Abstract
Iron is an essential element for the functioning of cellular processes. Ferritins, the major intracellular iron storage proteins, convert the free Fe2+ into the nontoxic Fe3+ which can be stored and transported where needed. To date, little is known about the iron metabolism in copepods; however, in these crustaceans, ferritins have been used as biomarkers of stress and diapause. A limiting factor of these studies has been the use of a single ferritin transcript as a biomarker. In this paper, we in silico mined the publicly available copepod transcriptomes to characterize the multiplicity of the ferritin transcripts in different orders and families. We also examined the expression of ferritin in three ecologically important copepods—Calanus finmarchicus, C. helgolandicus and Temora stylifera—during development and under stress conditions. A full-length transcript encoding ferritin heavy chain has been identified in all 27 mined transcriptomes, with 50% of the species possessing multiple transcripts. Ferritin expression increased in C. finmarchicus during the early–late development transition, and in T. stylifera females exposed to oxylipins at sea. Overall, our results suggest that copepod ferritins can be involved in iron storage, larval development and stress response, thus representing potential biomarker genes for ocean health status monitoring.
1. Introduction
In all animals, iron (Fe) is an essential element required for the functioning of many cellular processes and to meet nutritional requirements. It participates in various metabolic processes, including DNA synthesis, electron and oxygen transport and cellular oxidation mechanisms [1]. Iron must be absorbed from the diet into gut cells and then transported and stored for future use [2]. However, excess free Fe in living cells can be potentially toxic. In the presence of oxygen, Fe can be rapidly reduced to the ferrous ion (Fe2+) form and this may catalyze the generation of reactive oxygen species (ROS). High levels of ROS cause oxidative stress, which is harmful to cellular compounds such as lipids, DNA and proteins [3,4]. Thus, the maintenance of a balanced Fe metabolism is essential for the organism’s homeostasis [2].
In order to avoid cell harm, organisms possess iron storage proteins (transferrins and ferritins) that keep Fe in their cavity and transport it where needed [2,3]. Ferritins are globular proteins composed of multiple subunits, with 24 equivalent subunits assembled into a “cage-like” oligomer [2]. In the ferroxidase center, they convert the Fe2+ into a nontoxic, soluble and biologically non-reactive ferric ion (Fe3+). In vertebrates, ferritins have been classified in heavy (H) chain and light (L) chain subunits; in arthropods, ferritins are homologs to the vertebrate proteins (heavy-chain homolog, HCH, and light-chain homolog, LCH) [2,5]. The major difference is that H-chain ferritins possess the ferroxidase center and are capable of ferroxidase activity, while L-chain ferritins, lacking the ferroxidase center, are characterized by amino acid residues that induce nucleation of iron and may be responsible for the electron transfer across the protein cage [6]. A third type of ferritin, mitochondrial ferritin—a homopolymer with ferroxidase activity targeted to mitochondria—has been identified in mammals and Drosophila melanogaster (ferritin 3, heavy-chain homolog, Fer3HCH), (reviewed in [5]). Although intensively studied in mammals and yeast, much less is known about the structure and function of ferritins in arthropods. Three ferritins have been identified in the fruit fly D. melanogaster: the cytoplasmic ferritin 1 (HCH), the ferritin 2 (LCH) and the mitochondrial ferritin [2]. Their role in dietary iron efflux or delivery was confirmed by knockdown experiments of either HCH or LCH ferritin subunits by RNAi, which resulted in the downregulation of the protein level of both subunits and in Fe accumulation in the midgut (reviewed in [5]). In the cladoceran Daphnia pulex, genome annotation and phylogenetic analysis revealed that ferritins clearly expanded compared with insects with a total of seven distinct ferritin loci [7].
Scanty information is available on the iron metabolism in zooplankton organisms, particularly in copepods; however, in these crustaceans, ferritins have been intensively used as biomarkers of stress. In the neritic calanoid species Acartia tonsa, an increase in transcriptional expression of ferritin has been reported in individuals after exposure to nano-contaminants such as Ni and CdSe/ZnS quantum dots [8], after acclimation to high pCO2 conditions (1200 ppm) [9], and in response to infestation by the epibiotic euglenid Colacium vesiculosum [10]. Higher expression of a ferritin transcript was also reported in A. tonsa quiescent eggs compared with the subitaneous stage [11]. In the calanoid Calanus finmarchicus, ferritin has been suggested as a biomarker of stress associated with the diapause phase; a significant upregulation of ferritin was found in copepodites (CV) and females collected from deep water compared with individuals from the surface [12,13]. However, in all these studies, only a single ferritin gene has been used as a biomarker. In a recent study, genome analysis revealed the presence of a total of four ferritin genes (LsFer1–4) in the salmon louse Lepeophtheirus salmonis, with three encoding heavy chains (LsFer1,3,4) and one (LsFer2) encoding a light chain [14]. Based on these results, we might suggest that in copepods, the use of a single biomarker for ferritin might be limiting.
