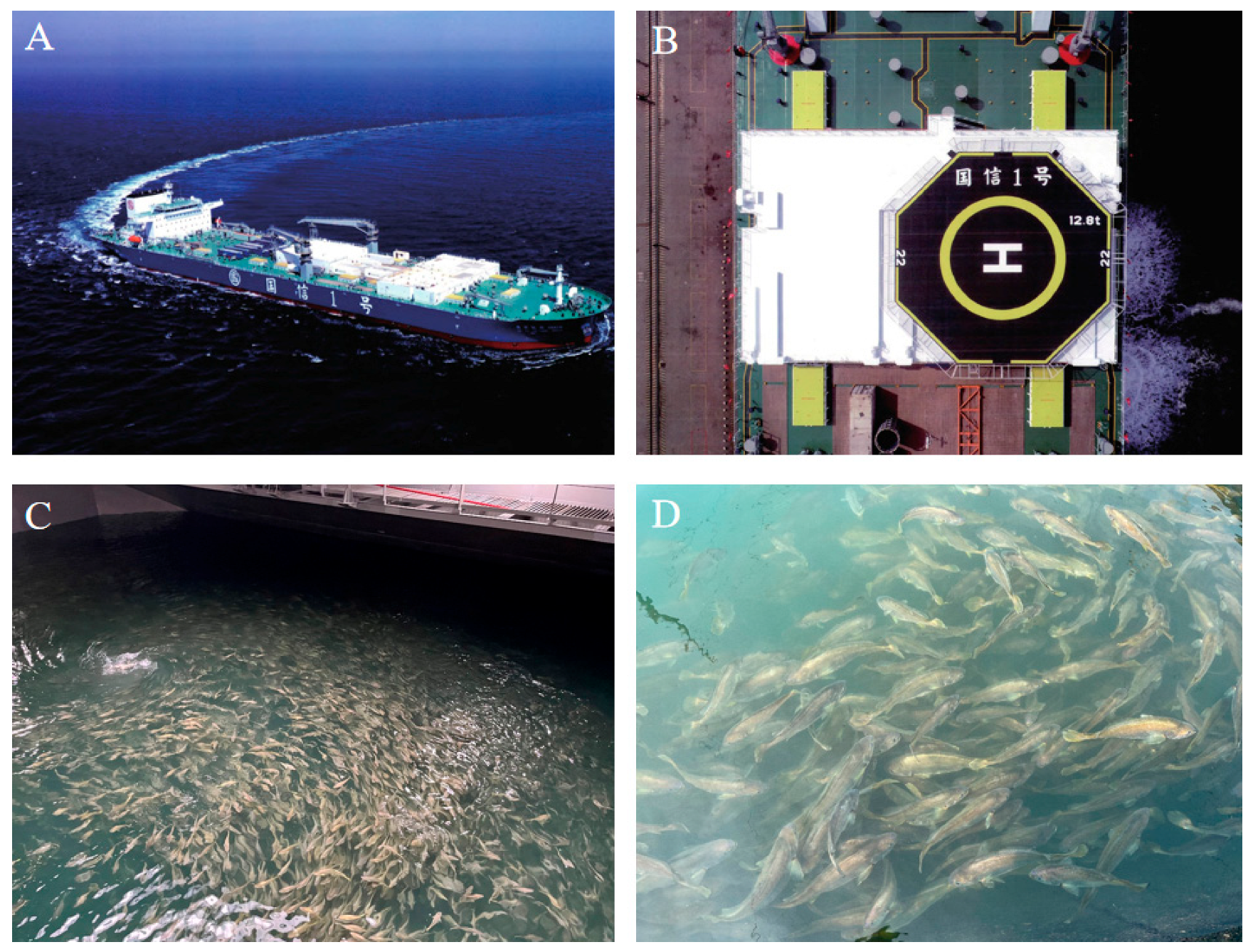
3.1. The Growth and Body Shape of L. crocea Under Two Aquiculture Modes
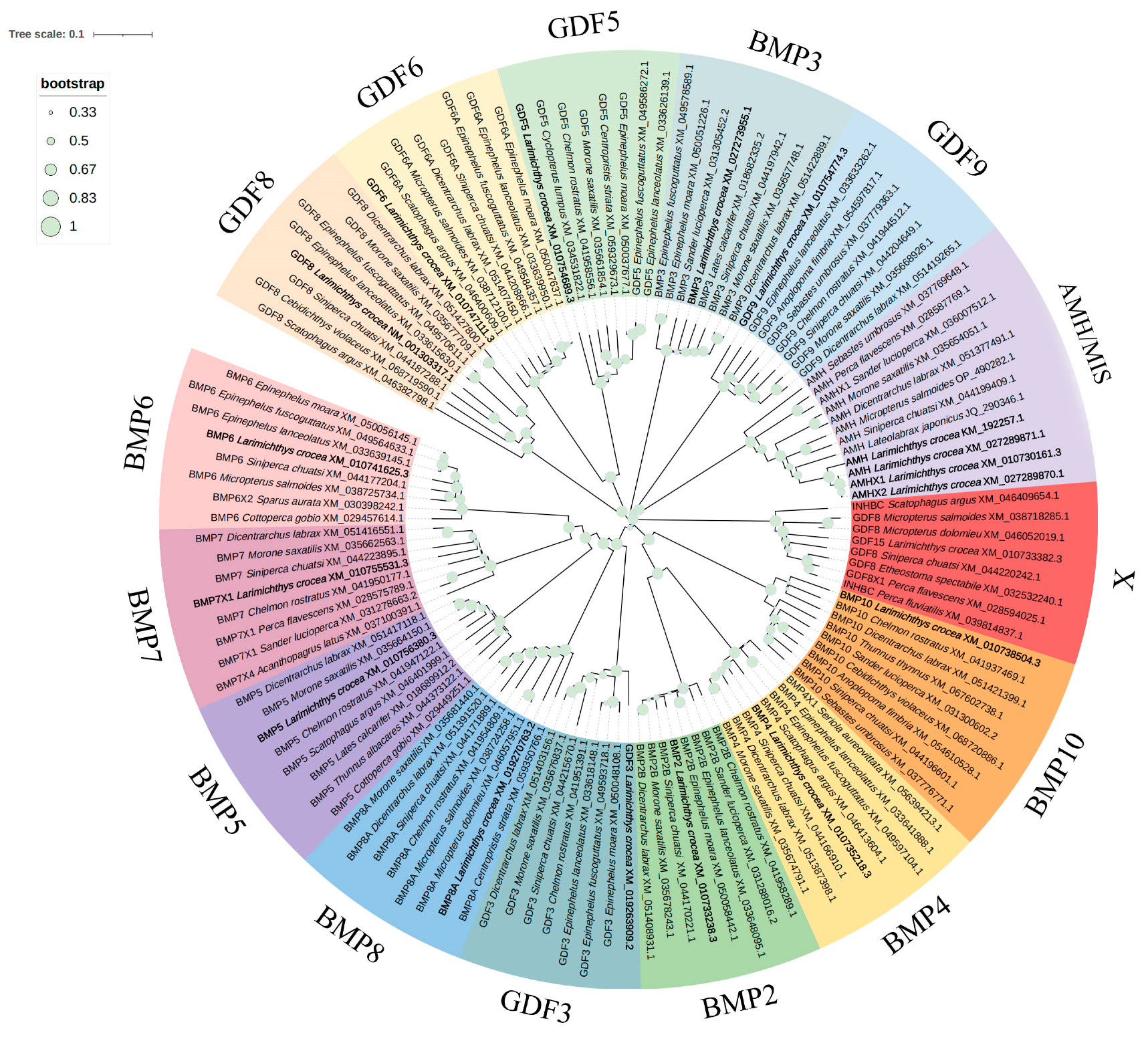
3.2. Phylogenetic Analysis of Ligand Sequences
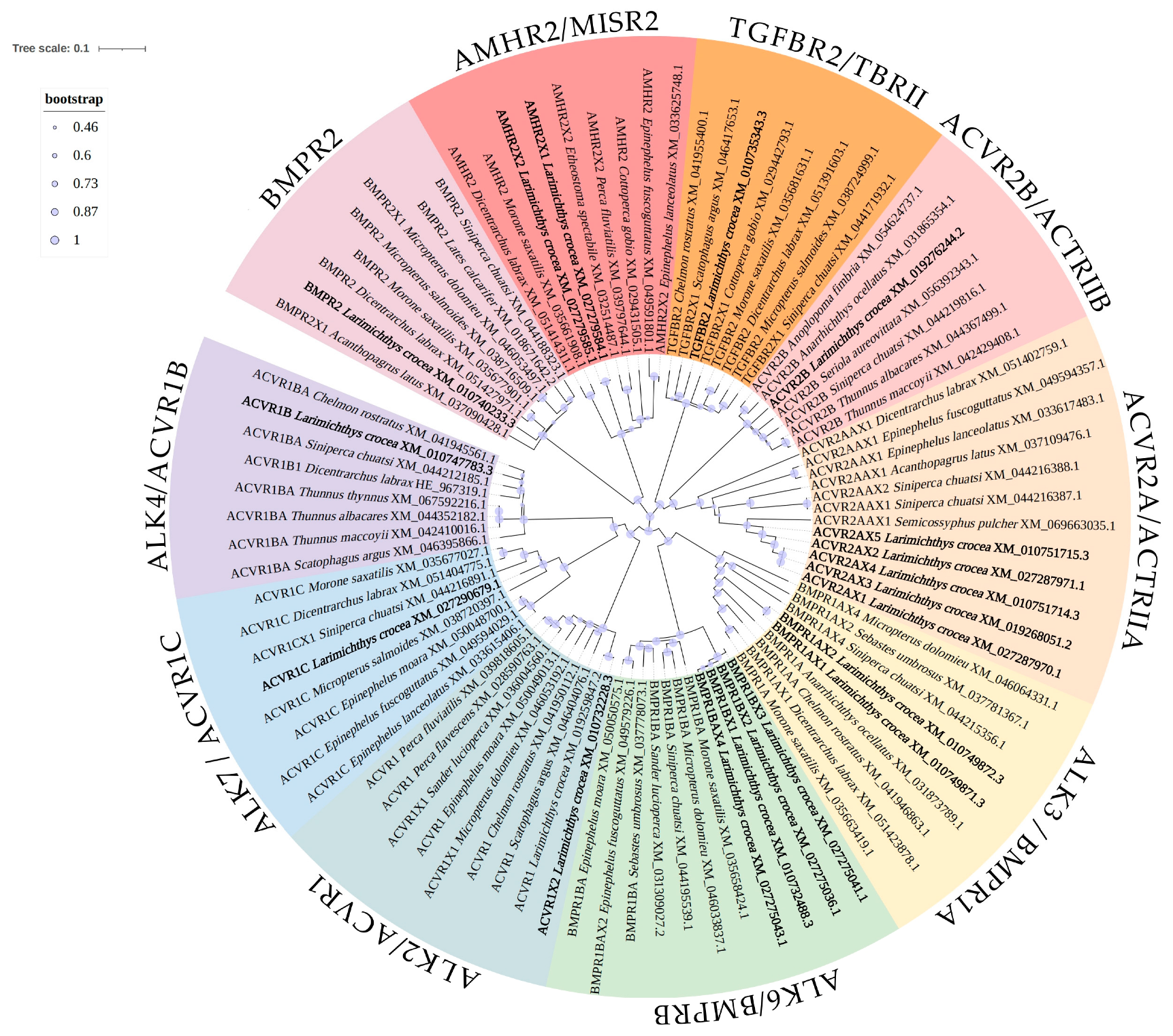
3.3. Phylogenetic Analysis of Receptors
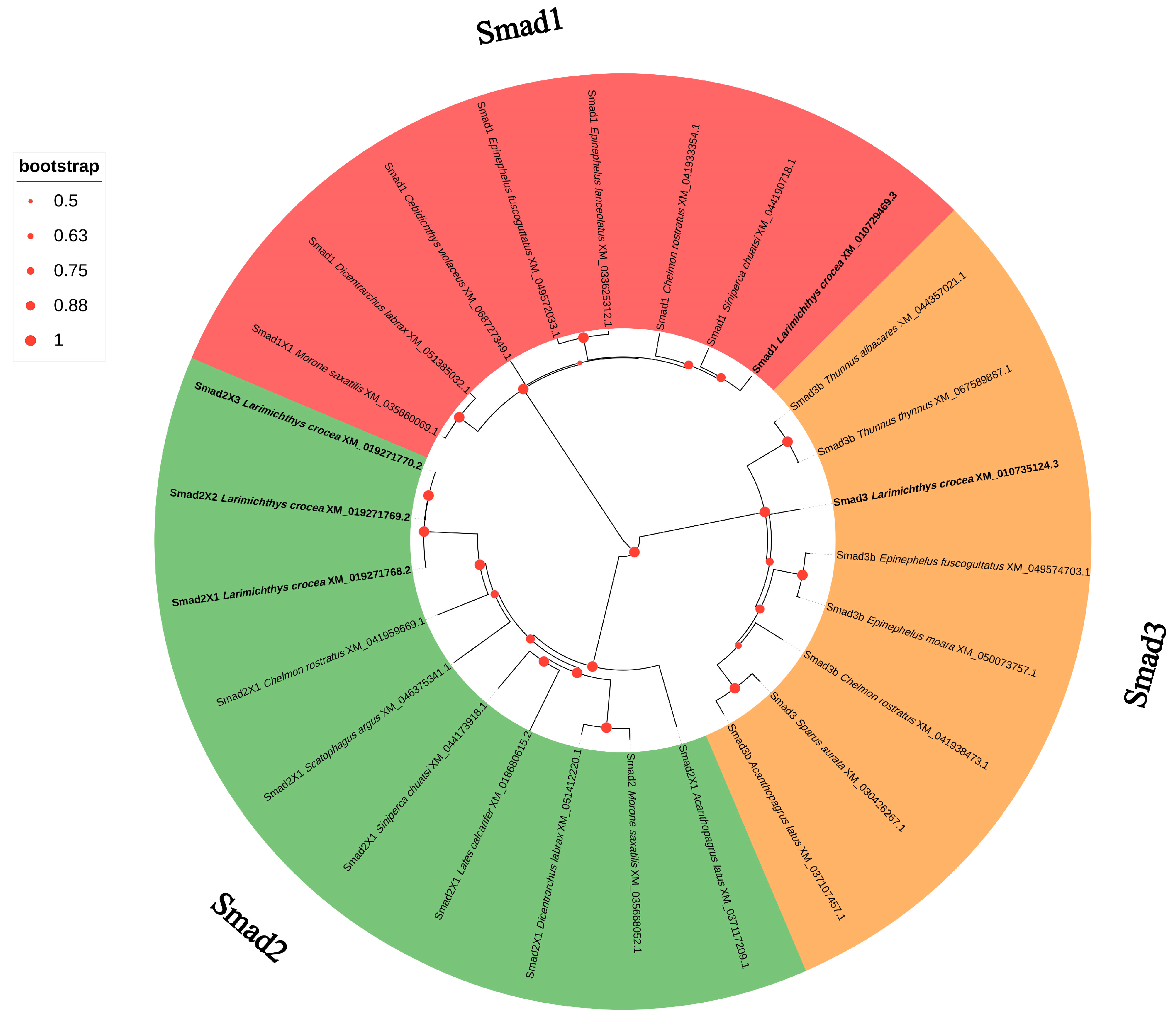
3.4. Phylogenetic Analysis of R-Smads
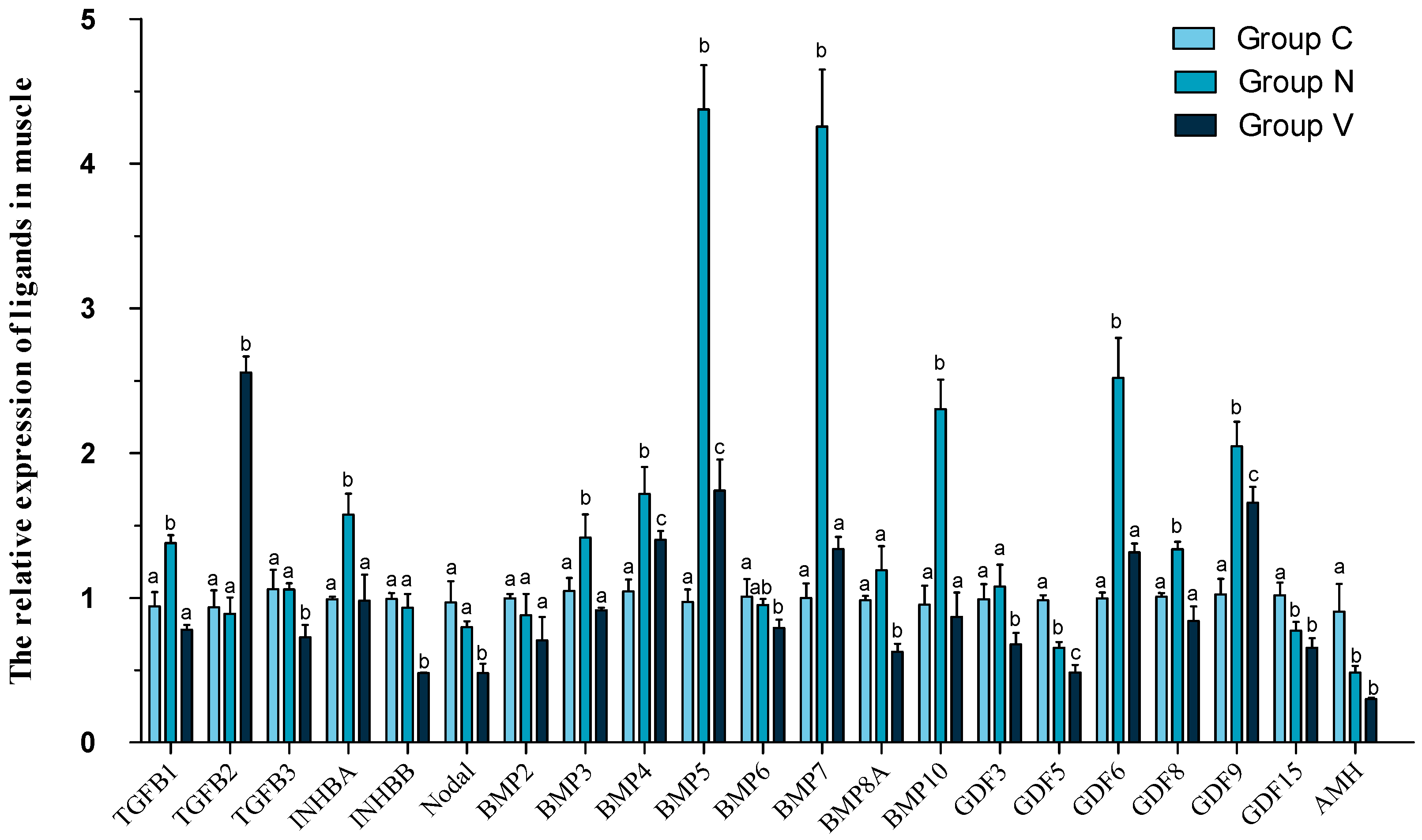
3.5. mRNA Levels of Ligands
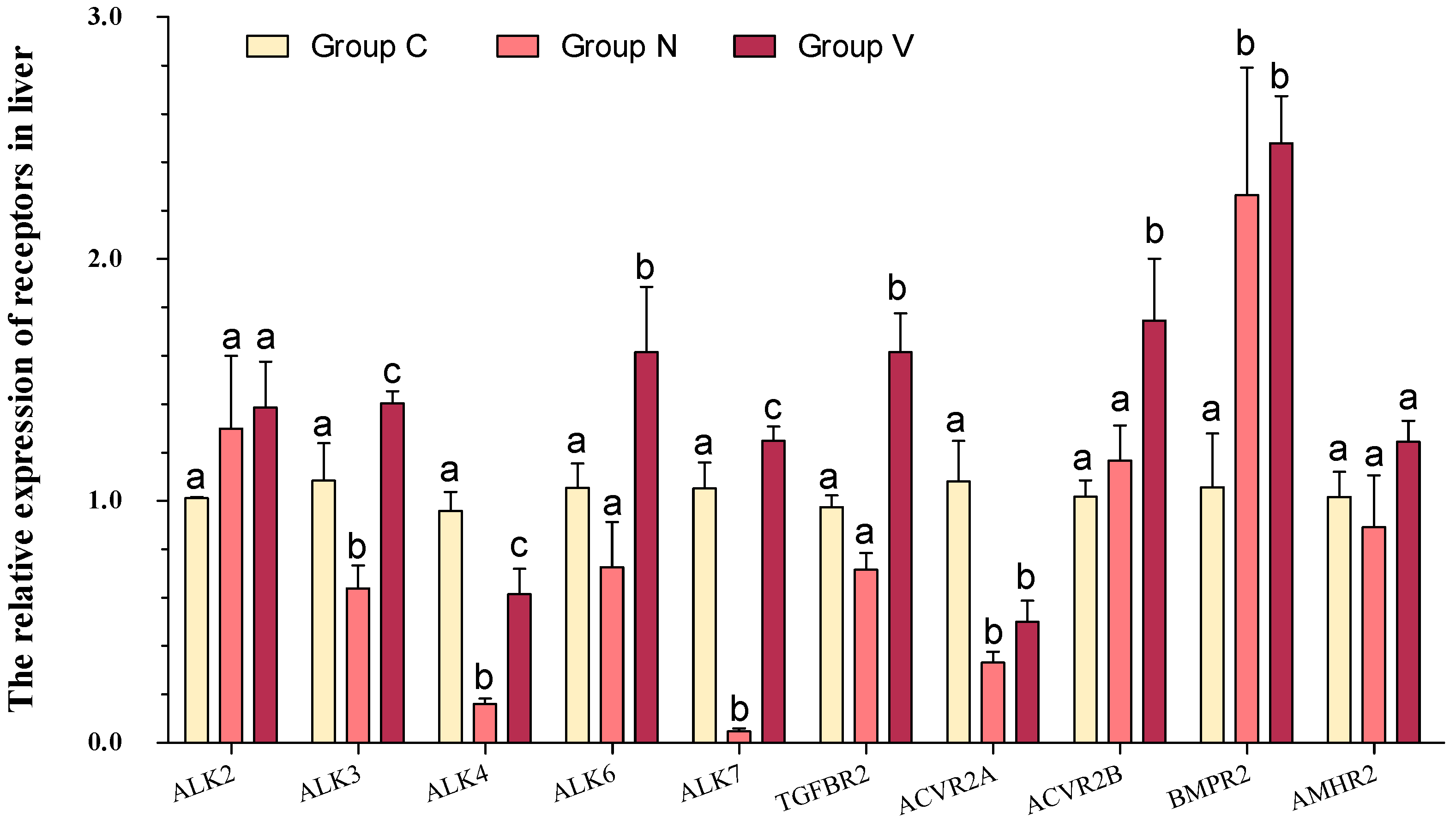
3.6. mRNA Levels of Receptors
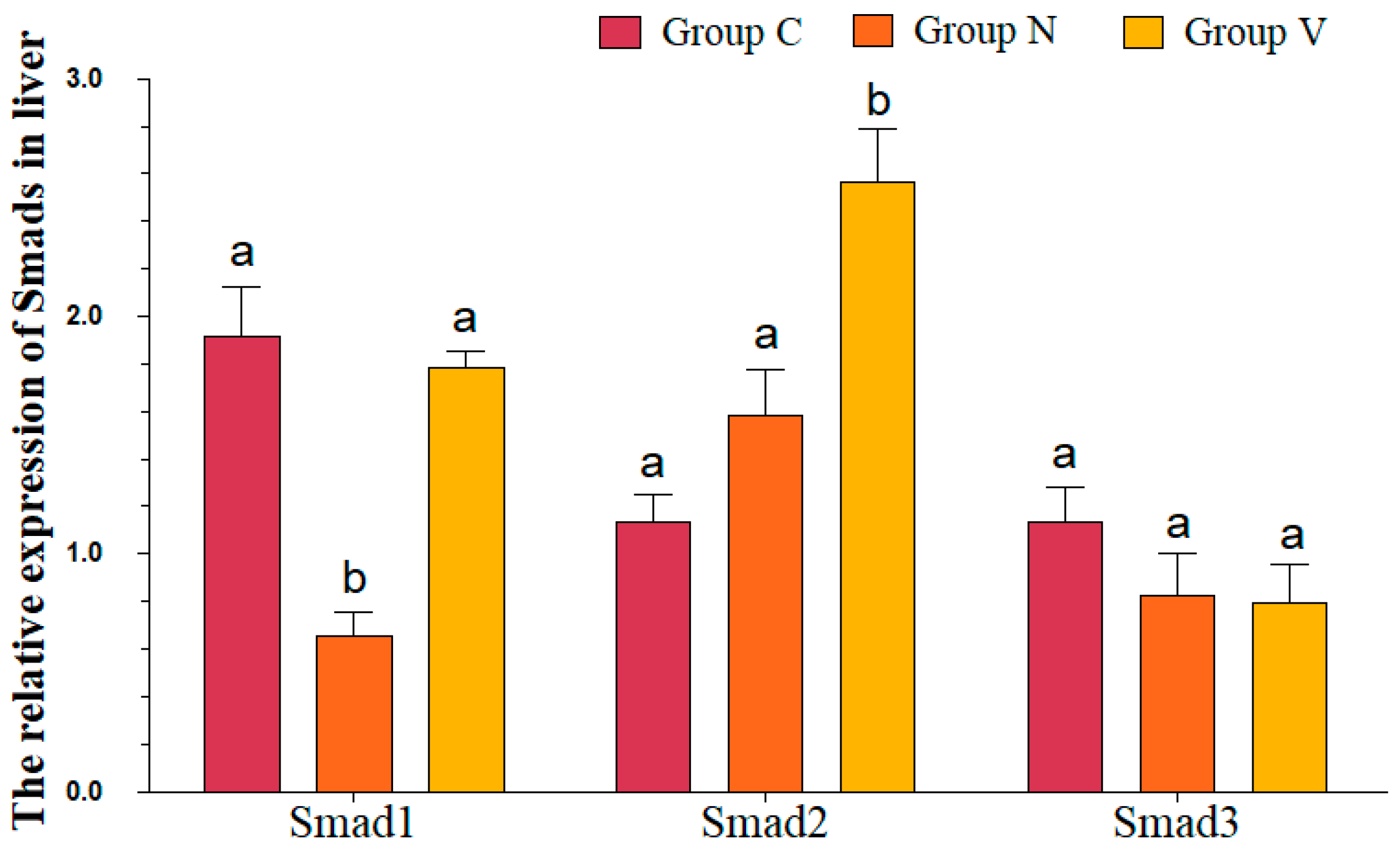
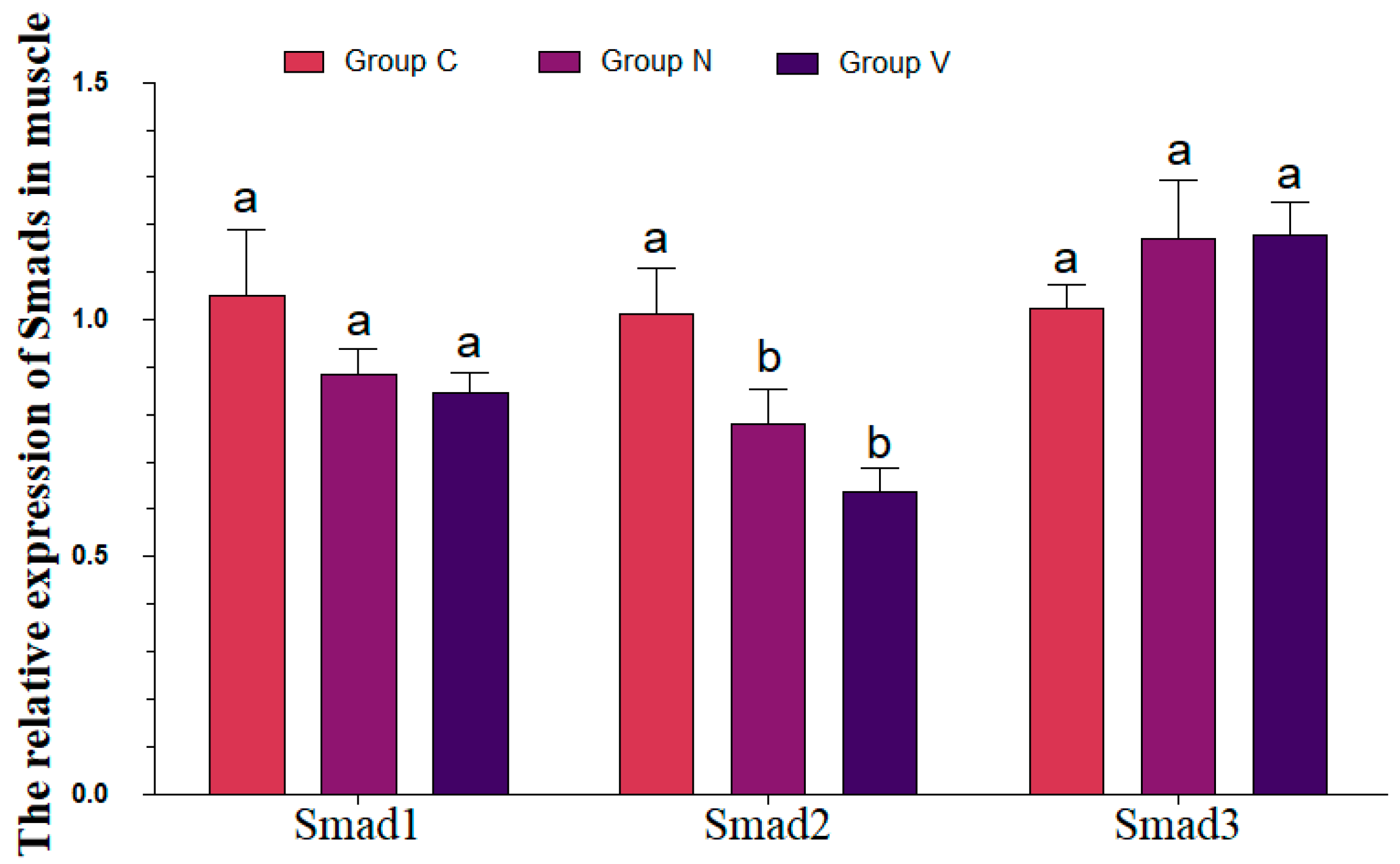
3.7. mRNA Levels of R-Smads
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Costello, C.; Cao, L.; Gelcich, S.; Cisneros-Mata, M.Á.; Free, C.M.; Froehlich, H.E.; Golden, C.D.; Ishimura, G.; Maier, J.; Macadam-Somer, I.; et al. The future of food from the sea. Nature 2020, 588, 95–100. [Google Scholar] [CrossRef]
- Asche, F.; Yang, B.; Gephart, J.A.; Smith, M.D.; Anderson, J.L.; Camp, E.V.; Garlock, T.M.; Love, D.C.; Oglend, A.; Straume, H.M. China’s seafood imports—Not for domestic consumption? Science 2022, 375, 386–388. [Google Scholar] [CrossRef]
- Crona, B.; Wassenius, E.; Troell, M.; Barclay, K.; Mallory, T.; Fabinyi, M.; Zhang, W.; Lam, V.W.Y.; Cao, L.; Henriksson, P.J.G.; et al. China at a Crossroads: An Analysis of China’s Changing Seafood Production and Consumption. One Earth 2020, 3, 32–44. [Google Scholar] [CrossRef]
- Wei, Y.; Wei, Q.; An, D. Intelligent monitoring and control technologies of open sea cage culture: A review. Comput. Electron. Agric. 2020, 169, 105119. [Google Scholar] [CrossRef]
- Xu, H.; Wu, T.; Budhathoki, M.; Fang, D.S.; Zhang, W.; Wang, X. Consumption Patterns and Willingness to Pay for Sustainable Aquatic Food in China. Foods 2024, 13, 2435. [Google Scholar] [CrossRef] [PubMed]
- Bureau of Fisheries and Fishery Administration, Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center, China Society of Fisheries. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2025. [Google Scholar]
- Stentiford, G.D.; Bateman, I.J.; Hinchliffe, S.J.; Bass, D.; Hartnell, R.; Santos, E.M.; Devlin, M.J.; Feist, S.W.; Taylor, N.G.H.; Verner-Jeffreys, D.W.; et al. Sustainable aquaculture through the One Health lens. Nat. Food 2020, 1, 468–474. [Google Scholar] [CrossRef]
