Abstract
Carotenoids are extensively used in drugs, cosmetics, nutrients, and foods, owing to their antioxidant and anti-inflammatory characteristics. Diverse marine heterotrophic prokaryotes can accumulate carotenoids and become promising alternatives for the advancement of carotenoids production. In this research, 55 strains were isolated and cultivated from tidal flat sediment in Zhoushan and classified into the phyla Pseudomonadota (n = 24), Bacillota (n = 18), Bacteroidota (n = 9), and Actinomycetota (n = 4). Nine of them accumulated carotenoids, and most of them belonged to the families Flavobacteriaceae (n = 4) and Erythrobacteraceae (n = 4). Among those carotenoid-producing strains, one strain, designated as D39T, was proposed as one novel species belonging to the genus Croceibacterium through polyphasic taxonomy approaches. Genomic annotations and carotenoid compound determinations revealed that strain D39T encoded crtEBIYZG genes and mainly accumulated zeaxanthin as major carotenoids. Furthermore, carotenoid biosynthesis pathway in the majority of Croceibacterium strains were identical with that in the strain D39T, implying that Croceibacterium members can be sources of producing zeaxanthin. This study enhances knowledge of microbial biodiversity in tidal flats, proposes a novel carotenoid-producing Croceibacterium species, and elucidates carotenoid biosynthesis pathway in the genus Croceibacterium, which contribute to enriching marine carotenoid-producing strains and promoting a comprehensive insight into genomic contents of them.
Keywords:
tidal flat; marine bacteria; Croceibacterium; polyphasic taxonomy; carotenoids; zeaxanthin 1. Introduction
Carotenoids are lipophilic bioactive triterpene pigments and are mainly divided into two categories based on their chemical structure: xanthophylls, which are oxidized carotenoids with oxygen as a functional group, and carotenes, which are hydrocarbon carotenoids composed of carbon and hydrogen atoms [1]. Carotenoids exhibit a wide distribution across plants, algae, fungi, and prokaryotes, showcasing various colors including red, orange, yellow, purple, and brown [2]. Currently, they are extensively used in drugs, cosmetics, nutrients, and foods, especially for treating ophthalmic diseases [3]. Compared to carotenoids extracted from plants, algae, and fungi or chemically synthesized, prokaryote-derived carotenoids are characterized by high efficiency and low environmental impact [4]. Consequently, there is a burgeoning interest in the exploration of novel sources of prokaryotic carotenoids and the advancement of their applications.
Apart from those phototrophic prokaryotes, diverse marine heterotrophic prokaryotes belonging to the phyla Bacteroidota (known as Bacteroidetes previously) and Pseudomonadota (known as Proteobacteria previously) can also accumulate carotenoids for defending against photooxidative damage and regulating membrane fluidity [5]. The biosynthesis of carotenoids is regarded as an adaptive response to certain marine environmental adversities, such as excess solar radiation and reactive oxygen species [6,7]. Recently, several carotenoids that are of significant commercial value—including astaxanthin, used as a dietary supplement for human, animal, and aquaculture consumption; β-cryptoxanthin, which prevents free radical damage to cells and DNA; and zeaxanthin, which relieves age-related macular degeneration—have been found to be biosynthesized by marine Bacteroidota and Pseudomonadota members [8,9,10,11,12]. Consequently, marine heterotrophic prokaryotes could be favorable alternatives for developing novel and sustainable carotenoid sources.
The family Erythrobacteraceae, belonging to the phylum Pseudomonadota, is a representative carotenoid-producing group [5,13]. Currently, the family Erythrobacteraceae accommodates 20 genera (https://lpsn.dsmz.de/family/erythrobacteraceae, accessed on 27 December 2023) [14]. Ecologically, the family Erythrobacteraceae strains are commonly isolated from multiple marine environments, including seawater [15,16], sediments [17,18], as well as marine fauna and flora [19,20,21]. The genus Croceibacterium, belonging to the family Erythrobacteraceae, was proposed by Liu et al. [22], describing Croceibacterium ferulae as the type species and reclassifying Porphyrobacter mercurialis as C. mercuriale. Subsequently, four Altererythrobacter species, including Altererythrobacter atlanticus Wu et al., 2014, and Altererythrobacter salegens Liang et al., 2017, were reclassified into the genus Croceibacterium based on the core-genomic phylogeny [23]. At the time of writing, this genus includes seven validly published species according to the List of Prokaryotic names with Standing in Nomenclature (https://lpsn.dsmz.de/genus/croceibacterium, accessed on 27 December 2023). In this study, 28 pigmented strains were isolated and cultivated during a prokaryotic investigation inhabiting the tidal flat of Zhoushan Archipelago, which is the largest archipelago of People’s Republic of China. Among these pigmented strains, one carotenoid-producing and yellow-pigmented colony, designated as D39T, was specifically characterized by polyphasic taxonomic approaches and carotenoid profile determinations to clarify its taxonomic status and carotenoid biosynthesis pathway. The aim of this study is to enrich marine carotenoid-producing prokaryotic strains and promote a comprehensive insight into genomic contents of marine carotenoid-producing prokaryotes.
2. Materials and Methods
2.1. Isolation, Cultivation, Preservation, and Identification of Carotenoid-Producing Strains
One tidal flat sediment sample was collected from Zhoushan Archipelago (29.92° N, 122.41° E) and stored at a temperature of 4 °C for short-term preservation. The sediment subsamples were repeatedly suspended with NaCl solution (3.0%, w/v), diluted by ten-fold dilution, and then coated on marine 2216 agar (MA; Difco, Sparks, NV, USA). After one month of aerobic cultivation at room temperature, the colonies were picked and incubated in marine 2216 broth (MB; Difco, Sparks, NV, USA) for an additional period of three days. Following a purity test, the mixture was cryopreserved with a solution add with sterilized glycerol of a same volume at −80 °C.
Taxonomic status of each isolated strain was distinguished using 16S ribosomal RNA gene sequence identity analysis. In brief, extracted genomic DNAs of each strain were used as templates for amplifying 16S ribosomal RNA genes by using 27F (5′-GAGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) prime pair. Then the 16S ribosomal RNA gene sequences were sequenced at Tsingke Biotechnology Co., Ltd. (Hangzhou, China). The EzBioCloud webserver [24] was applied to determine the sequence identity of all 16S ribosomal RNA sequences, thereby elucidating their taxonomic classification.
Furthermore, all pigmented strains were tested for the presence of carotenoids using spectrophotometry. Cells of MB medium culture solution was centrifuged, washed with NaCl solution (2.0%, w/v) once, and then resuspended in the acetone/methanol (7:2, v/v) solution with violent shaking of 10 min. After centrifugation, the acetone/methanol extracts were collected and analyzed for an absorbance spectrum in the visible range of 300–600 nm by using an ultraviolet spectrophotometer (UV-2700i, Shimadzu Instruments Manufacturing Co., Ltd., Suzhou, China) [25]. Additionally, the same acetone/methanol solution was used as a blank control.
2.2. Phylogenetic Relationship and Genomic Characteristics of Carotenoid-Producing Strain D39T
In the process of classifying and identifying these colonies, one colony, designated as D39T, was discovered, which belonged to the Erythrobacteriaceae family and exhibited yellow pigment production along with carotenoid synthesis. However, the specific taxonomic status remained unknown. To enhance our understanding of carotenoids produced by prokaryotic bacteria in tidal flats, we conducted an analysis on its classification, identification, and composition of carotenoid compounds.
The lyophilized sample of strain D39T was subsequently submitted to Guangdong Magigene Biotechnology Co., Ltd. (Guangzhou, China) for sequencing its genome on Illumina HiSeq 6000 platform (San Diego, CA, USA). After filtering low-quality reads, 6,000,410 high-quality clean reads with total size of 1,785,766,533 bp were generated.
SPAdes version 3.14.1 [26] and CheckM v.1.0.7 [27] were used to assemble the genome draft and estimate its assembled complete genome quality, respectively. Subsequently, Rapid Annotation using Subsystem Technology web server version 2.0 [28] was carried out to annotate open reading frames (ORFs) and RNA genes. Functional annotations were performed using eggNOG-mapper v2.0.5 [29]. In addition, the genomes of seven Croceibacterium type strains and other Erythrobacteraceae relatives were retrieved from NCBI GenBank database, with comprehensive details enumerated in Table S1.