In consideration of the critical role of ferritin in Fe homeostasis, the aim of this study is to expand the understanding of the ferritin diversity and function in copepods. Mining the publicly available high-quality transcriptomes for several calanoid families [15,16,17], we examined the presence of transcripts encoding ferritins and compared them to homologous ferritins in D. melanogaster and in salmon louse L. salmonis. To provide a better understanding of the functioning of these genes, the expression of ferritins was examined using existing RNASeq data in C. finmarchicus, Calanus helgolandicus and Temora stylifera. Expression was examined across development (C. finmarchicus) [18] and after exposure to toxic algae (C. finmarchicus, C. helgolandicus, T. stylifera) [19,20,21,22]. This study expands the knowledge of the diversity of the ferritin family in copepods and suggests species-specific and stage-specific functional roles in these organisms. Our results shed light on the need to characterize the gene family of interest in particular when the genes are widely used as biomarkers in eco-physiological studies.
2. Materials and Methods
2.1. In Silico Workflow
Searches for putative transcripts encoding ferritins (HCH and LCH) were performed using a well-established vetting workflow that includes a mining step, a reciprocal blast and an examination of the protein structural domain [23,24]. Query sequences from the fruit fly Drosophila melanogaster (NP_524873, AAF07876, NP_572854) were used to mine the transcriptome shotgun assembly (TSA) database on the National Center for Biotechnology Information (NCBI), limiting the searches (tblastn algorithm) to Copepoda (taxid:6830) (search February 2023). For Rhincalanus gigas, raw reads were downloaded from NCBI [25] and assembled as described in [26]. Additional mining was performed using queries from the copepod Lepeophtheirus salmonis (order Siphonostomatoida) (BT121711, BT121232, MK887318, BT077723, BT121164) [14]. The results of both searches were compared and integrated.
All resulting transcripts were fully translated using ExPASY [27] and blasted against the NCBI non-redundant (nr) protein database (blastp algorithm) limited to Arthropoda (taxid:6656). The presence of the expected protein structural domain for ferritin (Pfam:PF00210) was examined using SMART software [28]. In the species with multiple transcripts, amino acid sequences were aligned and amino acid identity was calculated between pairs. Sequences with ≥95% amino acid identity were considered the same protein, and among those the longest sequence was kept. A final alignment was performed for the copepod transcripts identified in this study encoding full-length proteins and having the expected ferritin domain, with HCHs and LCHs from D. melanogaster, L. salmonis and Homo sapiens, in order to verify the presence of amino acids characterizing the ferroxidase center [14]. Amino acid sequence alignment was performed using MAFFT software [29].
2.2. Cladogram of Copepod Ferritin Genes
A phylogenetic analysis was performed to confirm the annotation of transcripts identified in this study and to establish their relationship to each other, to other copepod species and arthropods. An unrooted phylogenetic tree was generated with amino acid sequences from this study, and sequences previously identified from the copepods Acartia pacifica and Calanus sinicus (order: Calanoida) [8], Caligus rogercresseyi, C. clemensi, L. salmonis (order: Siphonostomatoida) [14] and Tigriopus californicus (order: Harpacticoida) [30]. We also considered sequences from the insects D. melanogaster and Anopheles aegypti, homologs from Homo sapiens [14] and the water flea Daphnia pulex [7]. Among the seven ferritin sequences identified in D. pulex, two partial sequences (DQ983427, DQ983426) were excluded from the analysis. All sequences were first aligned using ClustalW software (Galaxy version 2.1), and then a maximum-likelihood phylogenetic tree was built using the evolution model Kimura computing bootstrap for 1000 samples (RapidNJ, Galaxy version 2.3.2).