- Reid, D.T.; Peichel, C.L. Perspectives on the Genetic Architecture of Divergence in Body Shape in Sticklebacks. Integr. Comp. Biol. 2010, 50, 1057–1066. [Google Scholar] [CrossRef]
- Kong, S.; Zhou, Z.; Zhou, T.; Zhao, J.; Chen, L.; Lin, H.; Pu, F.; Ke, Q.; Bai, H.; Xu, P. Genome-Wide Association Study of Body Shape-Related Traits in Large Yellow Croaker (Larimichthys crocea). Mar. Biotechnol. 2020, 22, 631–643. [Google Scholar] [CrossRef]
- Ye, H.; Liu, Y.; Liu, X.; Wang, X.; Wang, Z. Genetic Mapping and QTL Analysis of Growth Traits in the Large Yellow Croaker Larimichthys crocea. Mar. Biotechnol. 2014, 16, 729–738. [Google Scholar] [CrossRef]
- DeLorenzo, L.; Mathews, D.; Brandon, A.A.; Joglekar, M.; Baez, A.C.; Moore, E.C.; Ciccotto, P.J.; Roberts, N.B.; Roberts, R.B.; Powder, K.E. Genetic basis of ecologically relevant body shape variation among four genera of cichlid fishes. Mol. Ecol. 2023, 32, 3975–3988. [Google Scholar] [CrossRef]
- Zhou, Z.; Han, K.; Wu, Y.; Bai, H.; Ke, Q.; Pu, F.; Wang, Y.; Xu, P. Genome-Wide Association Study of Growth and Body-Shape-Related Traits in Large Yellow Croaker (Larimichthys crocea). Using ddRAD Sequencing. Mar. Biotechnol. 2019, 21, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Rehman, M.U.; Yatoo, A.M.; Arafah, A.; Khan, A.; Rashid, S.; Majid, S.; Ali, A.; Ali, M.N. TGF-β signaling pathway: Therapeutic targeting and potential for anti-cancer immunity. Eur. J. Pharmacol. 2023, 947, 175678. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C.P.; Benham-Pyle, B.W.; Lange, J.J.; Wood, C.J.; Alvarado, A.S. Wnt and TGFβ coordinate growth and patterning to regulate size-dependent behaviour. Nature 2019, 572, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Shen, B.; Loor, J.J.; Jiang, Q.; Yuan, Y.; Kong, Y.; Tan, P.; Zeng, F.; Zhao, C.; Zhu, X.; et al. Strontium Regulates the Proliferation and Differentiation of Isolated Primary Bovine Chondrocytes via the TGFβ/SMAD Pathway. Front. Pharmacol. 2022, 13, 925302. [Google Scholar] [CrossRef]
- Goebel, E.J.; Corpina, R.A.; Hinck, C.S.; Czepnik, M.; Castonguay, R.; Grenha, R.; Boisvert, A.; Miklossy, G.; Fullerton, P.T.; Matzuk, M.M.; et al. Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity. Proc. Natl. Acad. Sci. USA 2019, 116, 15505–15513. [Google Scholar] [CrossRef]
- Richardson, L.; Wilcockson, S.G.; Guglielmi, L.; Hill, C.S. Context-dependent TGFβ family signalling in cell fate regulation. Nat. Rev. Mol. Cell Biol. 2023, 24, 876–894. [Google Scholar] [CrossRef]