The complete 16S ribosomal RNA gene sequences of strain D39T were retrieved as described previously [30] and aligned with 16S ribosomal RNA gene sequences of other related type strains by using Clustal_W [31]. Phylogenetic trees based on three methods— including neighbor-joining [32], maximum likelihood [33], and maximun parsimony [34]—were reconstructed by MEGA X software v.10.2 [35]. The Kimura two-parameter model and 1000 repetitions were used as the nucleotide replacement model and bootstrap values.
The genome relatedness indices were determined by using OrthoANI Tool v.0.93.1 and Genome-to-Genome Distance Calculator 2.1 webserver [36,37]. Orthologous clusters (OCs) were searched by the Proteinortho v.5.16b [38] based on the amino acid sequences of ORFs, with command ‘-e 1e-5 -cov=50-identity=50’. Shared single-copy orthologous cluster amino acid sequences were aligned by MAFFT v.7.490 [39] and refined by trimAL v.2rev59 [40]. Then one phylogenomic tree was reconstructed employing IQ-TREE v.1.6.12 [41] with ultrafast bootstraps set to 1000.
2.3. Phenotypic Determinations of Strain D39T
In this study, reference strains Croceibacterium salegens MCCC 1K01500T and C. soli MCCC 1K02066T were employed for parallel comparisons. The NaCl concentration range for growth (0, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 4.0, 5.0, 6.0, 8.0, 10.0, 12.0%, w/v) was determined in sodium-free medium at 37 °C following the formulation of MB. The temperature and pH ranges for growth were examined by incubation at 4, 10, 25, 28, 30, 37, 40, and 45 °C as well as pH 5.0 to 10.0 (0.5 pH units per interval and buffer agents added). The motility was assessed by aseptically transferring a small aliquot of the fresh bacterial solution from the later stages of growth onto a pre-prepared semi-solid medium (MA, agar concentration halved), using inoculating needles, followed by incubation for five days under optimal growth conditions. Colony morphology was observed when strain D39T was cultivated on MA at 37 °C for four days. Cellular morphology was observed via transmission electron microscopy (JEM-1400Flash HC, JEOL Ltd., Tokyo, Japan). Gram-staining was performed as described previously [42]. Catalase activity was assessed through the production of bubbles upon the addition of hydrogen peroxide solution, while oxidase activity was detected by observing the oxidation of N,N-dimethyl-1,4-phenylenediamine oxalate. Anaerobic growth assay was performed, and the hydrolyses of casein, cellulose, starch, Tweens 20, 40, 60, and 80, as well as tyrosine were carried out according to Xu et al. [43]. Sole carbon, nitrogen, and energy source utilizations were carried out in oligotrophic MB containing 2.0% (w/v) NaCl, yeast extract (0.1 g/L), peptone (0.5 g/L), and carbohydrates (2 g/L), organic acids (1 g/L), or amino acids (1 g/L) as substrates. For acid production test, sugars or alcohols (10 g/L) were supplemented in MOF medium containing 2.0% (w/v) NaCl. Other phenotypic tests were investigated using API 20NE and API ZYM strips (bioMérieux, Marcy-l’Étoile, France). All API tests were performed in accordance with the manufacturer’s instructions, with some modifications aiming to establish experimental conditions that fostered optimal growth and viability of the cells.
Cells of strain D39T, Croceibacterium salegens MCCC 1K01500T, and C. soli MCCC 1K02066T were harvested after four days of cultivation on MA at the optimal temperature, and subsequently subjected to parallel analysis using the Microbial Identification System under identical conditions [44]. The extraction of intercellular polar lipids was performed as Minnikin et al. [45]. The polar lipid types corresponding to the points on the silica gel plate (10 × 10 cm; Merck Millipore, Burlington, MA, USA) were determined based on the color development results obtained from various lipid spray agents. Lyophilized cells of strain D39T were stirred overnight to extract the isoprenoid quinone with 45 mL chloroform/methanol (2:1, v/v) solution. Extracts were evaporated to dryness at 37 °C after filtering and resuspended in 1 mL chloroform. The isoprenoid quinone was separated on GF254 silica gel plates (20 × 10 cm; Merck Millipore, Burlington, MA, USA) using n-hexane/ether (34:6, v/v). The silica gel plate was positioned beneath a 254 nm ultraviolet lamp for observation, and the corresponding section of the silica gel strip was gently scraped off. Subsequently, 1ml of acetone was added overnight to facilitate the dissolution of respiratory quinones present in the silica gel, followed by analysis utilizing HPLC-MS (Agilent, Santa Clara, CA, USA) [43].
2.4. Carotenoid Determination and Biosynthetic Gene Analysis of Strain D39T
The bacterial solution of strain D39T, with a volume of 300 mL at the end of logarithmic growth phase, was centrifuged at 12,000× g for a half hour to harvest cells. The samples were subsequently subjected to freeze-drying and pulverization into a fine powder for subsequent carotenoid extraction. The resulting powder (50 mg) was extracted using a mixture of n-hexane/acetone/ethanol (1:1:1, v/v/v) under agitation at room temperature for 20 min, followed by centrifugation at 4 °C and 12,000× g for 5 min to collect the supernatant. The residue was then extracted under identical conditions, evaporated, and dried in methylene chloride (100 μL). Finally, the solution was filtered through a membrane filter with a pore size of 0.22 μm prior to analysis. Carotenoids contents were determined by MetWare based on the AB Sciex QTRAP 6500 LC-MS/MS platform (AB Sciex, Singapore) [46,47].
As described by Siddaramappa et al. [48], the Crt proteins responsible for carotenoid biosynthesis—including CrtB (15-cis-phytoene synthase, K02291), CrtI (phytoene desaturase, K10027), CrtW (β-carotene/zeaxanthin 4-ketolase, K09836), CrtY (lycopene β-cyclase, K22502), and CrtZ (β-carotene 3-hydroxylase, K15746)—were retrieved based on the Kyoto Encyclopedia of Genes and Genomes annotations. Furthermore, these genes were confirmed by using BLAST searches against NCBI UniProtKB/Swiss-Prot protein sequence database [49]. Additionally, another two key proteins CrtE (geranylgeranyl pyrophosphate synthase) and CrtG (2,2′-β-ionone ring hydroxylase) which amino acid sequences were searched using BLAST against the reference proteins obtained from UniProt database (Table S2). The Crt protein sequences were subjected to comparative analysis and refinement using MAFFT software v7.490 and trimAL software 1.2rev59, respectively. Then, a maximum-likelihood phylogenetic tree based on concatenated Crt protein sequences was obtained using IQ-Tree version 1.6.12 as described above [41].
3. Results
3.1. Bacterial Isolation of Tidal Flat Sediment
During isolation and cultivation, 55 bacterial strains were obtained from the tidal flat sediment sample. Based on the taxonomic identification of 16S ribosomal RNA gene sequences, these strains were classified into four phyla, with dominant one as Pseudomonadota (n = 24), followed by Bacillota (n = 18), Bacteroidota (n = 9), and Actinomycetota (n = 4). Meanwhile, 28 of 55 strains showed obvious colors such as red, orange, and yellow, and nine of these pigment extracts contained absorbance peaks at 435, 451, 452, 453, 454, 456, 460, 479, 480, 482, and/or 497 nm, suggesting the existence of carotenoid compounds (Figure 1). The detailed information of 55 strains was listed in Table S3.

Figure 1.
In vitro pigment-absorption spectrum of acetone/methanol extracts from strains D19 (A), D37 (B), D38 (C), D39 (D), H6 (E), H9 (F), H20 (G), H24 (H), and Z16 (I) extracts. The colonies of each strain grown on MA were displayed in the middle-right of each subfigure.