2.3. Expression of Ferritin in Calanus finmarchicus, Calanus helgolandicus and Temora stylifera
Relative expression of transcripts encoding for ferritins was examined in the copepods Calanus finmarchicus, Calanus helgolandicus and Temora stylifera (order: Calanoida), using existing RNASeq data. In C. finmarchicus, the expression of ferritins was examined across development [18] and in females exposed to a toxic dinoflagellate [19]. For the developmental dataset, adult C. finmarchicus and stage CV copepodites were collected from the Gulf of Maine in 2012. Wild-caught females were laboratory maintained to obtain the target developmental stages: embryos, early nauplii, early copepodites (CI) and late copepodites (CIV). From each stage, total RNA was extracted from three biological replicates (two for CI and CIV), and cDNA libraries were multiplexed and sequenced on an Illumina HiSeq 2000 platform (PE 100 bp) [18]. The second dataset includes C. finmarchicus females exposed to the saxitoxin-producing dinoflagellate Alexandrium fundyense [19]. Briefly, adult females collected from the Gulf of Maine (Mount Desert Rock, 2012) were laboratory incubated over a week with a low dose (LD: 50 cell mL−1) and high dose (HD: 200 cell mL−1) of the toxic dinoflagellate and with the non-toxic cryptophyte Rhodomonas baltica (8000 cells mL−1) as the control diet. Copepods were kept under 10 °C, on a 14:10 light:dark cycle and at two and five days, three biological replicates (15 females each) were harvested and processed for RNASeq. The two time points were chosen to test the hypothesis that the toxic algae would induce a detoxification response after two days that would persist over time (five days). Total RNA was extracted from each sample, followed by the generation of multiplexed cDNA libraries that were sequenced on an Illumina HiSeq 2000 platform (PE 100 bp) [19].
In C. helgolandicus, ferritin expression was derived from RNASeq data of laboratory-incubated females feeding on the oxylipin-producing toxic diatom Skeletonema marinoi (SKE) and the dinoflagellate control diet Prorocentrum minimum (PRO) for five days [20]. Briefly, females were collected in the Gulf of Naples (Central Tyrrhenian Sea, Western Mediterranean Sea, 2012) and laboratory incubated for five days with either S. marinoi (45,000 cells mL−1) or P. minimum (5000 cells mL−1) (three replicates per diet, 10 females each). Copepods were kept under 18 °C, on a 12:12 light:dark cycle, fed daily either with SKE or PRO and harvested after five days for RNASeq. Total RNA was extracted from each sample followed by the generation of multiplexed cDNA libraries that were sequenced on an Illumina platform HiSeq (PE 50 bp) [20]. Lastly, the T. stylifera dataset included females collected in the Gulf of Naples during two consecutive weeks in May 2017 (test: 23rd and control: 30th) when low–high reproductive fitness was associated to high–low oxylipin content in the natural phytoplankton assemblage, respectively [21]. Phytoplankton-derived oxylipins were measured from surface water samples collected on the two dates, as described in [22]. Wild-caught T. stylifera females were immediately flash-frozen in liquid nitrogen and stored for RNA extraction. Three replicates (10 females each) were harvested on both weeks, extracted for total RNA, processed for cDNA library preparation and sequenced on a HiSeq 2500 platform (PE 100 bp) [21]. For each copepod species, expression levels were quantified by mapping the RNASeq libraries against their species-specific reference transcriptome using Bowtie and reads were normalized by length using the RPKM methods reads per kilobase per million mapped reads (RPKM). RPKM in each species were compared among transcripts and among stages or treatments using two-way ANOVA (p < 0.0001) followed by Tukey’s post hoc test and multiple unpaired t-tests (p < 0.05). Statistical analyses were performed using GraphPad Prism, version 9 (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. Identification of Ferritins in Copepods
Ferritin-encoding transcripts were identified in 27 copepod species, including 22 from the order Calanoida, three from the order Harpacticoida and two from the order Cyclopoida (Table 1). Within the Calanoida order, transcripts were identified among different families, the majority within the Calanidae (e.g., Calanus finmarchicus, C. helgolandicus, C. propinquus, Calanoides acutus, Neocalanus flemingeri, N. cristatus and N. plumchrus), followed by the Temoridae (Temora stylifera, T. longicornis, Epischura baikalensis), the Acartiidae (Acartia clausi, A. tonsa), the Pontellidae (Labidocera madurae), the Pseudodiaptomidae (Pseudodiaptomus annandalei), the Centropagidae (Centropages hamatus) and the Rhincalanidae (Rhincalanus gigas) (Table 1). For all transcripts, reciprocal blast confirmed their annotation as a heavy chain (HCH) or light chain (LHC), with the majority being highly similar (top hit reciprocal blast) to homologs from L. salmonis (LS Fe1; BT121711), Calanus sinicus (APC62655) and Eurytemora affinis (XM_023489461). For the majority of the transcriptomes mined in this study, the resulting transcripts encoded full-length proteins with the typical conserved ferritin domain (Pfam00210; p-values < 0.05) (Table S1). Few partial transcripts, with no significant domain, were identified in N. flemingeri (2), C. helgolandicus (1), C. hamatus (1) and C. acutus (1), and these were not included in the downstream analyses.

Table 1.
Summary of ferritins in copepods. For each copepod, genus, species, order, family and number of transcripts encoding ferritin were listed. The table includes only ferritins that were identified as full length that passed the reciprocal blast and protein domain analysis (Table S1).