- Yadin, D.; Knaus, P.; Mueller, T.D. Structural insights into BMP receptors: Specificity, activation and inhibition. Cytokine Growth Factor Rev. 2016, 27, 13–34. [Google Scholar] [CrossRef]
- David, C.J.; Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 2018, 19, 419–435. [Google Scholar] [CrossRef]
- Massagué, J. How cells read TGF-β signals. Nat. Rev. Mol. Cell Biol. 2000, 1, 169–178. [Google Scholar] [CrossRef]
- Chen, W.; Ten Dijke, P. Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immunol. 2016, 16, 723–740. [Google Scholar] [CrossRef]
- Tzavlaki, K.; Moustakas, A. TGF-β Signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef]
- Ao, J.; Li, J.; You, X.; Mu, Y.; Ding, Y.; Mao, K.; Bian, C.; Mu, P.; Shi, Q.; Chen, X. Construction of the High-Density Genetic Linkage Map and Chromosome Map of Large Yellow Croaker (Larimichthys crocea). Int. J. Mol. Sci. 2015, 16, 26237–26248. [Google Scholar] [CrossRef]
- Xiao, S.; Li, J.; Ma, F.; Fang, L.; Xu, S.; Chen, W.; Wang, Z.Y. Rapid construction of genome map for large yellow croaker (Larimichthys crocea) by the whole-genome mapping in BioNano Genomics Irys system. BMC Genom. 2015, 16, 670. [Google Scholar] [CrossRef]
- Xiao, S.; Wang, P.; Zhang, Y.; Fang, L.; Liu, Y.; Li, J.-T.; Wang, Z.-Y. Gene map of large yellow croaker (Larimichthys crocea) provides insights into teleost genome evolution and conserved regions associated with growth. Sci. Rep. 2015, 5, 18661. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Dong, L.; Xiao, S.; Han, Z.; Wang, X.; Wang, Z. Genomewide association study for economic traits in the large yellow croaker with different numbers of extreme phenotypes. J. Genet. 2018, 97, 887–895. [Google Scholar] [CrossRef]
- Langerhans, R.B.; Reznick, D.N. Ecology and evolution of swimming performance in fishes: Predicting evolution with biomechanics. In Fish Locomotion: An Eco-Ethological Perspective, 1st ed.; CRC Press: Boca Raton, FL, USA, 2010; Volume 200, p. 248. [Google Scholar]
- Zhang, W. Guoxin-1 Vessel: A Mobile Fish Farm on the Sea. World 2022, 50, 38–41. [Google Scholar]
- de Caestecker, M. The transforming growth factor-β superfamily of receptors. Cytokine Growth Factor Rev. 2004, 15, 1–11. [Google Scholar] [CrossRef]