3.2. Phylogenetic Relationship and Genomic Comparison of Strain D39T
Strain D39T had the highest 16S ribosomal RNA gene sequence identities with C. salegens XY-R17T (97.3%) and C. atlanticum 26DY36T (97.0%) as well as sequence identities of <96.7% with other Croceibacterium members, which indicated that strain D39T could represent one novel Croceibacterium species based on those gene sequence identities below the species threshold of 98.65% [50]. Phylogenetic trees constructed based on the 16S ribosomal RNA genes illustrated that strain D39T formed a distinct and stable branch exclusively with C. salgenes XY-R17T (Figure S1). However, due to the low bootstrap value and limited recovery nodes, it is challenging to establish conclusive relationships between Croceibacterium species using the 16S ribosomal RNA gene-based phylogeny. Hence, it was imperative to construct a more robust phylogenomic tree to elucidate its evolutionary association with Croceibacterium species, as performed in other Erythrobacteraceae genera recently [23,51,52,53,54,55,56,57,58,59,60,61].
Genomic size of strain D39T genome was 3,347,658 bp, and genomic DNA G + C content of that was 65.8%. Furthermore, genomic completeness of this genome was determined to be 99.2% and its contamination was estimated to be 1.0%, indicating that this genome adheres to the high-quality standard as proposed by Bowers et al. [62]. The genome of strain D39T encoded 3247 ORFs and 50 RNA genes, respectively. In addition, 2702 (83.2%), 690 (21.2%), and 1904 (58.6%) ORFs were assigned to Clusters of Orthologous Groups of proteins, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes databases, respectively. Comparative genomic analysis revealed that genomes of strain D39T and other seven Croceibacterium type strains had a pan-genome of 10,408 OCs, among which 1149 OCs were common in all Croceibacterium members. A phylogenomic tree based on the shared single-copy OCs showed that strain D39T was classified within a clade comprising the genus Croceibacterium and formed a distinct branch with C. salegens MCCC 1K01500T, thereby indicating its affiliation to the genus Croceibacterium (Figure 2).
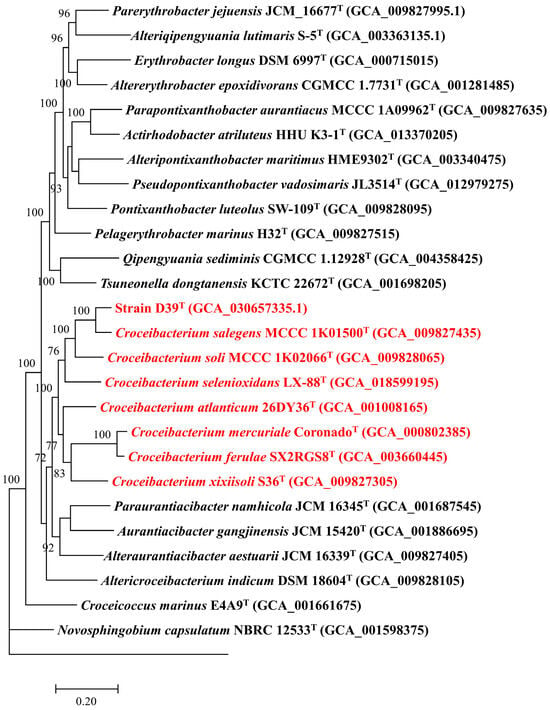
Figure 2.
Phylogenomic tree based on the partition of 133 single-copy orthologous cluster protein sequences depicting the phylogenetic relationship among strain D39T and Croceibacterium type strains, which are colored by red. Only bootstrap values higher than 70% are shown. Rhodospirillum rubrum ATCC 11170T served as an outgroup.
The average nucleotide identity and in silico DNA-DNA hybridization values of strain D39T, C. salegens MCCC 1K01500T, and C. soli MCCC 1K02066T were determined to be 83.0% and 44.2%, respectively. Moreover, strain D39T exhibited low overall genome relatedness index values of <77.2% (average nucleotide identity) and <20.5% (in silico DNA-DNA hybridization) when compared with the rest of Croceibacterium type strains. These findings show that overall genome relatedness indices between strain D39T and Croceibacterium type strains fell below the species demarcation thresholds for average nucleotide identity (95–96%) [50] and for in silico DNA-DNA hybridization (70%) [37], suggesting that strain D39T is a novel Croceibacterium species. Detailed overall genome relatedness index values for strain D39T and Croceibacterium type strains are presented in Table 1.

Table 1.
The overall genome relatedness index values among strain D39T and Croceibacterium type strains.
3.3. Phenotypic Characteristics of Strain D39T
The colony of strain D39T exhibited the formation of yellow, round, opaque cells with smooth edges after the incubaction on MA medium for four days. The rod-shaped cells of strain D39T measured 0.8–1.5 μm in length and 0.4–0.7 μm in width, exhibiting a Gram-negative phenotype and possessing flagella (Figure 3). The motility of the strain was evidenced by the presence of dendritic colonies at the site where the inoculation needle was inserted in a semi-solid medium.
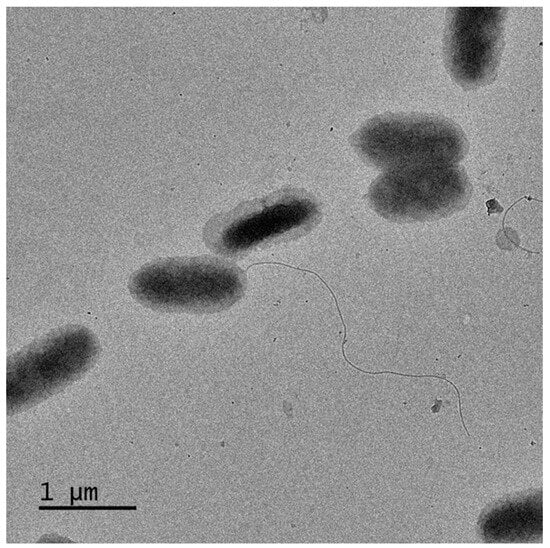
Figure 3.
Transmission electron micrographs of strain D39T cells incubated in MA medium containing 2% (w/v) NaCl at 37 °C for four days.
Compared with those two reference strains including C. salegens MCCC 1K01500T and C. soli MCCC 1K02066T, strain D39T could be differentiated from them based on optimal temperature and pH for growth, hydrolysis of Tween 60, utilizations of D-galactose and D-maltose, and leucine arylamidase activity and acid production from D-maltose and sucrose. Furthermore, strain D39T could be distinguished from its closest relative C. salegens MCCC 1K01500T by hydrolysis of Tween 20, utilizations of L-alanine and D-glucose, β-Galactosidase, β-Glucuronidase, α-Glucosidase and oxidase activities, as well as acid production from D-glucose and glycerol. Specific phenotypic characteristics were compared in Table 2.

Table 2.
Differential characteristics among strain D39T and its Croceibacterium reference strains. 1, strain D39T; 2, C. salegens MCCC 1K01500T; 3, C. soli MCCC 1K02066T. All results were determined in this study, with the exception of temperature, NaCl concentration and pH for growth and oxidase activities for reference strains, which were cited from Liang et al. [63] and Zhao et al. [64]. +, Positive; −, negative.
Strain D39T contained ten major polar lipids, including diphosphatidylglycerol (DPG), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), sphingoglycolipid (SGL), four unidentified glycolipids, and one unidentified lipid (Figure 4). Compared with reference strains, three unidentified glycolipids were absent in C. salegens MCCC 1K01500T [63] and C. soli MCCC 1K02066T [64]. Moreover, strain D39T exclusively contained ubiquinone-10 (Q-10) as its respiratory quinone, which was similar to Croceibacterium members [22,23]. Strain D39T, C. salegens MCCC 1K01500T, and C. soli MCCC 1K02066T exhibited similar fatty acid compositions, characterized by summed feature 8 (C18:1ω6c and/or C18:1ω7c) and C16:0 as major fatty acids (>10%). However, strain D39T had C18:1ω7c 11-methyl as a major fatty acid that was different from reference strains, especially for that not detected in C. salegens MCCC 1K01500T. Detailed fatty acid compositions were listed in Table S4.
