Fucanases Related to the GH107 Family from Members of the PVC Superphylum
Abstract
1. Introduction
2. Materials and Methods
2.1. Intertidal Sediment Metagenomic Datasets
2.2. Identification of GH107 Homolog Sequences in Metagenomes, MAGs, and Genomes
2.3. Taxonomic Classification
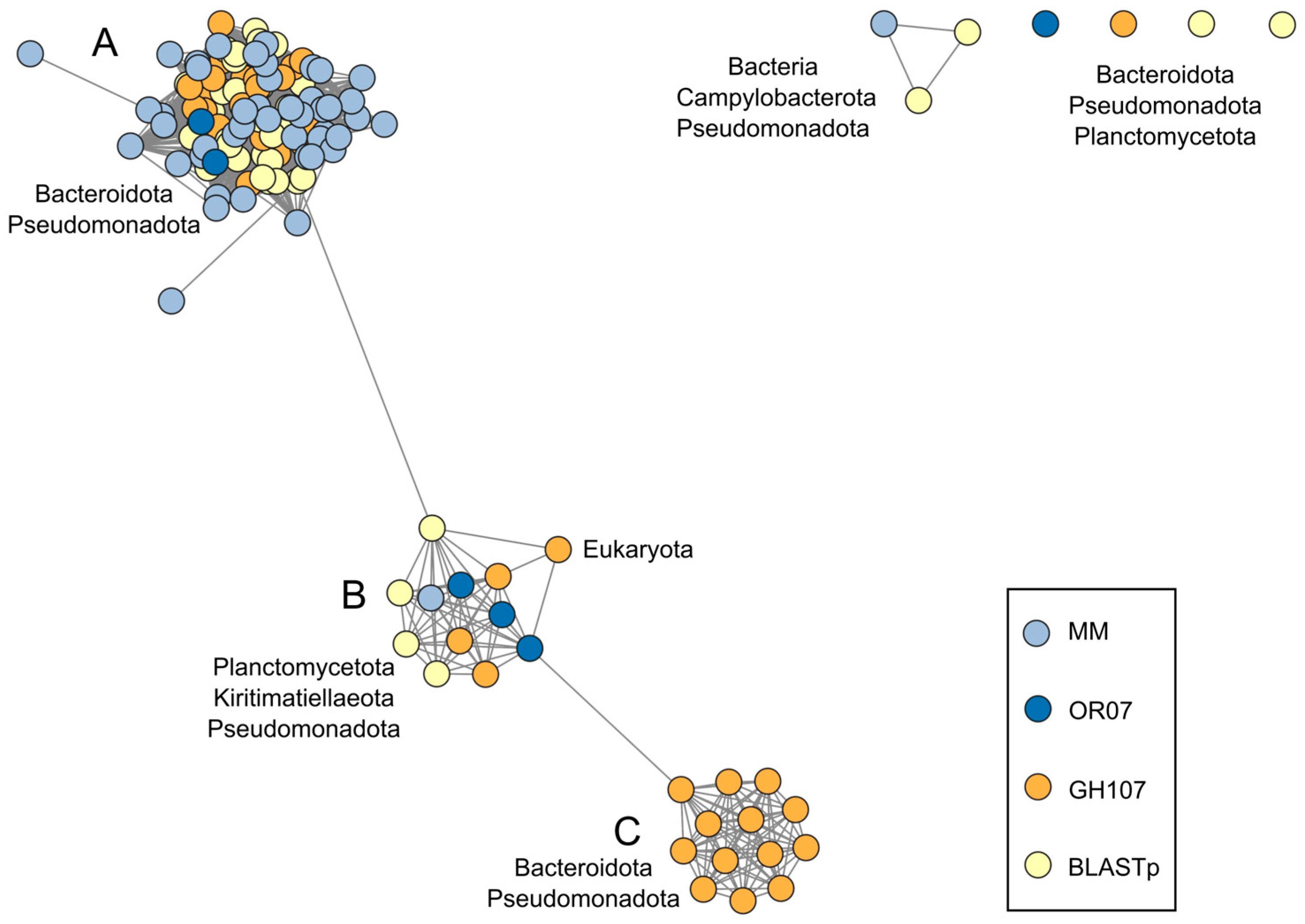
2.4. Sequence Similarity Network
2.5. Phylogenetic Analysis
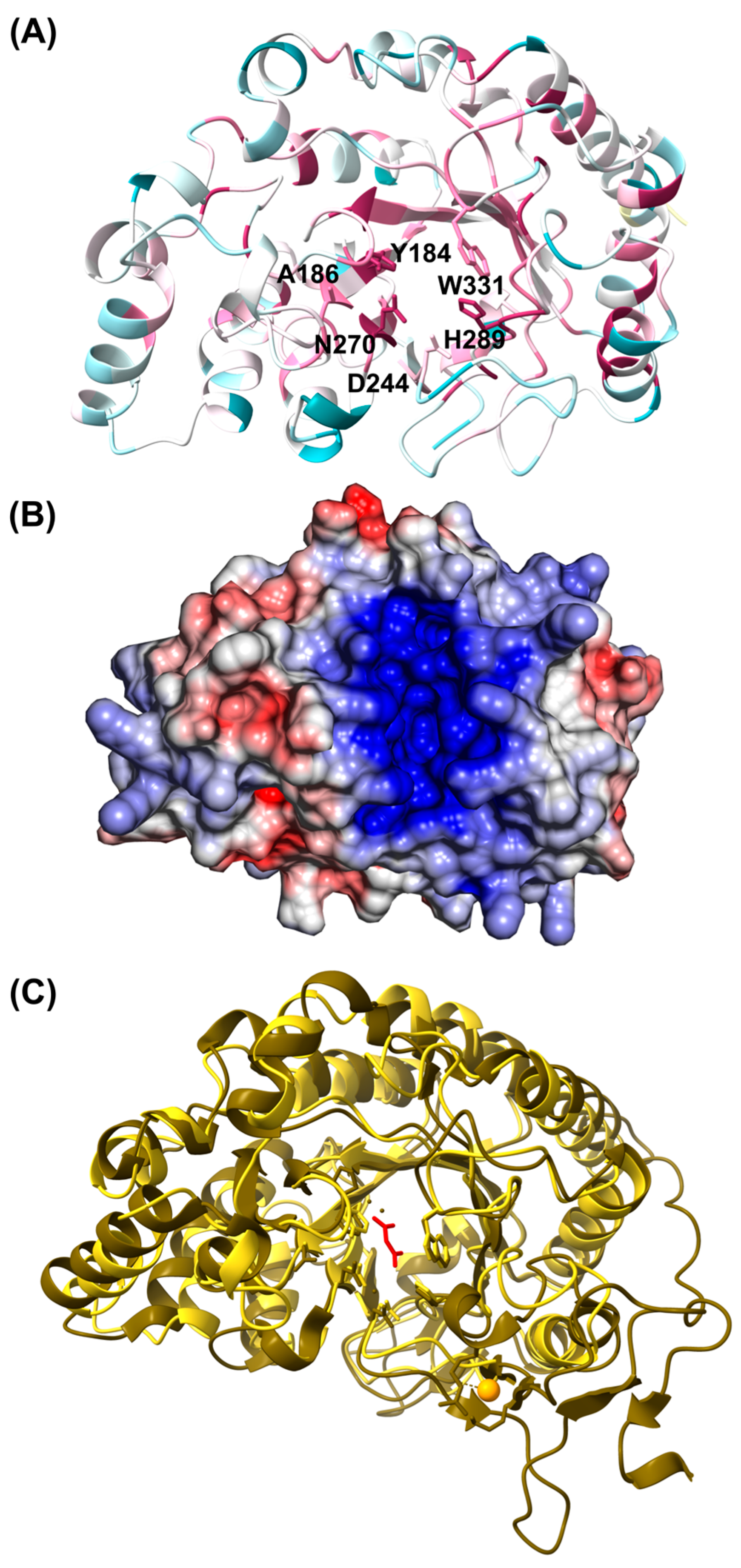
2.6. In Silico Structural Analyses
2.7. Cloning, Recombinant Expression, and Purification of a Putative Fucanase
2.8. Fucoidan Purification and Characterization
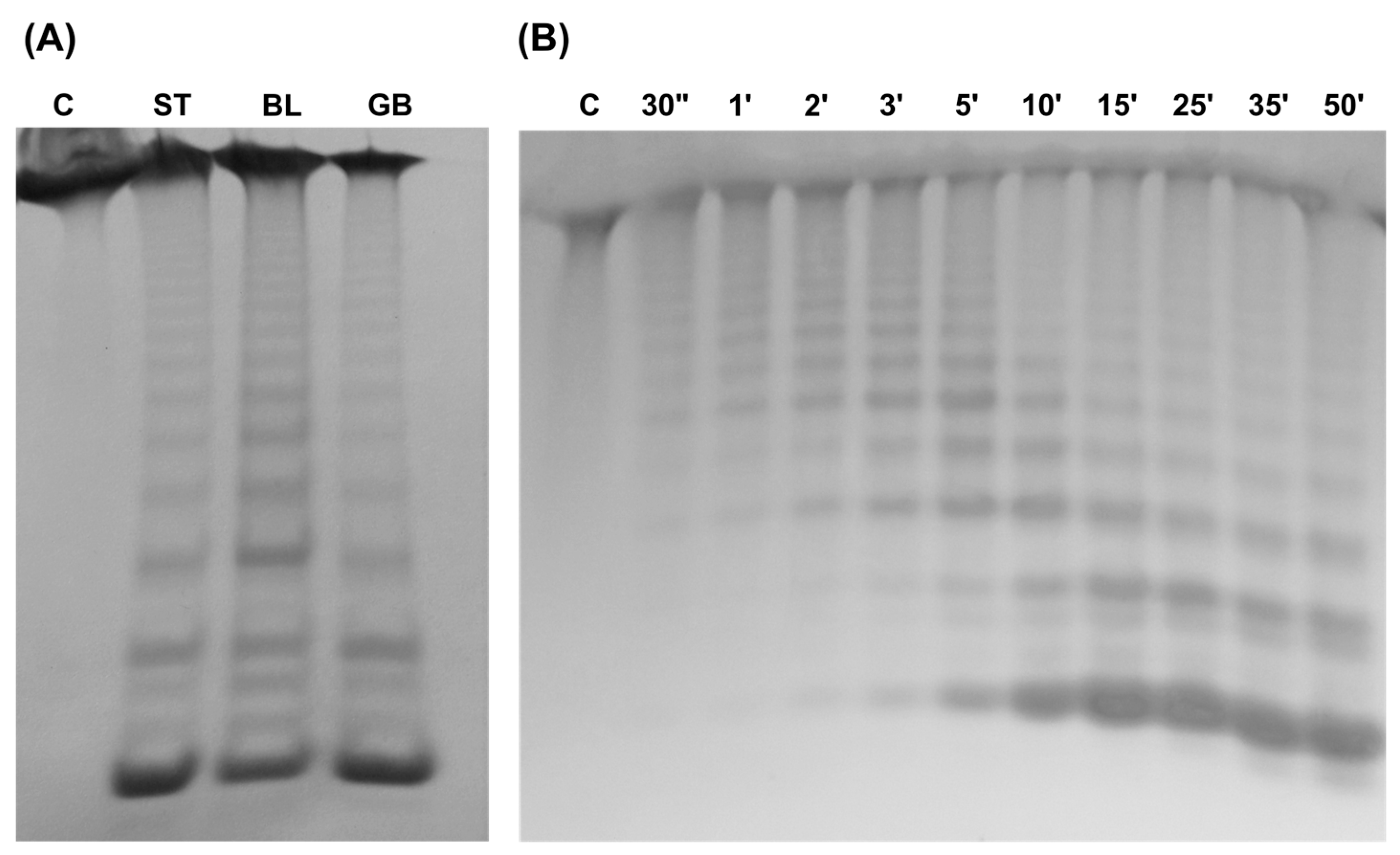
2.9. Fucanase Activity
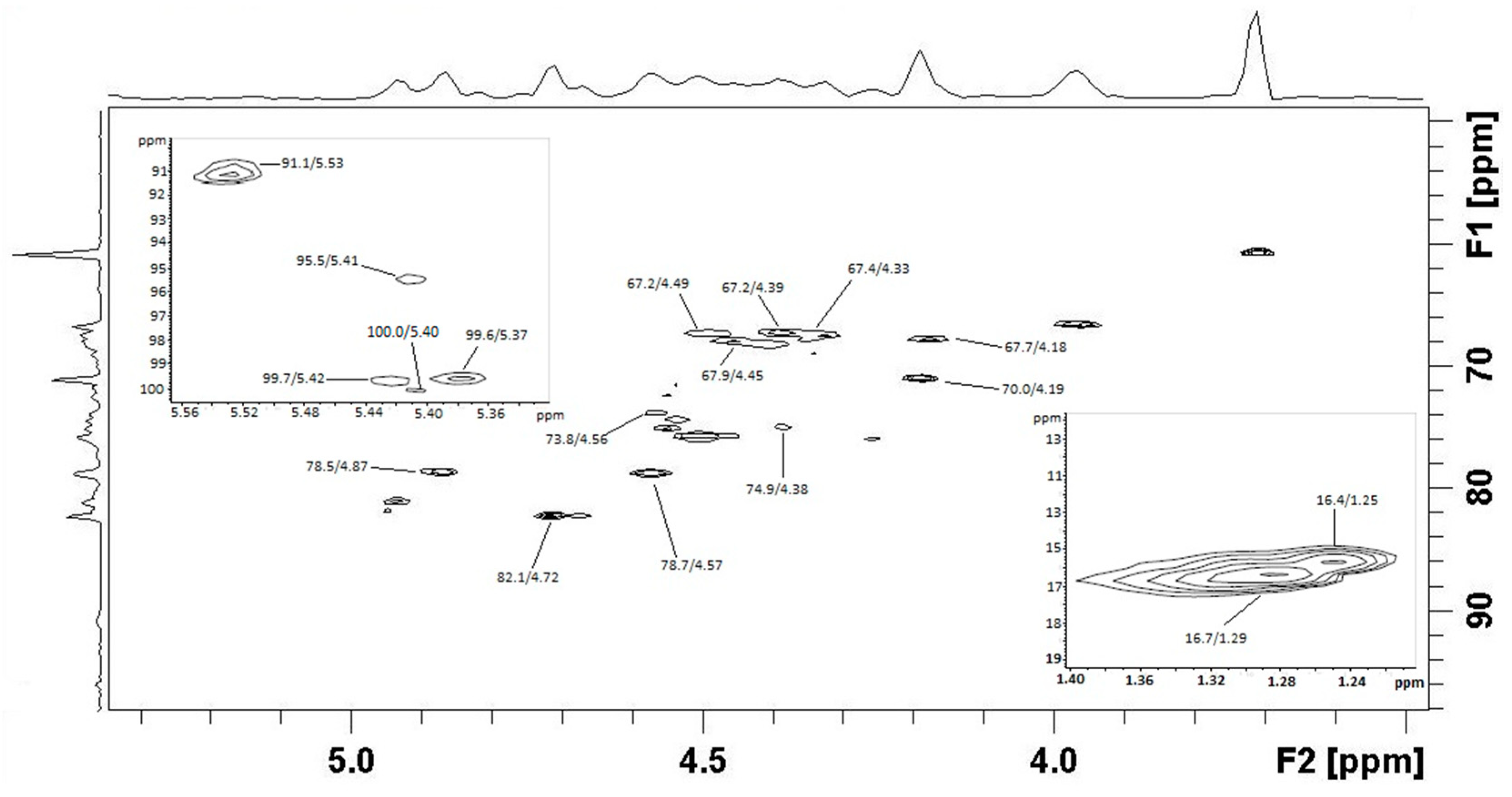
2.10. Preparation and Characterization of Enzymatic Products
3. Results
3.1. Diversity of GH107 Homolog Sequences in Intertidal Sediment Metagenomes
3.2. GH107 Homolog Sequences from Members of the PVC Superphylum
3.2.1. Sequence Identification
3.2.2. Phylogenetic Relationships
3.2.3. In Silico Structural Analyses of the D1 Domain
3.2.4. Domain Architecture
3.3. Characterization of a Fucanase from an Uncultured Planctomycete Bacterium
3.3.1. Recombinant Expression of OR07_113646
3.3.2. Enzyme Characterization
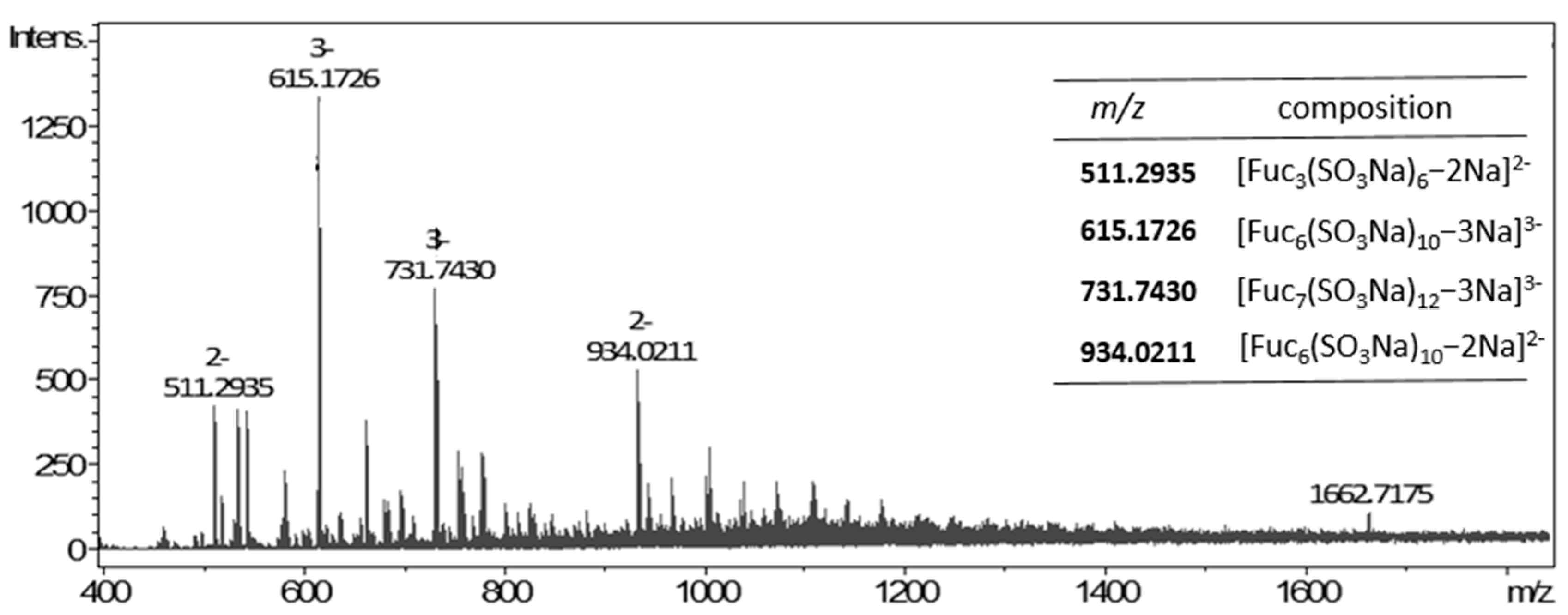
3.3.3. Composition and Structure of M. pyrifera Polysaccharide and Fuco-Oligosaccharides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Niftrik, L.; Devos, D.P. Planctomycetes-Verrucomicrobia-Chlamydiae bacterial superphylum: New model organisms for evolutionary cell biology. Front. Microbiol. 2017, 8, 1458. [Google Scholar]
- Vitorino, I.R.; Lage, O.M. The Planctomycetia: An overview of the currently largest class within the phylum Planctomycetes. Antonie Van Leeuwenhoek 2022, 115, 169–201. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, R.; Liu, R.; Li, R.; Sun, C. Cultivation and functional characterization of a deep-sea Lentisphaerae representative reveals its unique physiology and ecology. Front. Mar. Sci. 2022, 9, 458. [Google Scholar] [CrossRef]
- Sichert, A.; Corzett, C.H.; Schechter, M.S.; Unfried, F.; Markert, S.; Becher, D.; Fernandez-Guerra, A.; Liebeke, M.; Schweder, T.; Polz, M.F. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 2020, 5, 1026–1039. [Google Scholar] [CrossRef]
- Dharamshi, J.E.; Tamarit, D.; Eme, L.; Stairs, C.W.; Martijn, J.; Homa, F.; Jørgensen, S.L.; Spang, A.; Ettema, T.J. Marine sediments illuminate Chlamydiae diversity and evolution. Curr. Biol. 2020, 30, 1032–1048. [Google Scholar] [CrossRef]
- Freitas, S.; Hatosy, S.; Fuhrman, J.A.; Huse, S.M.; Mark Welch, D.B.; Sogin, M.L.; Martiny, A.C. Global distribution and diversity of marine Verrucomicrobia. ISME J. 2012, 6, 1499–1505. [Google Scholar] [CrossRef]
- Wiegand, S.; Jogler, M.; Jogler, C. On the maverick Planctomycetes. FEMS Microbiol. Rev. 2018, 42, 739–760. [Google Scholar]