The number of transcripts encoding ferritins changed across copepods, ranging from one to a maximum of six. A single ferritin transcript was identified in 12 copepod species, whereas the highest diversification was found in N. flemigeri and T. longicornis, both having six different transcripts (Table 1). C. finmarchicus, Calanus pacificus, C. helgolandicus and T. stylifera showed four different ferritins. Lastly, three transcripts were identified in Calanus marshallae, C. hamatus and R. gigas.
Alignment of the copepod sequences identified in this study with ferritins from D. melanogaster, H. sapiens and L. salmonis showed that 57 out of 64 transcripts identified in this study conserved all the amino acids of the ferroxidase site in HCH from the reference sequences (Figure S1). The exception to this is the single ferritin from C. finmarchicus (GAXK01169093), C. helgolandicus (GJFL01003552) and N. flemingeri (GFUD01021847), and two ferritins from T. longicornis (GINW01248260, GINW01100697) and C. pacificus (GJQY01004559, GJQY01232255). While in the N. flemingeri and C. pacificus ferritins only a few substitutions were observed, in the other sequences five (out of seven) of the amino acids making the ferroxidase center were not highly conserved; both C. finmarchicus and C. helgolandicus showed in all the sites the same substitutions (Figure S1). The amino acid variability found in the ferroxidase center for C. finmarchicus, C. helgolandicus and T. longicornicus sequences might suggest that these ferritins belong to the LCH category, although further analyses are needed.
The unrooted tree generated from a total of 100 transcripts showed that ferritins clustered into several phylogenetic groups (Figure 1). LCH clustered in a separate group (Figure 1 light blue, bootstrap 98) including LCHs from D. melanogaster, Aedes aegypti, the copepods C. clemensi and L. salmonis (previously known), two T. longicornis transcripts, one from C. finmarchicus and one from C. helgolandicus (this study). The sequences in this group were the ones showing changes in the ferroxidase center in Figure S1. HCH ferritins are separated into several groups. The largest group (Figure 1 yellow, bootstrap 85) included known HCHs from the insects D. melanogaster and A. aegypti, the cladoceran D. pulex, the copepods L. salmonis, C. clemensi, C. rogergressery, E. affinis, T. californicus and 17 sequences identified in this study; these included ferritins from several species such as C. marshallae, N. flemingeri, L. madurae, A. clausi and A. tonsa, P. annanadalei, P. nana, T. longicornis and T. stylifera (two out of four). The second largest group (Figure 1 green, bootstrap 81) included only ferritins identified in this study (23) from C. finmarchicus (two out of four), C. helgolandicus (one), C. marshallae, (two out of three), N. flemingeri (three out of six), T. furcata and Tisbe holoturiae, P. annandalei and Pleuromamma xiphias. The remaining HCHs clustered in smaller groups. Among those, there was a small group including all D. pulex sequences (four out of five) and a single ferritin from C. propinquus (Figure 1, orange, bootstrap 58), and another group including six T. longicornis ferritins and two H. sapiens homologs (Figure 1, purple, bootstrap 51). None of the ferritin identified in this study clustered with the D. melanogaster mitochondrial ferritin which was separated from all other sequences.
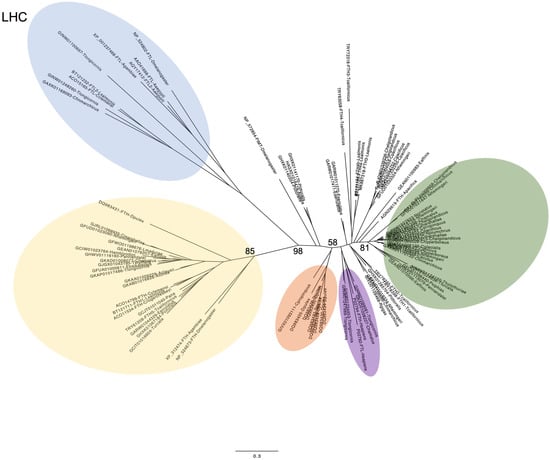
Figure 1.
Cladogram of ferritin-related genes identified in this study. The analysis includes sequences from this study (Table 1) from the insect D. melanogaster, the crustacean D. pulex and other copepods previously identified (see text). For the analysis, amino acid sequences were aligned using ClustalW and then a maximum-likelihood phylogenetic tree was built using the evolution model Kimura computing bootstrap for 1000 samples (RapidNJ, Galaxy version 2.3.2). Colors refer to the groups described in the text. Bootstrap is only indicated for the groups described. Scale bars: 0.3 estimated substitutions per site. LCH: light chain subunit.