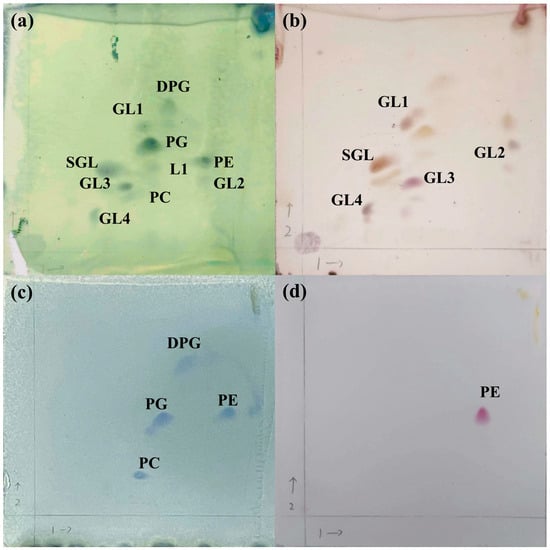
Figure 4.
Two-dimensional TLC chromatograms of the polar lipids in strain D39T. Lipids staining with molybdophosphoric acid (a), sulfuric acid and α-naphthol reagent (b), ammonium molybdate reagent (c), ninhydrin reagent (d). DPG, diphosphatidylglycerol; GL, glycolipid; L, lipid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; SGL, sphingoglycolipid. The arrow 1 indicates the first-dimensional TLC by using chloroform/methanol/water (13:5:0.8, v/v/v), and the arrow 2 indicates the second-dimensional TLC by using chloroform/methanol/acetic acid/water (16:2.4:3:0.8, v/v/v/v).
3.4. Carotenoid Composition and Biosynthesis Pathway of Strain D39T
In the carotenoid assay of strain D39T, α-carotene, β-carotene, lutein dimyristate, zeaxanthin, zeaxanthin dimyristate, and zeaxanthin dipalmitate were detected as major compounds; among these compounds, the highest content was found for zeaxanthin (412.4 ± 14.3 × 10−3 μg/g), followed by lutein dimyristate with a content of 58.4 ± 0.9 × 10−3 μg/g. Table 3 presented the classification, molecular weight, formula, and content of these compounds. The regression equation and peak review of six major carotenoids content in strain D39T are shown in Figure S2.

Table 3.
Major carotenoid contents in strain D39T.
Functional annotations revealed that the genus Croceibacterium members encoded crtEBIYZG genes, except for C. mercuriale CoronadoT (crtEBIYZGW) and C. soli MCCC 1K02066T (crtEBI). Phylogenetic reconstruction based on both indicated that their topologies were similar to their phylogenomic one, namely strain D39T, C. salegens MCCC 1K01500T, C. soli MCCC 1K02066T and forming clade A and clustered into clade B. (Figure 5).

Figure 5.
The maximum likelihood phylogenetic tree based on the genus Croceibacterium CrtEBI (A) and CrtEBIYZG (B) protein sequences with amino acid substitution models of LG+F+R4 and LG+F+I+G4, respectively. Only bootstrap values higher than 70% were shown. Filled circles indicate nodes that were also recovered in the phylogenomic tree. Croceibacterium strains are colored in red. Erythrobacter longus DSM 6997T was used as an outgroup.
During the carotenoid biosynthesis of the genus Croceibacterium members, primarily, a condensation reaction occurred between one molecule of isopentenyl pyrophosphate and one molecule of dimethylallyl pyrophosphate, resulting in the production of geranylpyrophosphate and elongation of farnesyl diphosphate, which was subsequently converted to geranylgeranyl pyrophosphate. Zeaxanthin synthesis was completed through the presence of crtEBIYZ. Based on this, crtWG genes were involved to form three other pathways to produce astaxanthin, nostaxanthin, and erythroxanthin, respectively. Comparative genomic analysis revealed that most of Croceibacterium members had identical carotenoid biosynthesis genes with strain D39T, implying that they had similar carotenoid profiles. The presumptive carotenoid biosynthesis pathways and their distributions in the Croceibacterium members were visualized in Figure 6.
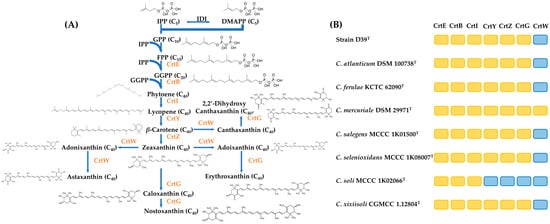
Figure 6.
Carotenoid biosynthesis pathways in Croceibacterium members were depicted. The shorthand notations of enzymes were highlighted in orange (A). The orange box indicates that the corresponding Crt protein was present in the Croceibacterium strains, while the blue box indicates that that protein was absent in it (B). The pathway scheme is drawn as designed by Siddaramappa et al. [48].
4. Discussion
4.1. Erythrobacteraceae and Flavobacteraceae as Dominant Carotenoid-Producing Taxa
The dominant taxon among bacterial strains isolated and cultivated from the tidal flat sediment sample of Zhoushan Archipelago is the phylum Pseudomonadota, accounting for 44% of them, which is similar to cultivation-independent bacterial investigations in other tidal flat regions, such as those found in China, Germany, Kenya, Republic of Korea, New Zealand, Oman, and Spain (Table S5) [65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80]. Moreover, nine strains—including D37T, D39T, D40T, H6T, H9T, H14T, H15T, H18T, and H20T—have low sequence identities (<98.65%) for species demarcation [50] to their top hit reference strains, indicating that they can represent novel bacterial species in the phyla Bacteroidota (the families Flavobacteriaceae and Prolixibacteraceae) and Pseudomonadota (the families Erythrobacteraceae, Roseobacteraceae and Sphingomonadaceae). Among those nine strains, strain D37T has been classified as one novel Maribacter species, for which the name Maribacter polysaccharolyticus has been proposed lately [81]. Thus, tidal flats contain numerous valuable and novel bacterial resources responsible for the stabilization of ecosystems.
The dominant taxa of nine carotenoid-producing strains vary obviously from that of isolated strains, with four Flavobacteriaceae and four Erythrobacteraceae strains belonging to the phyla Bacteroidota and Pseudomonadota, respectively, that are major marine carotenoid-producing prokaryotes [5]. Those carotenoid-producing Flavobacteriaceae strains are affiliated to the genera Maribacter, Christiangramia (known as Gramella previously), and Aequorivita, which are mainly reported to produce zeaxanthin [82,83]. Meanwhile, those carotenoid-producing Pseudomonadota strains are classified into the genera Croceibacterium and Qipengyuania, whose colony colors indicate different carotenoid compositions, with yellow-colored colonies mostly containing zeaxanthin, caloxanthin, and/or nostoxanthin, while orange-colored ones generally biosynthesize zeaxanthin, adonixanthin, and/or erythroxanthin [84]. Additionally, the remaining carotenoid-producing strain is classified as Mesobacillus subterraneus, whose colonies are yellow to orange on tryptic soy agar, whereas white on nutrient agar, indicating carotenoid production is dependent on substrate compositions [85,86]. Therefore, the urgent exploration and preservation of these tidal flat microbial resources is imperative to meet sustainable development needs for searching novel carotenoid microbial producers.
4.2. Carotenoid Synthesis in the Genus Croceibacterium
Functional annotations reveal that strain D39T is identical with most Croceibacterium members containing crtEBIYZG genes, that are also annotated in the majority of the order Sphingomonadales bacteria [48]. Phylogenetic reconstruction shows that those crt genes in the genus Croceibacterium are homologous genes, which have diverged through speciation rather than gene duplication [87]. Pathway reconstructions based on Siddaramappa et al. [49] point out that most Croceibacterium members have a complete pathway biosynthesizing from dimethylallyl pyrophosphate to nostoxanthin.
However, the carotenoid assay determination reveals that strain D39T accumulates zeaxanthin as the dominant carotenoid, rather than caloxanthin and nostoxanthin, indicating that carotenoid 2,2′-β-hydroxylase particulates less in introducing a hydroxyl group (−OH) into zeaxanthin in the strain D39T. Zeaxanthin, along with lutein, is one of the two carotenoids that accumulate in the retina, exhibiting higher concentrations in the macula compared to plasma and other tissues, according to Human Metabolome Database [88]. Zeaxanthin levels in the retina can lower the risk of age-related macular degeneration, owing to it serving as a protective blue light filter in the retina. Zeaxanthin also modulates antioxidant systems to protect proteins, lipids, and DNA from damage [83]. The global zeaxanthin market is projected to arrive at 210 million U.S. dollars by 2030, reflecting a Compound Annual Growth Rate of 8.2% [89], and further researches into the strain D39T, even the genus Croceibacterium, should be emphasized to promote accumulating zeaxanthin as a high-value carotenoid.