- Bondoso, J.; Godoy-Vitorino, F.; Balague, V.; Gasol, J.M.; Harder, J.; Lage, O.M. Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol. Ecol. 2017, 93, fiw255. [Google Scholar] [CrossRef]
- Lage, O.M.; Bondoso, J. Planctomycetes and macroalgae, a striking association. Front. Microbiol. 2014, 5, 267. [Google Scholar] [CrossRef]
- Vollmers, J.; Frentrup, M.; Rast, P.; Jogler, C.; Kaster, A.-K. Untangling genomes of novel planctomycetal and verrucomicrobial species from Monterey Bay kelp forest metagenomes by refined binning. Front. Microbiol. 2017, 8, 472. [Google Scholar] [CrossRef]
- Lozada, M.; Diéguez, M.C.; García, P.E.; Bigatti, G.; Livore, J.P.; Giarratano, E.; Gil, M.N.; Dionisi, H.M. Undaria pinnatifida exudates trigger shifts in seawater chemistry and microbial communities from Atlantic Patagonian coasts. Biol. Invasions 2021, 23, 1781–1801. [Google Scholar] [CrossRef]
- Reintjes, G.; Arnosti, C.; Fuchs, B.; Amann, R. Selfish, sharing and scavenging bacteria in the Atlantic Ocean: A biogeographical study of bacterial substrate utilisation. ISME J. 2019, 13, 1119–1132. [Google Scholar] [CrossRef]
- Van Vliet, D.M.; Palakawong Na Ayudthaya, S.; Diop, S.; Villanueva, L.; Stams, A.J.; Sánchez-Andrea, I. Anaerobic degradation of sulfated polysaccharides by two novel Kiritimatiellales strains isolated from Black Sea sediment. Front. Microbiol. 2019, 10, 253. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Alves, C.; Silva, J.; Pinteus, S.; Gaspar, H.; Pedrosa, R. Sulfated polysaccharides from macroalgae—A simple roadmap for chemical characterization. Polymers 2023, 15, 399. [Google Scholar] [CrossRef] [PubMed]
- Haggag, Y.A.; Abd Elrahman, A.A.; Ulber, R.; Zayed, A. Fucoidan in pharmaceutical formulations: A comprehensive review for smart drug delivery systems. Mar. Drugs 2023, 21, 112. [Google Scholar] [CrossRef]
- Kusaykin, M.I.; Silchenko, A.S.; Zakharenko, A.M.; Zvyagintseva, T.N. Fucoidanases. Glycobiology 2016, 26, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Colin, S.; Deniaud, E.; Jam, M.; Descamps, V.; Chevolot, Y.; Kervarec, N.; Yvin, J.-C.; Barbeyron, T.; Michel, G.; Kloareg, B. Cloning and biochemical characterization of the fucanase FcnA: Definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology 2006, 16, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Chang, Y.; Zhang, Y.; Mei, X.; Xue, C. Discovery and characterization of an endo-1, 3-fucanase from marine bacterium Wenyingzhuangia fucanilytica: A novel glycoside hydrolase family. Front. Microbiol. 2020, 11, 1674. [Google Scholar] [CrossRef]
- Liu, G.; Shen, J.; Chang, Y.; Mei, X.; Chen, G.; Zhang, Y.; Xue, C. Characterization of an endo-1, 3-fucanase from marine bacterium Wenyingzhuangia aestuarii: The first member of a novel glycoside hydrolase family GH174. Carbohydr. Polym. 2023, 306, 120591. [Google Scholar] [CrossRef]