3.2. Expression of ferritin in C. finmarchicus, Calanus helgolandicus and Temora stylifera
The expression of ferritin-encoding transcripts significantly changed in C. finmarchicus across the different developmental stages; a significant difference was also found among the different transcripts in each stage (two-way ANOVA, p < 0.001) (ta 2). Two ferritins, the HCH (GAXK01142559) and the LCH (GAXK01169093), showed a similar and low expression in all developmental stages, whereas HCH ferritin (GAXK01168686) was always highly expressed in copepodites (C1-C5) compared to embryos, copepodites and adults (568 RPKM vs. 82 RPKM, on average; Tukey’s multiple comparison test, p < 0.0001); similarly, the other HCH transcript (GAXK01170132) showed significantly higher expression in the later developmental stages (500 RPKM, on average, in C4 and AF), with a significant peak in expression in the C5 stage (Tukey’s multiple comparison test, p < 0.0001) (Figure 2).
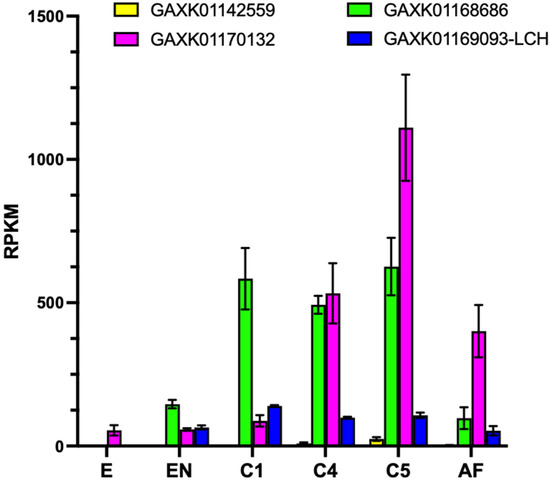
Figure 2.
Relative expression of ferritin transcripts in C. finmarchicus across development. Relative expression normalized by length (RPKM) across six developmental stages: embryos, early nauplii (NII-NIII), copepodites I, IV and V (C1, C4 and C5), and adult females (AF). Bars are mean ± standard deviation (n = 3 replicates, n = 2 C1 and C4). Colors indicate different transcripts and their NCBI accession number.
No significant differences were found in the expression of all ferritins in C. finmarchicus females feeding for two days on the toxic Alexandrium fundyense at different concentrations (low and high dose) compared to control conditions (Figure 3a,b). However, in spite of treatments and time points, the expression of the different transcripts was significantly different (two-way ANOVA, p < 0.0001). At two days, similarly to the previous dataset, two transcripts showed a null or very low expression in all treatments, whereas the transcripts GAXK01168686 and GAXK01170132 were significantly more abundant (average 300 RPKM) compared to the other two (Tukey’s multiple comparison test, p < 0.001) (Figure 3a). The same results were observed after five days of exposure, with the same two transcripts (GAXK01168686 and GAXK01170132) being significantly highly expressed in all conditions compared with the other two (Tukey’s multiple comparison test, p < 0.001) (Figure 3b).
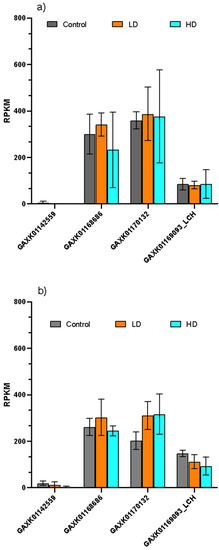
Figure 3.
Relative expression of ferritin transcripts in C. finmarchicus feeding on a toxic diet. Relative expression normalized by length (RPKM) in C. finmarchicus females fed the non-toxic cryptophyte Rhodomonas baltica (control) and the toxic dinoflagellate Alexandrium fundyense at low (orange) and high (cyan) doses, for 2 (a) and 5 (b) days. Different names indicate different transcripts. Bars are mean ± standard deviation (n = 3 replicates).
A similar result was also found in C. helgolandicus, where expression of the four ferritins did not change significantly between females feeding the toxic Skeletonema marinoi diet and the control treatment Prorocentrum minimum. However, similar to what was observed in C. finmarchicus, in spite of the treatments, one HCH ferritin (GJFL01006140) and the LCH ferritin (GJFL01003552) showed a significantly high expression (up to 20-fold and 60-fold difference) compared with the other two transcripts (Figure 4a). In T. stylifera females collected from the field during different weeks, the same transcript-dependent expression was observed: one transcript (GJGX01146428) showed ca. 100-fold higher expression compared to the other three; the same transcript was also significantly upregulated in females exposed to a high content of harmful oxylipins (test) (multiple unpaired t-test, p < 0.05) (Figure 4b).