4.3. Proposal of a Novel Species Croceibacterium aestuarii sp. nov.
Phylogenomic analysis revealed that carotenoid-producing strain D39T is clustered into a clade consisting of the genus Croceibacterium and forms a stable branch with C. salegens MCCC 1K01500T. Overall genome relatedness indices between strain D39T and its reference strains are obviously lower than species demarcation thresholds. In addition, strain D39T can be distinguished from its reference strains by optimal temperature and pH for growth, hydrolysis of Tween 60, sole carbon, nitrogen, and energy source utilization of D-galactose and D-maltose, leucine arylamidase activity and acid production from D-maltose and sucrose, C18:1ω7c 11-methyl as a major fatty acid, and the presence of three unidentified glycolipids. Therefore, strain D39T represents a novel species in the genus Croceibacterium, for which the name Croceibacterium aestuarii is proposed.
- Description of Croceibacterium aestuarii sp. nov.
Croceibacterium aestuarii (aes.tu.a’ri.i. L. gen. n. aestuarii, of a tidal flat, from where the type strain was isolated).
Gram-stain-negative. Rod-shaped with 0.8–1.5 μm in length and 0.4–0.7 μm in width. Aerobic and motile by flagella. Colonies are yellow-pigmented, convex, circular, and smooth, with a diameter of 0.5 mm after incubation on MA containing 2.0% (w/v) NaCl at 37 °C for four days. Growth is observed within the temperature range of 20 to 45 °C, with an optimal growth at 37 °C. The pH and NaCl concentration ranges span from 5.0 to 9.5 and from 0.5% to 5.0% (w/v), respectively, whereas the optimal ones are pH range of 7.5 to 8.5 and NaCl concentration of 2% (w/v). Produce carotenoids but not bacteriochlorophyll a. Catalase- and oxidase-positive. Nitrate is not reduced. Hydrolyze Tween 60, while not hydrolyze Tween 20, 40, and 80, casein, cellulose, tyrosine, and starch. Positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), valine arylamidase, trypsin, chymotrypsin, acid phosphatase, napthol-AS-BI-phosphopydrase activities, β-glucosidase, and hydrolysis of gelatine (API ZYM and API 20NE). Acid is produced from D-galactose, D-glucose, glycerol, D-maltose, and sucrose. Utilize L-alanine, D-galactose, D-glucose and D-maltose as sole carbon, nitrogen, and energy sources. Respiratory ubiquinone is Q-10. Major fatty acids (>10%) are summed feature 8 (C18:1ω6c and/or C18:1ω7c), C18:1ω7c 11-methyl, and C16:0. Polar lipids include diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, glycosphingolipid, four unidentified glycolipid, and one unidentified lipid.
The type strain is D39T (= KCTC 82757T = MCCC 1K07008T) and was isolated from tidal flat in Zhoushan Archipelago, People’s Republic of China. The DNA G+C content of the type strain is 65.8% (by genome). The GenBank accession numbers for the 16S ribosomal RNA gene and genome sequences of strain D39T are OP132876 and GCA_030657335.1, respectively.
5. Conclusions
Fifty-five strains were isolated and cultured from the sediment of Zhoushan tidal flat, and the dominant phylum was identified as Pseudomonadota. Carotenoids were detected in nine out of the twenty-eight pigment-containing strains, including strain D39T. The predominant families of carotenoid-producing strains were Erythrobacteriaceae (n = 4) and Flavobacteriaceae (n = 4). Based on polyphasic taxonomic methods including phenotypic characteristics, phylogenetic relationship, and genomic comparison, strain D39T was proposed as one novel Croceibacterium species belonging to the family Erythrobacteraceae. Genomic analysis revealed that strain D39T encoded crtEBIYZG genes, which were also commonly found in most Croceibacterium strains. Detection results of carotenoid compounds indicated that strain D39T primarily accumulated zeaxanthin. Furthermore, all Croceibacterium members except for Croceibacterium soli MCCC 1K02066T possessed a complete biosynthesis pathway from dimethylpropenyl pyrophosphate to nostoxanthin, supposing that they can also accumulate zeaxanthin. This study enhances microbial biodiversity in tidal flats, summarizes the carotenoid biosynthesis pathway in the genus Croceibacterium, mainly zeaxanthin, and proposes one novel carotenoid-producing species Croceibacterium aestuarii sp. nov., which contribute to enriching marine carotenoid-producing prokaryotic strains and promoting a comprehensive insight into genomic contents of marine carotenoid-producing prokaryotes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jmse12010099/s1, Figure S1: Neighbor-joining (A), maximum-likelihood (B) and maximum-parsimony (C) phylogenetic trees based on 16S rRNA gene sequences showing he phylogenetic relationship of strain D39T and its similar type strains. Only bootstrap values higher than 70% are shown. Figure S2. The regression equation and peak review of six major carotenoids content in strain D39T are shown. Table S1: Genomic information of strain D39T and other related Erythrobacteraceae type strains used in this study. Table S2: All Crt protein sequences of strain D39T, seven Croceibacterium type strains and Erythrobacter longus DSM 6997T. Table S3: The 16S ribosomal RNA gene sequence identities (by using EzBioCloud websever) and colonial color of isolated bacterial strains in this study; Table S4: Comparison of cellular fatty acid compositions (%) in strain D39T and its related reference strains. 1, strain D39T; 2, C. salegens MCCC 1K01500T; 3, C. soli MCCC 1K02066T. Summed feature 3 contains C16:1ω6c and/or C16:1ω7c; summed feature 6 contains C19:1ω9c and/or C19:1ω11c; summed feature 8 contains C18:1ω6c and/or C18:1ω7c. ND, not detected. TR, traces (<0.5%). Table S5: Summary of relative abundance of Pseudomonadota members in other tidal flat regions.
Author Contributions
Conceptualization, X.-Y.S., C.S. and L.X.; methodology, X.-Y.S., H.D. and L.X.; formal analysis, X.-Y.S. and H.D.; data curation, X.-Y.S., Y.Z. and J.-W.G.; writing—original draft, X.-Y.S., P.Z. and L.X.; visualization, X.-Y.S. and Y.Z.; supervision, L.X.; project administration, L.X.; funding acquisition, P.Z., C.S. and L.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32000001, U23A2034), the Zhejiang Provincial Natural Science Foundation of China (LY24D060001), the National Science and Technology Fundamental Resources Investigation Program of China (2021FY100900), the Fundamental Research Funds of Zhejiang Sci-Tech University (23042128-Y) and the Open Fund of Zhejiang Sci-Tech University Shaoxing Academy of Biomedicine Co., Ltd. (SXAB202217).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The 16S ribosomal RNA gene sequences are deposited into the GenBank/EMBL/DDBJ under accession numbers of OR871657 (strain D1), OR835564 (strain D3), OR871658 (strain D6), OR871659 (strain D7), OR871660 (strain D12), OR603115 (strain D15), OR871661 (strain D16), OR871664 (strain D17), OR875941 (strain D19), OR871663 (strain D21), OR603134 (strain D24), OR603136 (strain D25), OR871665 (strain D31), OR871669 (strain D32), OR871666 (strain D33), OR871667 (strain D34), OP132875 (strain D37), OR603149 (strain D38), OP132876 (strain D39), OR603151 (strain D40), OR603153 (strain D42), OR871668 (strain D44), OR871671 (strain D46), OR656552 (strain H3), OR656553 (strain H4), OR656554 (strain H6), OP132826 (strain H9), OR656561 (strain H11), OR656562 (strain H13), OP132872 (strain H14), OP132873 (strain H15), OR656565 (strain H16), OR656566 (strain H18), OP132874 (strain H20), OR656569 (strain H22), OR656568 (strain H23), OR885467 (strain H24), OR656570 (strain H25), OR612978 (strain Z5), OR612977 (strain Z6), OR612979 (strain Z8), OR612980 (strain Z10), OR612981 (strain Z12), OR612983 (strain Z14), OR612982 (strain Z15), OR612984 (strain Z16), OR612985 (strain Z17), OR612986 (strain Z19), OR612990 (strain Z21), OR885466 (strain Z24), OR871671 (strain Z28), OR612989 (strain Z29), OR612987 (strain Z30), and OR612988 (strain Z33), respectively. The genome sequences of strain D39T are deposited into the GenBank/EMBL/DDBJ under an accession number of GCA_030657335.