- Shen, J.; Zheng, L.; Zhang, Y.; Chen, G.; Mei, X.; Chang, Y.; Xue, C. Discovery of a catalytic domain defines a new glycoside hydrolase family containing endo-1,3-fucanase. Carbohydr. Polym. 2024, 323, 121442. [Google Scholar]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Schultz-Johansen, M.; Cueff, M.; Hardouin, K.; Jam, M.; Larocque, R.; Glaring, M.A.; Hervé, C.; Czjzek, M.; Stougaard, P. Discovery and screening of novel metagenome-derived GH 107 enzymes targeting sulfated fucans from brown algae. FEBS J. 2018, 285, 4281–4295. [Google Scholar]
- Silchenko, A.S.; Ustyuzhanina, N.E.; Kusaykin, M.I.; Krylov, V.B.; Shashkov, A.S.; Dmitrenok, A.S.; Usoltseva, R.V.; Zueva, A.O.; Nifantiev, N.E.; Zvyagintseva, T.N. Expression and biochemical characterization and substrate specificity of the fucoidanase from Formosa algae. Glycobiology 2017, 27, 254–263. [Google Scholar]
- Zueva, A.; Silchenko, A.; Rasin, A.; Kusaykin, M.; Usoltseva, R.; Kalinovsky, A.; Kurilenko, V.; Zvyagintseva, T.; Thinh, P.; Ermakova, S. Expression and biochemical characterization of two recombinant fucoidanases from the marine bacterium Wenyingzhuangia fucanilytica CZ1127T. Int. J. Biol. Macromol. 2020, 164, 3025–3037. [Google Scholar] [PubMed]
- Vuillemin, M.; Silchenko, A.S.; Cao, H.T.T.; Kokoulin, M.S.; Trang, V.T.D.; Holck, J.; Ermakova, S.P.; Meyer, A.S.; Mikkelsen, M.D. Functional characterization of a new GH107 endo-α-(1,4)-fucoidanase from the marine bacterium Formosa haliotis. Mar. Drugs 2020, 18, 562. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Jiang, H.; Dong, Y.; Wang, Y.; Hamouda, H.I.; Balah, M.A.; Mao, X. Expression and biochemical characterization of a novel fucoidanase from Flavobacterium algicola with the principal product of fucoidan-derived disaccharide. Foods 2022, 11, 1025. [Google Scholar] [PubMed]
- Trang, V.T.D.; Mikkelsen, M.D.; Vuillemin, M.; Meier, S.; Cao, H.T.T.; Muschiol, J.; Perna, V.; Nguyen, T.T.; Tran, V.H.N.; Holck, J. The endo-α (1,4) specific fucoidanase Fhf2 from Formosa haliotis releases highly sulfated fucoidan oligosaccharides. Front. Plant Sci. 2022, 13, 823668. [Google Scholar] [CrossRef]
- Tran, V.H.N.; Nguyen, T.T.; Meier, S.; Holck, J.; Cao, H.T.T.; Van, T.T.T.; Meyer, A.S.; Mikkelsen, M.D. The endo-α (1,3)-fucoidanase Mef2 releases uniquely branched oligosaccharides from Saccharina latissima fucoidans. Mar. Drugs 2022, 20, 305. [Google Scholar]
- Sakai, T.; Kawai, T.; Kato, I. Isolation and characterization of a fucoidan-degrading marine bacterial strain and its fucoidanase. Mar. Biotechnol. 2004, 6, 335–346. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, Z.; Ren, L.; Jiao, S.; Zhang, X.; Wang, Q.; Li, Z.; Du, Y.; Li, J.-J. Overexpression and biochemical characterization of a truncated endo-α (1→3)-fucoidanase from Alteromonas sp. SN-1009. Food Chem. 2021, 353, 129460. [Google Scholar] [CrossRef]