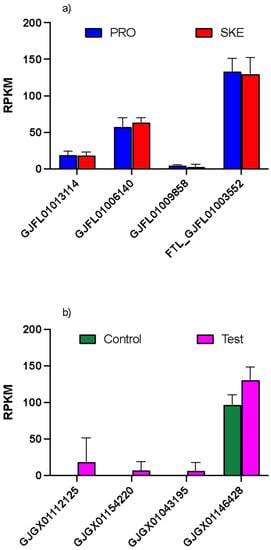
Figure 4.
Relative expression of ferritin transcripts in C. helgolandicus and T. stylifera. (a) Relative expression normalized by length (RPKM) in C. helgolandicus females fed the dinoflagellate Prorocentrum minimum (PRO) and the toxic diatom Skeletonema marinoi (SKE) for 5 days. (b) Relative expression normalized by length (RPKM) in T. stylifera collected in the Gulf of Naples when low (control) and high (test) content of harmful oxylipins were measured in the phytoplankton. Different names indicate different transcripts. Bars are mean ± standard deviation (n = 3 replicates).
4. Discussion
The evolution of life on Earth has been dependent on the availability of iron, leading to the evolution of ferritins, multimeric iron storage proteins that are ubiquitous in the living kingdom [1]. Most cells express ferritin genes, but protein concentration can vary among cell types. Ferritin sequence and structures are highly conserved among plants and animals, suggesting evolutionary convergence in eukaryotes, while bacterial ferritins diverge in sequence but not in structure (as reviewed in [31]).
Ferritins are also present in copepods, the most abundant metazoans on the planet [32]. Gene expression changes in ferritin have been reported in studies in the literature (Table 2) when copepods are exposed to abiotic stress, such as metals [8,33], heat shocks [10], high pCO2 levels [9] or water-accommodated fractions [34]. Changes in gene expression have also been recorded in response to biological drivers, namely, infestation by an epibiotic parasite [10], during different phases of dormancy, such as induction and recovery from quiescence [11], or during the diapause “stationary” phase [12,13]. In addition, it has been reported an impairment of feeding and reproduction upon the knockdown of ferritin, as well as a reduction in the protein level during starvation [14]. Interestingly, no changes in ferritin expression were associated with different crowding levels [35]. Overall, the reported differences in the regulation of ferritins opened questions related to stressor-specific responses associated with different transcripts or species-specific responses.

Table 2.
Summary of ferritin gene expression changes in copepods reported in the literature and our study. For each study, the species tested, the stressor and the regulation (+ = upregulation, − = downregulation and ns = non-significant change) of the ferritin gene are shown.
Indeed, the role of ferritin in the metabolism and physiology of copepods is presently understudied. Based on the relatively sparse available evidence (as summarized in Table 2), we speculate about different mechanisms potentially at play, which may differ not only among stressors but also among species. In this context, we interpret our results also by direct comparison with insect ferritins, considering their closeness in terms of absolute success [36].
Insect ferritins can be divided into three groups depending on their structure, location and function [37]. HCH and LCH differentiate based on the presence in HCH of the ferroxidase center, which confers the role of reducing the ferrous ion (Fe2+) to its non-reactive ferric ion (Fe3+). Insects also present a third type of protein, mitochondrial ferritin (HCH). Endoplasmic reticulum ferritins (HCH) act as iron transporters, mitochondrial ferritins act as antioxidants, while hemolymph ones (LCH) have a dual function [37]. In copepods, a full-length transcript encoding ferritin HCH has been identified in all 27 mined transcriptomes, with 50% of the species possessing multiple transcripts. Consistently with what was reported in the genome of Lepeophtheirus salmonis [14], mitochondrial ferritins were not found in any of the mined copepod transcriptomes. As opposed to insects [37], the location of ferritins has not been explored yet in copepods. From our results, mining transcripomes generated from whole or even pool of individuals, it is not possible to infer the specific location of the identified ferritins. Nonetheless, by analogy with insects, we can speculate that copepod ferritins are associated with the endoplasmic reticulum (HCH) and the hemolymph (LCH).
Arthropod immunity is based solely on an innate system [38]. In this framework, ferritin plays a key role, acting as a stress-induced protein and permitting the accumulation of iron which is no longer available to pathogens [38]. This process is compliant with the upregulation of ferritin in Acartia tonsa infested by the epibiotic euglenid Colacium vesiculosum [10]. Blood-sucking arthropods need further protection to reduce the risks associated with the massive ingestion of iron and heme [39]. In the yellow fever mosquito Aedes aegypti, ferritin expression is upregulated upon iron uptake, likely as cytotoxic protection [39,40]. Consistently, in the parasitic copepod L. salmonis ferritin levels in the midgut decrease during starvation [14], evidencing the direct link between gene expression and blood-feeding.