- Derynck, R.; Budi, E.H. Specificity, versatility, and control of TGF-β family signaling. Sci. Signal. 2019, 12, eaav5183. [Google Scholar] [CrossRef]
- Massague, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef]
- Pennison, M.; Pasche, B. Targeting transforming growth factor-β signaling. Curr. Opin. Oncol. 2007, 19, 579–585. [Google Scholar] [CrossRef]
- Yao, L.; Zhao, B.; Wang, Q.; Jiang, X.; Han, S.; Hu, W.; Li, C. Contribution of the TGFβ signaling pathway to pigmentation in sea cucumber (Apostichopus japonicus). Front. Mar. Sci. 2023, 10, 1101725. [Google Scholar] [CrossRef]
- Yao, L.; Liu, Y.; Li, L.; Jiang, X. Contribution of the TGF β signaling pathway to growth of Pacific oyster (Crassostrea gigas). Aquaculture 2024, 590, 740983. [Google Scholar] [CrossRef]
- Bazioti, V.; Triantafyllidou, V.; Chen, P.Y.; Simons, M.; Santovito, D.; Atzler, D.; Lutgens, E. Myeloid Tgfbr1/2-deficiency leads to a pro-atherogenic environment in ApoE-/- mice. Eur. Heart J. 2024, 45, ehae666-3889. [Google Scholar] [CrossRef]
- Ge, R.; Huang, G.M. Targeting transforming growth factor beta signaling in metastatic osteosarcoma. J. Bone Oncol. 2023, 43, 100513. [Google Scholar] [CrossRef]
- Hinck, A.P.; Mueller, T.D.; Springer, T.A. Structural Biology and Evolution of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2016, 8, a022103. [Google Scholar] [CrossRef]
- Li, S.; Yan, L.; Li, C.; Lou, L.; Cui, F.; Yang, X.; He, F.; Jiang, Y. NPC1 controls TGFBR1 stability in a cholesterol transport-independent manner and promotes hepatocellular carcinoma progression. Nat. Commun. 2025, 16, 439. [Google Scholar] [CrossRef]
- White, S.E.; Schwartze, T.A.; Mukundan, A.; Schoenherr, C.; Singh, S.P.; van Dinther, M.; Cunningham, K.T.; White, M.P.J.; Campion, T.; Pritchard, J.; et al. TGM6 is a helminth secretory product that mimics TGF-β binding to TGFBR2 to antagonize signaling in fibroblasts. Nat. Commun. 2025, 16, 1847. [Google Scholar] [CrossRef]
- Lee, J.; Yon, D.K.; Choi, Y.S.; Lee, J.; Yeo, J.H.; Kim, S.S.; Lee, J.M.; Yeo, S.G. Roles of SMAD and SMAD-Associated Signaling Pathways in Nerve Regeneration Following Peripheral Nerve Injury: A Narrative Literature Review. Curr. Issues Mol. Biol. 2024, 46, 7769–7781. [Google Scholar] [CrossRef]
- Wu, D.; Kong, X.; Zhang, W.; Di, W. Reconstruction of the TGF-β signaling pathway of Fasciola gigantica. Parasitol. Res. 2023, 123, 51. [Google Scholar] [CrossRef]