- Vickers, C.; Liu, F.; Abe, K.; Salama-Alber, O.; Jenkins, M.; Springate, C.M.; Burke, J.E.; Withers, S.G.; Boraston, A.B. Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to α-L-fucosidases from GH29. J. Biol. Chem. 2018, 293, 18296–18308. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.H.N.; Nguyen, T.T.; Meier, S.; Holck, J.; Cao, H.T.T.; Van, T.T.T.; Meyer, A.S.; Mikkelsen, M.D. Structural and functional characterization of the novel endo-α(1,4)-fucoidanase Mef1 from the marine bacterium Muricauda eckloniae. Acta Crystallogr. D Struct. Biol. 2023, 79, 1026–1043. [Google Scholar]
- Sakai, T.; Ishizuka, K.; Kato, I. Isolation and characterization of a fucoidan-degrading marine bacterium. Mar. Biotechnol. 2003, 5, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Ohshiro, T.; Harada, N.; Kobayashi, Y.; Miki, Y.; Kawamoto, H. Microbial fucoidan degradation by Luteolibacter algae H18 with deacetylation. Biosci. Biotechnol. Biochem. 2012, 76, 620–623. [Google Scholar] [CrossRef]
- Lozada, M.; Diéguez, M.C.; García, P.E.; Dionisi, H.M. Microbial communities associated with kelp detritus in temperate and subantarctic intertidal sediments. Sci. Total Environ. 2023, 857, 159392. [Google Scholar] [CrossRef]
- Dionisi, H.M.; Lozada, M.; Campos, E. Diversity of GH51 α-L-arabinofuranosidase homolog sequences from subantarctic intertidal sediments. Biologia 2023, 78, 1899–1918. [Google Scholar]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef]
- Tamames, J.; Puente-Sánchez, F. SqueezeMeta, A Highly Portable, Fully Automatic Metagenomic Analysis Pipeline. Front. Microbiol. 2019, 9, 3349. [Google Scholar]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar]
- Commendatore, M.G.; Nievas, M.L.; Amin, O.; Esteves, J.L. Sources and distribution of aliphatic and polyaromatic hydrocarbons in coastal sediments from the Ushuaia Bay (Tierra del Fuego, Patagonia, Argentina). Mar. Environ. Res. 2012, 74, 20–31. [Google Scholar] [PubMed]
- Gil, M.N.; Torres, A.I.; Amin, O.; Esteves, J.L. Assessment of recent sediment influence in an urban polluted subantarctic coastal ecosystem. Beagle Channel (Southern Argentina). Mar. Pollut. Bull. 2011, 62, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [PubMed]
- Barrett, K.; Hunt, C.J.; Lange, L.; Meyer, A.S. Conserved unique peptide patterns (CUPP) online platform: Peptide-based functional annotation of carbohydrate active enzymes. Nucleic Acids Res. 2020, 48, W110–W115. [Google Scholar] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R. The IMG/M data management and analysis system v. 6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Oberg, N.; Zallot, R.; Gerlt, J.A. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) web resource for genomic enzymology tools. J. Molec. Biol. 2023, 435, 168018. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv 2022, preprint. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [PubMed]
- Holm, L. Dali server: Structural unification of protein families. Nucleic Acids Res. 2022, 50, W210–W215. [Google Scholar] [CrossRef] [PubMed]