Acknowledgments
We appreciate Aharon Oren (The Hebrew University of Jerusalem, Israel) for nomenclature issues.
Conflicts of Interest
Authors X.-Y.S., H.D., Y.Z., J.-W.G., C.S. and L.X. were employed by the company Shaoxing Biomedical Research Institute of Zhejiang Sci-Tech University Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declare that this study received funding from Zhejiang Sci-Tech University Shaoxing Academy of Biomedicine Co., Ltd. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Abbreviations
DMAPP, dimethylallyl pyrophosphate; DPG, diphosphatidylglycerol; FPP, farnesyl diphosphate; GGPP, geranylgeranyl pyrophosphate; GL, glycolipid; GPP, geranylpyrophosphate; IPP, isopentenyl pyrophosphate; KCTC, Korean Collection of Type Cultures; MA, marine 2216 agar; MB, marine 2216 broth; MCCC, Marine Culture Collection of China; OC, orthologous clusters; PC phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; Q-10, ubiquinone-10; SGL sphingoglycolipid.
References
- Soni, N.; Dhandhukia, P.; Thakker, J.N. Carotenoid from marine Bacillus infantis: Production, extraction, partial characterization, and its biological activity. Arch. Microbiol. 2023, 205, 161. [Google Scholar] [CrossRef]
- Maj, A.; Dziewit, L.; Drewniak, L.; Garstka, M.; Krucon, T.; Piatkowska, K.; Gieczewska, K.; Czarnecki, J.; Furmanczyk, E.; Lasek, R.; et al. In vivo creation of plasmid pCRT01 and its use for the construction of carotenoid-producing Paracoccus spp. strains that grow efficiently on industrial wastes. Microb. Cell Fact. 2020, 19, 141. [Google Scholar] [CrossRef]
- Britton, G. Carotenoid research: History and new perspectives for chemistry in biological systems. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158699. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Mumtaz, R.; Tayyab, S.; Rehman, N.U.; Khan, A.L.; Shinwari, Z.K.; Al-Harrasi, A. Therapeutic applications of bacterial pigments: A review of current status and future opportunities. 3 Biotech 2018, 8, 207. [Google Scholar] [CrossRef]
- Mapelli-Brahm, P.; Gómez-Villegas, P.; Gonda, M.L.; León-Vaz, A.; León, R.; Mildenberger, J.; Rebours, C.; Saravia, V.; Vero, S.; Vila, E.; et al. Microalgae, seaweeds and aquatic bacteria, archaea, and yeasts: Sources of carotenoids with potential antioxidant and anti-inflammatory health-promoting actions in the sustainability era. Marine Drugs 2023, 21, 340. [Google Scholar] [CrossRef]
- Béjà, O.; Spudich, E.N.; Spudich, J.L.; Leclerc, M.; DeLong, E.F. Proteorhodopsin phototrophy in the ocean. Nature 2001, 411, 786–789. [Google Scholar] [CrossRef]
- Dieser, M.; Greenwood, M.; Foreman, C.M. Carotenoid pigmentation in antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct. Antarct. Alp. Res. 2010, 42, 396–405. [Google Scholar] [CrossRef]
- Bernstein, P.S.; Li, B.; Vachali, P.P.; Gorusupudi, A.; Shyam, R.; Henriksen, B.S.; Nolan, J.M. Lutein, zeaxanthin, and meso-zeaxanthin: The basic and clinical science underlying carotenoid-based nutritional interventions against ocular disease. Prog. Retin. Eye Res. 2016, 50, 34–66. [Google Scholar] [CrossRef]
- Vila, E.; Hornero-Méndez, D.; Lareo, C.; Saravia, V. Biotechnological production of zeaxanthin by an Antarctic Flavobacterium: Evaluation of culture conditions. J. Biotechnol. 2020, 319, 54–60. [Google Scholar] [CrossRef]
- Lorenzo, Y.; Azqueta, A.; Luna, L.; Bonilla, F.; Domínguez, G.; Collins, A.R. The carotenoid beta-cryptoxanthin stimulates the repair of DNA oxidation damage in addition to acting as an antioxidant in human cells. Carcinogenesis 2009, 30, 308–314. [Google Scholar] [CrossRef]
- Setiyono, E.; Heriyanto; Pringgenies, D.; Shioi, Y.; Kanesaki, Y.; Awai, K.; Brotosudarmo, T.H.P. Sulfur-containing carotenoids from a marine coral symbiont Erythrobacter flavus strain KJ5. Mar. Drugs 2019, 17, 349. [Google Scholar] [CrossRef]
- Martín, J.F.; Gudiña, E.; Barredo, J.L. Conversion of β-Carotene into astaxanthin: Two separate enzymes or a bifunctional hydroxylase-ketolase protein? Microb. Cell Fact. 2008, 7, 3. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Xu, X.W. Erythrobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; Wiley: Hoboken, NJ, USA, 2023; pp. 1–18. ISBN 978-1-118-96060-8. [Google Scholar]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Park, S.C.; Baik, K.S.; Choe, H.N.; Lim, C.H.; Kim, H.J.; Ka, J.-O.; Seong, C.N. Altererythrobacter namhicola sp. nov. and Altererythrobacter aestuarii sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 2011, 61, 709–715. [Google Scholar] [CrossRef][Green Version]
- Gao, Y.; Li, G.; Fang, C.; Shao, Z.; Wu, Y.-H.; Xu, X.-W. Tsuneonella suprasediminis sp. nov., isolated from the Pacific Ocean. Int. J. Syst. Evol. Microbiol. 2019, 71, 004678. [Google Scholar] [CrossRef]
- He, Y.; Liu, Y.; Pei, T.; Duan, J.; Du, J.; Deng, X.; Zhu, H. Tsuneonella litorea sp. nov., a novel carotenoid-producing bacterium isolated from coastal sediment. Int. J. Syst. Evol. Microbiol. 2023, 73, 5785. [Google Scholar] [CrossRef]
- Wu, Y.-H.; Xu, L.; Meng, F.-X.; Zhang, D.-S.; Wang, C.-S.; Oren, A.; Xu, X.-W. Altererythrobacter atlanticus sp. nov., isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 2014, 64, 116–121. [Google Scholar] [CrossRef]
- Fidalgo, C.; Rocha, J.; Martins, R.; Proença, D.N.; Morais, P.V.; Henriques, I.; Alves, A. Altererythrobacter halimionae sp. nov. and Altererythrobacter endophyticus sp. nov., two endophytes from the salt marsh plant Halimione portulacoides. Int. J. Syst. Evol. Microbiol. 2017, 67, 3057–3062. [Google Scholar] [CrossRef]
- Nedashkovskaya, O.I.; Cho, S.-H.; Joung, Y.; Joh, K.; Kim, M.N.; Shin, K.-S.; Oh, H.W.; Bae, K.S.; Mikhailov, V.V.; Kim, S.B. Altererythrobacter troitsensis sp. nov., isolated from the sea urchin Strongylocentrotus intermedius. Int. J. Syst. Evol. Microbiol. 2013, 63, 93–97. [Google Scholar] [CrossRef]
- Tareen, S.; Risdian, C.; Müsken, M.; Wink, J. Alteriqipengyuania abyssalis sp. nov., a novel member of the class Alphaproteobacteria isolated from sponge, and emended description of the genus Alteriqipengyuania. Diversity 2021, 13, 670. [Google Scholar] [CrossRef]
- Liu, Y.-H.; Fang, B.-Z.; Dong, Z.-Y.; Li, L.; Mohamad, O.A.A.; Zhang, Y.-G.; Egamberdieva, D.; Xiao, M.; Li, W.-J. Croceibacterium gen. nov., with Description of Croceibacterium ferulae sp. nov., an endophytic bacterium isolated from Ferula sinkiangensis K. M. Shen and reclassification of Porphyrobacter mercurialis as Croceibacterium mercuriale comb. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 2547–2554. [Google Scholar] [CrossRef]