- Bisgrove, B.W.; Su, Y.C.; Yost, H.J. Maternal Gdf3 is an obligatory cofactor in Nodal signaling for embryonic axis formation in zebrafish. eLife 2017, 6, e28534. [Google Scholar] [CrossRef]
- Rodriguez, K.F.; Brown, P.R.; Amato, C.M.; Nicol, B.; Liu, C.F.; Xu, X.; Yao, H.H. Somatic cell fate maintenance in mouse fetal testes via autocrine/paracrine action of AMH and activin B. Nat. Commun. 2022, 13, 4130. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Grinberg, A.V.; Li, H.; Kuo, T.H.; Sako, D.; Krishnan, L.; Liharska, K.; Li, J.; Grenha, R.; Maguire, M.C.; et al. Functionally diverse heteromeric traps for ligands of the transforming growth factor-β superfamily. Sci. Rep. 2021, 11, 18341. [Google Scholar] [CrossRef] [PubMed]
- Kriseman, M.L.; Tang, S.; Liao, Z.; Jiang, P.; Parks, S.E.; Cope, D.I.; Yuan, F.; Chen, F.; Masand, R.P.; Castro, P.D.; et al. SMAD2/3 signaling in the uterine epithelium controls endometrial cell homeostasis and regeneration. Commun. Biol. 2023, 6, 261. [Google Scholar] [CrossRef]
- Heldin, C.-H.; Moustakas, A. Signaling receptors for TGF-β family members. Cold Spring Harb. Perspect. Biol. 2016, 8, a022053. [Google Scholar] [CrossRef]




| Gene Name/Another Name | Accession No. | Primer Sequence |
|---|---|---|
| β-actin/ACTB | FJ936563.1 | 5′-GACCTGACAGACTACCTCATG-3′ |
| 5′-AGTTGAAGGTGGTCTCGTGGA-3′ | ||
| TGFB1/TGFβ1 | XM_027280465.1 | 5′-GGTGGTTTTCACAGCAGCAG-3′ |
| 5′-GGGATGCTCGGGGAATAGTG-3′ | ||
| TGFB2/TGFβ2 | XM_010752699.3 | 5′-GCTGCCTACTGCTCCAGAAA-3′ |
| 5′-TTGGCCTCATAGCCCTTTGG-3′ | ||
| TGFB2/TGFβ3 | XM_010733855.3 | 5′-AAGACGACCCCAAAGCCAAA-3′ |
| 5′-ACTGATCTCCAGGCCCAGAT-3′ | ||
| INHBA/Activin A | XM_010747963.3 | 5′-TCATCAGGAGGACAGAGCGA-3′ |
| 5′-CGGGACTGGCTGTGTTACAT-3′ | ||
| INHBB/Activin BNodal | XM_010746279.3 | 5′-GAGGATGGGAGGGTGGAGAT-3′ |
| 5′-CAGATTCTGGTTGCCCTCGT-3′ | ||
| Nodal | XM_010743837.3 | 5′-CTTTTGCGCTGCACACATCT-3′ |
| 5′-CATGTAGGTTGGCAGGTGGT-3′ | ||
| BMP2 | XM_010733238.3 | 5′-CATGGCTTCATGGTGGAGGT-3′ |
| 5′-TCACCCTGACGTGCCTACTA-3′ | ||
| BMP3 | XM_027273955.1 | 5′-TGCTCGTGCTGCTTTATGGA-3′ |
| 5′-TGGTCGTCTTTCGTGAGGTG-3′ | ||
| BMP4 | XM_010735218.3 | 5′-CAGCTCTTGGATACGCGACT-3′ |
| 5′-GTGTCTGGTTGAGGTGCAGA-3′ | ||
| BMP5 | XM_010756380.3 | 5′-CACCTTTGAACCGTGCTGTG-3′ |
| 5′-GATCTCCCTGCGTTCGTGAT-3′ | ||