- Yariv, B.; Yariv, E.; Kessel, A.; Masrati, G.; Chorin, A.B.; Martz, E.; Mayrose, I.; Pupko, T.; Ben-Tal, N. Using evolutionary data to make sense of macromolecules with a “face-lifted” ConSurf. Protein Sci. 2023, 32, e4582. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Gilles, K.; Hamilton, J.; Rebers, P.; Smith, F. A colorimetric method for the determination of sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef]
- Filisetti-Cozzi, T.M.; Carpita, N.C. Measurement of uronic acids without interference from neutral sugars. Anal. Biochem. 1991, 197, 157–162. [Google Scholar]
- Dodgson, K.; Price, R. A note on the determination of the ester sulphate content of sulphated polysaccharides. Biochem. J. 1962, 84, 106. [Google Scholar] [CrossRef]
- Ponce, N.M.; Pujol, C.A.; Damonte, E.B.; Flores, M.a.L.; Stortz, C.A. Fucoidans from the brown seaweed Adenocystis utricularis: Extraction methods, antiviral activity and structural studies. Carbohydr. Res. 2003, 338, 153–165. [Google Scholar] [CrossRef]
- Ponce, N.M.; Flores, M.L.; Pujol, C.A.; Becerra, M.B.; Navarro, D.A.; Córdoba, O.; Damonte, E.B.; Stortz, C.A. Fucoidans from the phaeophyta Scytosiphon lomentaria: Chemical analysis and antiviral activity of the galactofucan component. Carbohydr. Res. 2019, 478, 18–24. [Google Scholar]
- Conesa, A.L.; Dellatorre, F.G.; Latour, E.; Ponce, N.M.A.; Stortz, C.A.; Scolaro, L.A.; Álvarez, V.A.; Lassalle, V.L. Potential of Fucoidan From Myriogloea Major Asensi as Antiviral Against Herpes Simplex Type 1 and 2 and Bovine Coronavirus. Res. Sq. 2023, preprint. [Google Scholar] [CrossRef]
- Latour, E.; Dellatorre, F.G.; Ponce, N.M.A. Cuantificación y caracterización de fucoidanos de distintas algas pardas del Golfo Nuevo, Chubut, Argentina. Actas De Jorn. Y Even. Académicos De UTN 2022, 15, 1–6. [Google Scholar] [CrossRef]
- Atkinson, H.J.; Morris, J.H.; Ferrin, T.E.; Babbitt, P.C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE 2009, 4, e4345. [Google Scholar]
- Ferrelli, M.L.; Pidre, M.L.; García-Domínguez, R.; Alberca, L.N.; del Saz-Navarro, D.; Santana-Molina, C.; Devos, D.P. Prokaryotic membrane coat-like proteins: An update. J. Struct. Biol. 2023, 215, 107987. [Google Scholar] [CrossRef]
- Zou, P.; Yang, X.; Yuan, Y.; Jing, C.; Cao, J.; Wang, Y.; Zhang, L.; Zhang, C.; Li, Y. Purification and characterization of a fucoidan from the brown algae Macrocystis pyrifera and the activity of enhancing salt-stress tolerance of wheat seedlings. Int. J. Biol. Macromol. 2021, 180, 547–558. [Google Scholar] [CrossRef]
- Arijón, M.; Ponce, N.M.; Solana, V.; Dellatorre, F.G.; Latour, E.A.; Stortz, C.A. Monthly fluctuations in the content and monosaccharide composition of fucoidan from Undaria pinnatifida sporophylls from northern Patagonia. J. Appl. Phycol. 2021, 33, 2433–2441. [Google Scholar] [CrossRef]