- Xu, L.; Sun, C.; Fang, C.; Oren, A.; Xu, X.-W. Genomic-based taxonomic classification of the family Erythrobacteraceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 4470–4495. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S ribosomal RNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Hildebrand, D.; Palleroni, N.; Hendson, M.; Toth, J.; Johnson, J. Pseudomonas flavescens sp. nov., isolated from walnut blight cankers. Int. J. Syst. Bacteriol. 1994, 44, 410–415. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Zhang, Y.; Hua, J.; Ying, J.-J.; Dong, H.; Li, H.; Xamxidin, M.; Han, B.-N.; Sun, C.; Xu, L. Erythrobacter aurantius sp. nov., isolated from intertidal seawater in Taizhou. Int. J. Syst. Evol. Microbiol. 2022, 72, 5616. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from dna sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Kannan, L.; Wheeler, W.C. Maximum Parsimony on Phylogenetic Networks. Algorithms Mol. Biol. 2012, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Findeiss, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Xu, L.; Wu, Y.-H.; Jian, S.-L.; Wang, C.-S.; Wu, M.; Cheng, L.; Xu, X.-W. Pseudohongiella nitratireducens sp. nov., isolated from seawater, and emended description of the genus Pseudohongiella. Int. J. Syst. Evol. Microbiol. 2016, 66, 5155–5160. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huo, Y.-Y.; Li, Z.-Y.; Wang, C.-S.; Oren, A.; Xu, X.-W. Chryseobacterium profundimaris sp. nov., a new member of the family Flavobacteriaceae isolated from deep-sea sediment. Antonie Van. Leeuwenhoek 2015, 107, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Xu, L.; Yu, X.-Y.; Zhao, Z.; Wu, Y.-H.; Oren, A.; Wang, C.-S.; Xu, X.-W. Minwuia thermotolerans gen. nov., sp. nov., a marine bacterium forming a deep branch in the Alphaproteobacteria, and proposal of Minwuiaceae fam. nov. and Minwuiales ord. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 3856–3862. [Google Scholar] [CrossRef] [PubMed]
- Minnikin, D.E. Chemical principles in the organization of lipid components in the mycobacterial cell envelope. Res. Microbiol. 1991, 142, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Bartley, G.E.; Scolnik, P.A. Plant carotenoids: Pigments for photoprotection, visual attraction, and human health. Plant. Cell 1995, 7, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Inbaraj, B.S.; Lu, H.; Hung, C.F.; Wu, W.B.; Lin, C.L.; Chen, B.H. Determination of carotenoids and their esters in fruits of Lycium barbarum Linnaeus by HPLC-DAD-APCI-MS. J. Pharm. Biomed. Anal. 2008, 47, 812–818. [Google Scholar] [CrossRef]
- Siddaramappa, S.; Viswanathan, V.; Thiyagarajan, S.; Narjala, A. Genomewide characterisation of the genetic diversity of carotenogenesis in bacteria of the order Sphingomonadales. Microb. Genom. 2018, 4, e000172. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt knowledgebase: How to use the entry view. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16s ribosomal RNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Kim, D.; Yoo, Y.; Khim, J.S.; Yang, D.; Pathiraja, D.; Park, B.; Choi, I.-G.; Kim, J.-J. Altererythrobacter lutimaris sp. nov., a marine bacterium isolated from a tidal flat and reclassification of Altererythrobacter deserti, Altererythrobacter estronivorus and Altererythrobacter muriae as Tsuneonella deserti comb. nov., Croceicoccus estronivorus comb. nov. and Alteripontixanthobacter muriae comb. nov. Int. J. Syst. Evol. Microbiol. 2021, 71, 005134. [Google Scholar] [CrossRef]
- Zhang, K.; Han, C.H.; Ling, J.; Yin, J. Alteriqipengyuania flavescens sp. nov., isolated from pearl river estuary sediment. Int. J. Syst. Evol. Microbiol. 2023, 73, 6161. [Google Scholar] [CrossRef]
- Lee, S.D.; Kim, I.S. Aurantiacibacter rhizosphaerae sp. nov., isolated from a rhizosphere mudflat of a halophyte and proposal to reclassify Erythrobacter suaedae Lee et al. 2019. and Erythrobacter flavus Yoon et al. 2003 as Aurantiacibacter suaedae comb. nov. and Qipengyuania flava comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2020, 70, 6257–6265. [Google Scholar] [CrossRef]
- Kim, C.-H.; Yoo, Y.; Kim, D.; Khim, J.S.; Pathiraja, D.; Kim, B.; Choi, I.-G.; Kim, J.-J. Aurantiacibacter sediminis sp. nov., a marine bacterium isolated from a tidal flat. Int. J. Syst. Evol. Microbiol. 2022, 72, 5406. [Google Scholar] [CrossRef]
- Pei, T.; Liu, Y.; Du, J.; Huang, K.; Deng, M.-R.; Zhu, H. Croceicoccus gelatinilyticus sp. nov., isolated from a tidal flat sediment. Arch. Microbiol. 2021, 204, 93. [Google Scholar] [CrossRef]
- Chen, B.; Ye, Y.; Lin, D.; Zhang, M.; Sun, J.; Tang, K. Croceicoccus hydrothermalis sp. nov., isolated from shallow-sea hydrothermal system off Kueishantao Island. Int. J. Syst. Evol. Microbiol. 2022, 72, 5472. [Google Scholar] [CrossRef]
- Park, S.; Chen, S.; Yoon, J.-H. Erythrobacter insulae sp. nov., isolated from a tidal flat. Int. J. Syst. Evol. Microbiol. 2020, 70, 1470–1477. [Google Scholar] [CrossRef]
- Hu, W.; Li, Z.; Ou, H.; Wang, X.; Wang, Q.; Tao, Z.; Huang, S.; Huang, Y.; Wang, G.; Pan, X. Novosphingobium album sp. nov., Novosphingobium organovorum sp. nov. and Novosphingobium mangrovi sp. nov. with the organophosphorus pesticides degrading ability isolated from mangrove sediments. Int. J. Syst. Evol. Microbiol. 2023, 73, 5843. [Google Scholar] [CrossRef]
- Chaudhary, D.K.; Dahal, R.H.; Kim, D.-U.; Kim, J. Novosphingobium olei sp. nov., with the ability to degrade diesel oil, isolated from oil-contaminated soil and proposal to reclassify Novosphingobium stygium as a later heterotypic synonym of Novosphingobium aromaticivorans. Int. J. Syst. Evol. Microbiol. 2021, 71, 4628. [Google Scholar] [CrossRef]
- He, X.; Lu, H.; Hu, W.; Deng, T.; Gong, X.; Yang, X.; Song, D.; He, M.; Xu, M. Novosphingobium percolationis sp. nov. and Novosphingobium huizhouense sp. nov., isolated from landfill leachate of a domestic waste treatment plant. Int. J. Syst. Evol. Microbiol. 2022, 72, 5394. [Google Scholar] [CrossRef]
- Pira, H.; Risdian, C.; Müsken, M.; Schupp, P.J.; Wink, J. Winogradskyella luteola sp. nov., Erythrobacter ani sp. nov., and Erythrobacter crassostrea sp. nov., isolated from the hemolymph of the pacific oyster Crassostrea gigas. Arch. Microbiol. 2022, 204, 488. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef]
- Liang, X.; Lin, H.; Wang, K.; Liao, Y.; Lai, Q.; Xu, Y.; Wang, C. Altererythrobacter salegens sp. nov., a slightly halophilic bacterium isolated from surface sediment. Int. J. Syst. Evol. Microbiol. 2017, 67, 909–913. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, H.-R.; Han, Q.-Q.; He, A.-L.; Nie, C.-Y.; Wang, S.-M.; Zhang, J.-L. Altererythrobacter soli sp. nov., isolated from desert sand. Int. J. Syst. Evol. Microbiol. 2017, 67, 454–459. [Google Scholar] [CrossRef]