| BMP6 | XM_010741625.3 | 5′-GAAACTCCAGGCCAGACACA-3′ |
| 5′-TCCACCACAATCCGACCAAG-3′ | ||
| BMP7 | XM_010755531.3 | 5′-CACGGCAGCAGAGTTTAGGA-3′ |
| 5′-ACCTGGTAGACGCTGACTCT-3′ | ||
| BMP8A | XM_019270763.2 | 5′-TTGAGGACCGCAGGAGAAAC-3′ |
| 5′-TTCATCCATTCACGCCTGCT-3′ | ||
| BMP10 | XM_010738504.3 | 5′-GGGACAGTTGGATTCTCGCA-3′ |
| 5′-GTTTGGTGGGTGTGACATGC-3′ | ||
| GDF3 | XM_019263909.2 | 5′-CTCTCTGGACCCTCATGGGA-3′ |
| 5′-CGGGCCAATCAGAACCTCAT-3′ | ||
| GDF5/BMP14 | XM_010754689.3 | 5′-CACTTCAACGTCAGCTCCCT-3′ |
| 5′-GGGATGAACCTCCTCCTCCT-3′ | ||
| GDF6/BMP13 | XM_010747111.3 | 5′-CCTGGATTACGAGGCGTACC-3′ |
| 5′-CTGATGGGGCTGAGTTTGGT-3′ | ||
| GDF8 | NM_001303317.1 | 5′-AGAGTCCGCTCCCTGAAGAT-3′ |
| 5′-CTCTGCTGAAGTGACAGCCA-3′ | ||
| GDF9 | XM_010754774.3 | 5′-ACAAGCAGATGGCGTTCAGA-3′ |
| 5′-AATCGTACAAGGCGCAGTCA-3′ | ||
| GDF15 | XM_010733382.3 | 5′-GTGGACAGAATGGACAGCCA-3′ |
| 5′-TGGGCTCCCTGTCTATACCC-3′ | ||
| AMH/MIS | XM_010730161.3 | 5′-GGACTCTGCACGGTTTCTGA-3′ |
| 5′-GCAGACTGTGGGAGGTCAAA-3′ | ||
| ALK2/ACVR1 | XM_019259847.2 | 5′-CACTGTGGAGCTGCCTGTTA-3′ |
| 5′-CTCGTGACGTAAGCCTCTCC-3′ | ||
| ALK3/BMPR1A | XM_010749871.3 | 5′-GAGAGAAGGTGGCCGTCAAA-3′ |
| 5′-TGAGGAAGAGCTGCGTGAAG-3′ | ||
| ALK4/ACVR1B | XM_010747783.3 | 5′-GGCGTGTTCCTGTTCCAGTA-3′ |
| 5′-ACAAACAGGGGCAAACCAGA-3′ | ||
| ALK6/BMPR1B | XM_010732488.3 | 5′-GACATCCCTCCCAACACGAG-3′ |
| 5′-CCCTCCTGAGACACAACGTC-3′ | ||
| ALK7/ACVR1C | XM_027290679.1 | 5′-CCTGCCATTGCTCACAGAGA-3′ |
| 5′-TGTCGATGGTGTTGGTCCTG-3′ | ||
| TBRII/TGFBR2 | XM_010735343.3 | 5′-ATGAAGGTGAGACGGCTGTG-3′ |
| 5′-GGCAGATGGAGGTGATCTCG-3′ | ||
| ACVR2A/ACTR2A | XM_027287970.1 | 5′-TGAAGGCAAACGTCCTCTCC-3′ |
| 5′-GGTCAGGTTCGACTTCAGCA-3′ | ||
| ACVR2B/ACTR2B | XM_019276244.2 | 5′-CTGGCTGGACGACTTCAACT-3′ |
| 5′-TGGAGGCCGGATTTTCACTC-3′ | ||
| BMPR2 | XM_010740233.3 | 5′-ACCACCCTGCACAATGAGAG-3′ |
| 5′-CGTGTTCTTAGCCACACCCT-3′ | ||
| AMHR2/MISR2 | XM_027279584.1 | 5′-GTGGCTGATTTTGGCTGTGG-3′ |
| 5′-GTTCTGGCGAGTCACTTGGA-3′ | ||
| Smad1/MADR1 | XM_010729469.3 | 5′-AATCGTGTTGGAGAGGCGTT-3′ |
| 5′-CGGGTGTTCTCAATGGTGGA-3′ | ||
| Smad2/MADR2 | XM_019271768.2 | 5′-TACATCGGAGGGGAGGTGTT-3′ |
| 5′-GTGCAGCAAACTCCTGGTTG-3′ | ||
| Smad3/MADH3 | XM_010735124.3 | 5′-CCACTACCAGAGGGTGGAGA-3′ |
| 5′-AGGGATGGATGGGCTGTAGT-3′ |
| Indexes | Initial Data (in November 2023) for Group A | Finish Data for Group N | Finish Data for Group V |
|---|---|---|---|