- Bilan, M.I.; Grachev, A.A.; Ustuzhanina, N.E.; Shashkov, A.S.; Nifantiev, N.E.; Usov, A.I. A highly regular fraction of a fucoidan from the brown seaweed Fucus distichus L. Carbohydr. Res. 2004, 339, 511–517. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kusaykin, M.I.; Kurilenko, V.V.; Zakharenko, A.M.; Isakov, V.V.; Zaporozhets, T.S.; Gazha, A.K.; Zvyagintseva, T.N. Hydrolysis of fucoidan by fucoidanase isolated from the marine bacterium, Formosa algae. Mar. Drugs 2013, 11, 2413–2430. [Google Scholar]
- Silchenko, A.S.; Rasin, A.B.; Kusaykin, M.I.; Kalinovsky, A.I.; Miansong, Z.; Changheng, L.; Malyarenko, O.; Zueva, A.O.; Zvyagintseva, T.N.; Ermakova, S.P. Structure, enzymatic transformation, anticancer activity of fucoidan and sulphated fucooligosaccharides from Sargassum horneri. Carbohydr. Polym. 2017, 175, 654–660. [Google Scholar]
- Satoh, T.; Sato, K.; Kanoh, A.; Yamashita, K.; Yamada, Y.; Igarashi, N.; Kato, R.; Nakano, A.; Wakatsuki, S. Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p. J. Biol. Chem. 2006, 281, 10410–10419. [Google Scholar] [CrossRef]
- Sakka, M.; Kunitake, E.; Kimura, T.; Sakka, K. Function of a laminin_G_3 module as a carbohydrate-binding module in an arabinofuranosidase from Ruminiclostridium josui. FEBS Lett. 2019, 593, 42–51. [Google Scholar]
- Gupta, G.; Gupta, R.K.; Gupta, G. R-type lectin families. In Animal Lectins: Form, Function and Clinical Applications; Gupta, G.S., Ed.; Springer: Vienna, Austria, 2012; pp. 313–330. [Google Scholar]
- Boraston, A.B.; Tomme, P.; Amandoron, E.A.; Kilburn, D.G. A novel mechanism of xylan binding by a lectin-like module from Streptomyces lividans xylanase 10A. Biochem. J. 2000, 350, 933–941. [Google Scholar]
- Notenboom, V.; Boraston, A.B.; Williams, S.J.; Kilburn, D.G.; Rose, D.R. High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding. Biochemistry 2002, 41, 4246–4254. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Tian, X.; Li, X.; She, Q. Identification and structural analysis of a carbohydrate-binding module specific to alginate, a representative of a new family, CBM96. J. Biol. Chem. 2023, 299, 102854. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, S.; Jogler, M.; Boedeker, C.; Pinto, D.; Vollmers, J.; Rivas-Marín, E.; Kohn, T.; Peeters, S.H.; Heuer, A.; Rast, P. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat. Microbiol. 2020, 5, 126–140. [Google Scholar] [CrossRef]
- Friedlander, A.M.; Ballesteros, E.; Bell, T.W.; Caselle, J.E.; Campagna, C.; Goodell, W.; Hüne, M.; Muñoz, A.; Salinas-de-León, P.; Sala, E. Kelp forests at the end of the earth: 45 years later. PLoS ONE 2020, 15, e0229259. [Google Scholar] [CrossRef] [PubMed]
- Mahadev, R.K.; Di Pietro, S.M.; Olson, J.M.; Piao, H.L.; Payne, G.S.; Overduin, M. Structure of Sla1p homology domain 1 and interaction with the NPFxD endocytic internalization motif. EMBO J. 2007, 26, 1963–1971. [Google Scholar] [PubMed]
- Santarella-Mellwig, R.; Franke, J.; Jaedicke, A.; Gorjanacz, M.; Bauer, U.; Budd, A.; Mattaj, I.W.; Devos, D.P. The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 2010, 8, e1000281. [Google Scholar] [CrossRef]