Involvement of ferritin has also been suggested during dormancy in the fly D. melanogaster, in the crustacean branchiopod Artemia sp., and in the copepods C. finmarchicus and A. tonsa. High expression of ferritins in Artemia sp. and A. tonsa embryos was hypothesized as preventing embryogenesis to continue and inhibiting development during the resting state [11,41]. Indeed, an increase in ferritin corresponds to higher chelation of iron stores and a reduction of iron available for processes such as embryogenesis [42]. Consistent with its role as an enhancer of stress resistance, high expression of ferritin has been found in both D. melanogaster and C. finmarchicus (C5, females) when at diapause [12,13,42,43]. Diapause, a type of dormancy, is characterized by a delay in development, a decrease in metabolism and an increase in stress resistance [44]. In Calanus spp., it has been reported that ferritin expression has its highest value during the early stages of diapause and it decreases at the beginning of the maintenance phase [12,13,42]. However, in another study, ferritin was not found among the genes differentiating C5s preparing for diapause from the ones on the reproductive program [45]. Furthermore, it seems that the ferritin role could be species-specific, as suggested by the fact that in Neocalanus flemingeri diapausing females ferritins were not among the genes characterizing the diapause phenotype [46].
In six out of the eleven papers published in the literature on copepod ferritins, the target species is the calanoid A. tonsa [8,9,10,11,34,35]. A renowned global-scale invasive species (e.g., [47,48]), A. tonsa is an established model organism in ecotoxicology studies (e.g., [49]) and it has proven valuable as feed for fish larvae in mariculture (e.g., [50]). The remaining papers surveyed are centered on two pelagic (Calanus spp.: [12,13,42]) and one parasitic (L. salmonis: [14]) species, relevant for their ecological (Calanus spp.) and economic (L. salmonis) impacts. Ferritin expression studies in these species allow the development of sensitive biomarkers for marine pollution monitoring, but also for the implementation of ecological indicators. The present study surveys the ferritin distribution in 27 copepods from the Calanoida, Harpacticoida and Cyclopoida orders, and it explores the variability of ferritin expression in three key ecological species: C. finmarchicus, C. helgolandicus and T. stylifera. Our study highlights the fact that the use of a single transcript as a biomarker can be highly limiting. In more than 50% of the examined copepods, in fact, we found more than one transcript encoding ferritin and these included C. finmarchicus, C. helgolandicus and T. stylifera. Moreover, our expression studies highlighted the fact that in spite of the stressor or developmental stage, two transcripts (out of four) were always more abundant compared with the others in all three species. Most protein-coding genes have dominant transcripts that are expressed at a considerably higher level than any minor transcripts across different conditions [51,52]. It is likely that these dominant transcripts are the main contributors to the proteome of the individual; thus, they might be used as good candidates for physiological and ecological studies on copepods.
Based on our results, we suggest that the role of ferritin in stress is still unclear. Changes in the expression of ferritins were only found in T. stylifera in response to different levels of the cytotoxic oxylipins in the natural phytoplankton assemblage; in contrast to the two Calanus species exposed to toxic algae, expression of ferritin did not change compared with a control diet. However, in C. helgolandicus, exposure to the harmful S. marinoi was associated with the upregulation of detoxification (GSTs), protein repair (HSP60 and HSP70), and immune system (prophenoloxidase activating enzyme, PPAE) genes [20]. Thus, we can speculate that the lack of a significant regulation of the ferritins could be associated with the fact that the concerted over-expression of detoxification genes could be sufficient to protect the copepod from the direct toxic effect of the diatom. In contrast, in T. stylifera, a high concentration of oxylipins induced the downregulation of stress response and oxidation–reduction genes [21]. Therefore, the upregulation of ferritin transcripts in T. stylifera could act as a compensatory defensive mechanism protecting the copepods from the oxidative damage and apoptosis associated with the ingestion of oxylipins [48]. Concerning C. finmarchicus, the differences in the expression found across development and not in response to the toxic algae could suggest a role of ferritin as a developmental marker. Two dominant ferritins were always poorly expressed in embryos and nauplii, compared to copepodites, confirming that iron mobilization due to low ferritin expression might stimulate embryogenesis and early larval development in copepods [43]. These ferritin-encoding dominant transcripts, thus, could be used as potential biomarkers of early–late development transition in C. finmarchicus.