| Weight (Wt, g) | 350.4 ± 34.24 c | 508 ± 29.02 b | 535.33 ± 22.24 a |
| Total Length (TL, cm) | 32.41 ± 1.16 c | 35.31 ± 1.22 b | 37.28 ± 0.92 a |
| Body Length (BL, cm) | 28.29 ± 1.15 c | 31.35 ± 0.93 b | 33.78 ± 1.24 a |
| Body Depth (BD, cm) | 7.9 ± 0.49 b | 9.04 ± 0.35 a | 9.06 ± 0.44 a |
| Caudal Peduncle Length (CPL, cm) | 7.49 ± 0.55 c | 8.07 ± 0.69 b | 9.34 ± 0.42 a |
| Caudal Peduncle Width (CPW, cm) | 2.03 ± 0.13 c | 2.19 ± 0.15 b | 2.3 ± 0.08 a |
| Caudal Peduncle Length/Caudal Peduncle Width (CPL/CPW) | 3.7 ± 0.28 b | 3.7 ± 0.32 b | 4.07 ± 0.24 a |
| Body Length/Body Depth (BL/BD) | 3.59 ± 0.21 b | 3.47 ± 0.18 b | 3.73 ± 0.17 a |
| Caudal Peduncle Length/Body Length (CPL/BL) | 0.26 ± 0.01 b | 0.26 ± 0.02 b | 0.28 ± 0.01 a |
| Fullness (g/cm3) | 1.55 ± 0.15 a | 1.66 ± 0.17 a | 1.4 ± 0.15 b |
| Subfamily | Ligand | Receptor I | Receptor II | R-Smad |
|---|---|---|---|---|
| TGFβ/activin/Nodal | INHBA/Activin A | ALK4/ACVR1B | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad/MADR2, 3 |
| INHBB/Activin B | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad/MADR2, 3 | |
| Nodal | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad/MADR2, 3 | |
| BMP/GDF/AMH | BMP2 | ALK3/BMPR1A/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 |
| BMP4 | ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| BMP5 | ALK2/ACVR1, ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| BMP6 | ALK2/ACVR1, ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| BMP7 | ALK2/ACVR1, ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| BMP8A | ALK2/ACVR1, ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| GDF3 | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad/MADR2, 3 | |
| GDF5/BMP14 | ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| GDF6/BMP13 | ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| GDF8 | ALK4/ACVR1B | ACVR2B/ACTRIIB | Smad/MADR2, 3 | |
| GDF9 | ALK4/ACVR1B | BMPR2 | Smad1/MADR1 | |
| AMH/MIS | ALK2/ACVR1 and ALK3/BMPR1A | AMHR2/MISR2 | Smad1/MADR1 |
| Subfamily | Ligand | Receptor I | Receptor II | R-Smad |
|---|---|---|---|---|
| TGFβ/activin/Nodal | INHBA/Activin A | ALK4/ACVR1B | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad2/MADR2 |
| INHBB/ActivinB | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad2/MADR2 | |
| Nodal | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad2/MADR2 | |