- Ye, Y.-L.; Ma, K.-J.; Fu, Y.-H.; Wu, Z.-C.; Fu, G.-Y.; Sun, C.; Xu, X.-W. The heterogeneity of microbial diversity and its drivers in two types of sediments from tidal flats in Beibu Gulf, China. Front. Mar. Sci. 2023, 10, 1256393. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, B.X.; Ren, H.; Zhang, J. Composition and functional diversity of microbial community across a mangrove-inhabited mudflat as revealed by 16S rDNA gene sequences. Sci. Total Environ. 2018, 633, 518–528. [Google Scholar] [CrossRef]
- Yue, Y.; Rong, H.; Yang, Z.; Pan, X.; Chen, Y.; Yang, M. Microbial diversity and functional profiling in coastal tidal flat sediment with pollution of nutrients and potentially toxic elements. J. Soils Sediments 2023, 23, 2935–2950. [Google Scholar] [CrossRef]
- Zhou, Z.; Meng, H.; Liu, Y.; Gu, J.-D.; Li, M. Stratified bacterial and archaeal community in mangrove and intertidal wetland mudflats revealed by high throughput 16S ribosomal RNA gene sequencing. Front. Microbiol. 2017, 8, 2148. [Google Scholar] [CrossRef]
- Jiang, X.-T.; Peng, X.; Deng, G.-H.; Sheng, H.-F.; Wang, Y.; Zhou, H.-W.; Tam, N.F.-Y. Illumina sequencing of 16S ribosomal RNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb. Ecol. 2013, 66, 96–104. [Google Scholar] [CrossRef]
- Wang, C.; Sun, D.; Junaid, M.; Xie, S.; Xu, G.; Li, X.; Tang, H.; Zou, J.; Zhou, A. Effects of tidal action on the stability of microbiota, antibiotic resistance genes, and microplastics in the Pearl River Estuary, Guangzhou, China. Chemosphere 2023, 327, 138485. [Google Scholar] [CrossRef]
- Luo, T.; Huang, Z.; Li, X.; Zhang, Y. Anaerobic microbe mediated arsenic reduction and redistribution in coastal wetland soil. Sci. Total Environ. 2020, 727, 138630. [Google Scholar] [CrossRef]
- Lv, X.; Ma, B.; Yu, J.; Chang, S.X.; Xu, J.; Li, Y.; Wang, G.; Han, G.; Bo, G.; Chu, X. Bacterial community structure and function shift along a successional series of tidal flats in the Yellow River Delta. Sci. Rep. 2016, 6, 36550. [Google Scholar] [CrossRef]
- Dinter, T.; Geihser, S.; Gube, M.; Daniel, R.; Kuzyakov, Y. Impact of sea level change on coastal soil organic matter, priming effects and prokaryotic community assembly. FEMS Microbiol. Ecol. 2019, 95, fiz129. [Google Scholar] [CrossRef]
- Muwawa, E.M.; Obieze, C.C.; Makonde, H.M.; Jefwa, J.M.; Kahindi, J.H.P.; Khasa, D.P. 16S ribosomal RNA gene amplicon-based metagenomic analysis of bacterial communities in the rhizospheres of selected mangrove species from Mida Creek and Gazi Bay, Kenya. PLoS ONE 2021, 16, e0248485. [Google Scholar] [CrossRef]
- Lee, H.; Lee, D.W.; Kwon, S.L.; Heo, Y.M.; Jang, S.; Kwon, B.-O.; Khim, J.S.; Kim, G.-H.; Kim, J.-J. Importance of functional diversity in assessing the recovery of the microbial community after the Hebei Spirit Oil spill in Korea. Environ. Int. 2019, 128, 89–94. [Google Scholar] [CrossRef]
- Choi, H.J.; Jeong, T.-Y.; Yoon, H.; Oh, B.Y.; Han, Y.S.; Hur, M.J.; Kang, S.; Kim, J.-G. Comparative microbial communities in tidal flats sediment on Incheon, South Korea. J. Gen. Appl. Microbiol. 2018, 64, 232–239. [Google Scholar] [CrossRef]
- Lee, D.W.; Lee, H.; Lee, A.H.; Kwon, B.-O.; Khim, J.S.; Yim, U.H.; Kim, B.S.; Kim, J.-J. Microbial community composition and PAHs removal potential of indigenous bacteria in oil contaminated sediment of Taean coast, Korea. Environ. Pollut. 2018, 234, 503–512. [Google Scholar] [CrossRef]
- Boey, J.S.; Mortimer, R.; Couturier, A.; Worrallo, K.; Handley, K.M. Estuarine microbial diversity and nitrogen cycling increase along sand-mud gradients independent of salinity and distance. Environ. Microbiol. 2022, 24, 50–65. [Google Scholar] [CrossRef]
- Vogt, J.C.; Abed, R.M.M.; Albach, D.C.; Palinska, K.A. Bacterial and archaeal diversity in hypersaline cyanobacterial mats along a transect in the intertidal flats of the Sultanate of Oman. Microb. Ecol. 2018, 75, 331–347. [Google Scholar] [CrossRef] [PubMed]
- Leontidou, K.; Abad-Recio, I.L.; Rubel, V.; Filker, S.; Däumer, M.; Thielen, A.; Lanzén, A.; Stoeck, T. Simultaneous analysis of seven 16S ribosomal RNA hypervariable gene regions increases efficiency in marine bacterial diversity detection. Environ. Microbiol. 2023, 25, 3484–3501. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.-W.; Ying, J.-J.; Dong, H.; Liu, W.-J.; He, D.-Y.; Xu, L.; Sun, C. Characterization of Maribacter polysaccharolyticus sp. nov., Maribacter huludaoensis sp. nov., and Maribacter zhoushanensis sp. nov. and illumination of the distinct adaptative strategies of the genus Maribacter. Front. Mar. Sci. 2023, 10, 1248754. [Google Scholar] [CrossRef]
- Takagi, T.; Aoyama, K.; Motone, K.; Aburaya, S.; Yamashiro, H.; Miura, N.; Inoue, K. Mutualistic interactions between dinoflagellates and pigmented bacteria mitigate environmental stress. Microbiol. Spectr. 2023, 11, e0246422. [Google Scholar] [CrossRef] [PubMed]
- Steven, R.; Humaira, Z.; Natanael, Y.; Dwivany, F.M.; Trinugroho, J.P.; Dwijayanti, A.; Kristianti, T.; Tallei, T.E.; Emran, T.B.; Jeon, H.; et al. Marine microbial-derived resource exploration: Uncovering the hidden potential of marine carotenoids. Mar. Drugs 2022, 20, 352. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Pei, T.; Du, J.; Yao, Q.; Deng, M.-R.; Zhu, H. Comparative genomics reveals genetic diversity and metabolic potentials of the genus Qipengyuania and suggests fifteen novel species. Microbiol. Spectr. 2022, 10, e0126421. [Google Scholar] [CrossRef] [PubMed]
- Kanso, S.; Greene, A.C.; Patel, B.K.C. Bacillus subterraneus sp. nov., an iron- and manganese-reducing bacterium from a deep subsurface Australian thermal aquifer. Int. J. Syst. Evol. Microbiol. 2002, 52, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Igreja, W.S.; Maia, F.d.A.; Lopes, A.S.; Chisté, R.C. Biotechnological production of carotenoids using low cost-substrates is influenced by cultivation parameters: A review. Int. J. Mol. Sci. 2021, 22, 8819. [Google Scholar] [CrossRef]
- Castresana, J. Topological variation in single-gene phylogenetic trees. Genome Biol. 2007, 8, 216. [Google Scholar] [CrossRef][Green Version]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Zafar, J.; Aqeel, A.; Shah, F.I.; Ehsan, N.; Gohar, U.F.; Moga, M.A.; Festila, D.; Ciurea, C.; Irimie, M.; Chicea, R. Biochemical and Immunological implications of lutein and zeaxanthin. Int. J. Mol. Sci. 2021, 22, 10910. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).