- Krause-Jensen, D.; Duarte, C.M. Substantial role of macroalgae in marine carbon sequestration. Nat. Geosci. 2016, 9, 737–742. [Google Scholar] [CrossRef]
- Matos, M.N.; Lozada, M.; Anselmino, L.E.; Musumeci, M.A.; Henrissat, B.; Jansson, J.K.; Mac Cormack, W.P.; Carroll, J.; Sjöling, S.; Lundgren, L. Metagenomics unveils the attributes of the alginolytic guilds of sediments from four distant cold coastal environments. Environ. Microbiol. 2016, 18, 4471–4484. [Google Scholar] [CrossRef]
- Ponce, N.M.; Stortz, C.A. A comprehensive and comparative analysis of the fucoidan compositional data across the Phaeophyceae. Front. Plant Sci. 2020, 11, 556312. [Google Scholar]
- Amin, O.; Comoglio, L.; Spetter, C.; Duarte, C.; Asteasuain, R.; Freije, R.H.; Marcovecchio, J. Assessment of land influence on a high-latitude marine coastal system: Tierra del Fuego, southernmost Argentina. Environ. Monit. Assess. 2011, 175, 63–73. [Google Scholar] [PubMed]
- Li, P.; Wang, S.; Samo, I.A.; Zhang, X.; Wang, Z.; Wang, C.; Li, Y.; Du, Y.; Zhong, Y.; Cheng, C. Common-ion effect triggered highly sustained seawater electrolysis with additional NaCl production. Research 2020, 2020, 872141. [Google Scholar] [CrossRef] [PubMed]
- Niiranen, L.; Altermark, B.; Brandsdal, B.O.; Leiros, H.K.S.; Helland, R.; Smalås, A.O.; Willassen, N.P. Effects of salt on the kinetics and thermodynamic stability of endonuclease I from Vibrio salmonicida and Vibrio cholerae. FEBS J. 2008, 275, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Z.; Pan, X.; Wang, N.; Li, L.; Du, Y.; Li, J.; Li, M. Structural and biochemical analysis reveals catalytic mechanism of fucoidan lyase from Flavobacterium sp. SA-0082. Mar. Drugs 2022, 20, 533. [Google Scholar] [CrossRef] [PubMed]
- Ropartz, D.; Marion, L.; Fanuel, M.; Nikolic, J.; Jam, M.; Larocque, R.; Ficko-Blean, E.; Michel, G.; Rogniaux, H. In-depth structural characterization of oligosaccharides released by GH107 endofucanase Mf FcnA reveals enzyme subsite specificity and sulfated fucan substructural features. Glycobiology 2022, 32, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Arai, Y.; Shingu, Y.; Yagi, H.; Suzuki, H.; Ohshiro, T. Occurrence of different fucoidanase genes in Flavobacterium sp. SW and enzyme characterization. J. Biosci. Bioeng. 2022, 134, 187–194. [Google Scholar] [CrossRef]
- Sichert, A.; Le Gall, S.; Klau, L.J.; Laillet, B.; Rogniaux, H.; Aachmann, F.L.; Hehemann, J.-H. Ion-exchange purification and structural characterization of five sulfated fucoidans from brown algae. Glycobiology 2021, 31, 352–357. [Google Scholar] [CrossRef]
- Lorbeer, A.J.; Charoensiddhi, S.; Lahnstein, J.; Lars, C.; Franco, C.M.; Bulone, V.; Zhang, W. Sequential extraction and characterization of fucoidans and alginates from Ecklonia radiata, Macrocystis pyrifera, Durvillaea potatorum, and Seirococcus axillaris. J. Appl. Phycol. 2017, 29, 1515–1526. [Google Scholar]