| BMP/GDF/MIS | BMP2 | ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 |
| BMP4 | ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| BMP6 | ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| BMP8A | ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| GDF3 | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad/MADR2, 3 | |
| GDF5/BMP14 | ALK3/BMPR1A and ALK6/BMPR1B | ACVR2A/ACTRIIA, ACVR2B/ACTRIIB and BMPR2 | Smad1/MADR1 | |
| GDF8 | ALK4/ACVR1B | ACTRIIB and BMPR2 | Smad2/MADR2 | |
| GDF9 | ALK4/ACVR1B | BMPR2 | Smad1/MADR1 |
| Subfamily | Ligand | Receptor I | Receptor II | R-Smad |
|---|---|---|---|---|
| TGFβ/activin/Nodal | INHBA/Activin A | ALK4/ACVR1B | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad2/MADR2 |
| INHBB/ActivinB | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad2/MADR2 | |
| Nodal | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad2/MADR2 | |
| BMP/GDF/MIS | GDF3 | ALK4/ACVR1B and ALK7/ACVR1C | ACVR2A/ACTRIIA and ACVR2B/ACTRIIB | Smad2/MADR2 |
| GDF8 | ALK4/ACVR1B | ACVR2B/ACTRIIB | Smad2/MADR2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Zhang, L.; Hu, X.; Cui, M.; Li, J.; Liu, H.; Yao, L. Characterization of TGFβ Signaling Components in Large Yellow Croaker (Larimichthys crocea) and Their Role in Growth and Body Shape Regulation. J. Mar. Sci. Eng. 2025, 13, 1716. https://doi.org/10.3390/jmse13091716
Jiang X, Zhang L, Hu X, Cui M, Li J, Liu H, Yao L. Characterization of TGFβ Signaling Components in Large Yellow Croaker (Larimichthys crocea) and Their Role in Growth and Body Shape Regulation. Journal of Marine Science and Engineering. 2025; 13(9):1716. https://doi.org/10.3390/jmse13091716
Chicago/Turabian StyleJiang, Xuyang, Lu Zhang, Xin Hu, Mingchao Cui, Jie Li, Huang Liu, and Linlin Yao. 2025. "Characterization of TGFβ Signaling Components in Large Yellow Croaker (Larimichthys crocea) and Their Role in Growth and Body Shape Regulation" Journal of Marine Science and Engineering 13, no. 9: 1716. https://doi.org/10.3390/jmse13091716
APA StyleJiang, X., Zhang, L., Hu, X., Cui, M., Li, J., Liu, H., & Yao, L. (2025). Characterization of TGFβ Signaling Components in Large Yellow Croaker (Larimichthys crocea) and Their Role in Growth and Body Shape Regulation. Journal of Marine Science and Engineering, 13(9), 1716. https://doi.org/10.3390/jmse13091716