- Zhang, W.; Oda, T.; Yu, Q.; Jin, J.-O. Fucoidan from Macrocystis pyrifera has powerful immune-modulatory effects compared to three other fucoidans. Mar. Drugs 2015, 13, 1084–1104. [Google Scholar] [CrossRef]
- Koh, H.S.A.; Lu, J.; Zhou, W. Structure characterization and antioxidant activity of fucoidan isolated from Undaria pinnatifida grown in New Zealand. Carbohydr. Polym. 2019, 212, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, M.G.; López Gappa, J.; Piriz, M.L. Latitudinal gradients in macroalgal biodiversity in the Southwest Atlantic between 36 and 55 S. Hydrobiologia 2011, 673, 205–214. [Google Scholar] [CrossRef]



| Species | Order, Family | Activity | Reference |
|---|---|---|---|
| * Macrocystis pyrifera | Laminariales; Laminariaceae | yes | [75] |
| Undaria pinnatifida | Laminariales; Alariaceae | nd | [76] |
| * Adenocystis utricularis | Ectocarpales; Adenocystaceae | nd | [69] |
| * Scytosiphon lomentaria | Ectocarpales; Scytosiphonaceae | nd | [70] |
| * Colpomenia sinuosa | Ectocarpales; Scytosiphonaceae | nd | [72] |
| Eudesme virescens | Ectocarpales; Chordariaceae | nd | [72] |
| Myriogloea major | Ectocarpales; Chordariaceae | nd | [71] |
| Asperococcus ensiformis | Ectocarpales; Chordariaceae | nd | [72] |
| Extract/ Fraction | MW (kDa) * | Yield (%) | Carboh. (% anh.) | Uronate (% anh.) | Sulfate (% SO3Na) | Neutral Monosaccharides (mol/100 mols) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rha | Fuc | Ara | Xyl | Man | Gal | Glc | ||||||
| BL | 864 | 6.0 | 50.2 | 5.7 | 29.4 | Tr. | 84 | 2 | 2 | 2 | 10 | Tr. |
| F4BL | 709.5 | 14.3 | 53.6 | Tr. | 41.4 | Tr. | 92 | Tr. | Tr. | Tr. | 7 | 1 |
| F1-P2 | 1.82 | 11.6 | - | - | 46.7 | Nd | 100 | nd | nd | nd | nd | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez, J.A.; Ponce, N.M.A.; Lozada, M.; Daglio, Y.; Stortz, C.A.; Dionisi, H.M. Fucanases Related to the GH107 Family from Members of the PVC Superphylum. J. Mar. Sci. Eng. 2024, 12, 181. https://doi.org/10.3390/jmse12010181
Gonzalez JA, Ponce NMA, Lozada M, Daglio Y, Stortz CA, Dionisi HM. Fucanases Related to the GH107 Family from Members of the PVC Superphylum. Journal of Marine Science and Engineering. 2024; 12(1):181. https://doi.org/10.3390/jmse12010181
Chicago/Turabian StyleGonzalez, Jessica A., Nora M. A. Ponce, Mariana Lozada, Yasmín Daglio, Carlos A. Stortz, and Hebe M. Dionisi. 2024. "Fucanases Related to the GH107 Family from Members of the PVC Superphylum" Journal of Marine Science and Engineering 12, no. 1: 181. https://doi.org/10.3390/jmse12010181
APA StyleGonzalez, J. A., Ponce, N. M. A., Lozada, M., Daglio, Y., Stortz, C. A., & Dionisi, H. M. (2024). Fucanases Related to the GH107 Family from Members of the PVC Superphylum. Journal of Marine Science and Engineering, 12(1), 181. https://doi.org/10.3390/jmse12010181