5. Conclusions
Ferritins are the major intracellular iron storage proteins, highly conserved and present in most prokaryotes and eukaryotes including fungi, algae, plants, and animals [1]. Their expression in copepods is regulated by numerous endogenous and exogenous factors and may also present species-specific modulations. Our study expands the knowledge on the ferritin diversity in copepods; in all species, we found a high chain ferritin (HCH) and we confirm the lack of mitochondrial ferritin in crustaceans. We suggest that copepod ferritins can be involved in multiple processes such as iron storage, larval development and stress, highlighting that ferritin regulation can be species-specific, stressor-specific and stage-specific.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jmse11061187/s1. Table S1: Summary of reciprocal blast and protein domain analysis for the investigated ferritins. For each species, NCBI accession number (BioProject and the single transcript), E-value of the reciprocal blast, annotation result, top hit (species) and its accession number. Additionally, information includes the presence of the Pfam domain (PF00210) and its E value. The list only includes full-length sequences. Figure S1: Alignment of ferritin sequences identified for copepods in this study with known sequences from D. melanogaster (NP_524873, NP_524802, NP_572854), L. salmonis (LsFer1–4) (BT121711, BT121232, MK887318, BT077723, BT121164) and homologs from H. Sapiens (P02794, P02792). Copepod sequences are in alphabetical order as in Table 1. Light chain subunits (LCH) are indicated in green. Amino acids that make up the ferroxidase center of the heavy chain in H. Sapiens are bolded in red. Highlighted in yellow are the amino acids which were not conserved.
Author Contributions
Conceptualization, V.R.; writing—original draft preparation, V.R., M.U. and Y.C.; writing—review and editing, V.R., M.U. and Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
M.U. acknowledges the support of NBFC to Stazione Zoologica Anton Dohrn, funded by the Italian Ministry of University and Research, PNRR, Missione 4 Componente 2, “Dalla ricerca all’impresa”, Investimento 1.4, Project CN00000033.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The National Center for Biotechnology Information (NCBI) Bioproject numbers for the datasets examined in the present study are indicated in Table S1. Supplementary File S1 includes FASTA files for the transcript-encoding protein identified in this study.
Acknowledgments
We would like to thank Hartline and Lenz from the University of Hawaii at Manoa for the intellectual discussion.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Theil, E.C. Ferritin: Structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu. Rev. Biochem. 1987, 56, 289–315. [Google Scholar] [CrossRef] [PubMed]
- Nichol, H.; Law, J.H.; Winzerling, J.J. Iron metabolism in insects. Annu. Rev. Entomol. 2002, 47, 535–559. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Ingrassia, R.; Cavadini, P. Ferritins: A family of molecules for iron storage, antioxidation and more. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2009, 1790, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Emerit, J.; Beaumont, C.; Trivin, F. Iron metabolism, free radicals, and oxidative injury. Biomed. Pharmacother. 2001, 55, 333–339. [Google Scholar] [CrossRef]
- Tang, X.; Zhou, B. Ferritin is the key to dietary iron absorption and tissue iron detoxification in Drosophila melanogaster. FASEB J. 2013, 27, 288–298. [Google Scholar] [CrossRef]
- Hamburger, A.E.; West, A.P., Jr.; Hamburger, Z.A.; Hamburger, P.; Bjorkman, P.J. Crystal structure of a secreted insect ferritin reveals a symmetrical arrangement of heavy and light chains. J. Mol. Biol. 2005, 349, 558–569. [Google Scholar] [CrossRef]
- Colbourne, J.K.; Eads, B.D.; Shaw, J.; Bohuski, E.; Bauer, D.J.; Andrews, J. Sampling Daphnia’s expressed genes: Preservation, expansion and invention of crustacean genes with reference to insect genomes. BMC Genom. 2007, 8, 217. [Google Scholar] [CrossRef]
- Zhou, C.; Hou, J.; Lin, D. A ferritin gene in the marine copepod Acartia tonsa as a highly sensitive biomonitor for nano-contamination. Aquat. Toxicol. 2022, 253, 106353. [Google Scholar] [CrossRef]
- Aguilera, V.M.; Vargas, C.A.; Lardies, M.A.; Poupin, M.J. Adaptive variability to low-pH river discharges in Acartia tonsa and stress responses to high PCO2 conditions. Mar. Ecol. 2016, 37, 215–226. [Google Scholar] [CrossRef]
- Petkeviciute, E.; Kania, P.W.; Skovgaard, A. Genetic responses of the marine copepod Acartia tonsa (Dana) to heat shock and epibiont infestation. Aquac. Rep. 2015, 2, 10–16. [Google Scholar] [CrossRef]
- Nilsson, B.; Jepsen, P.M.; Rewitz, K.; Hansen, B.W. Expression of hsp70 and ferritin in embryos of the copepod Acartia tonsa (Dana) during transition between subitaneous and quiescent state. J. Plankton Res. 2014, 36, 513–522. [Google Scholar] [CrossRef]
- Aruda, A.M.; Baumgartner, M.F.; Reitzel, A.M.; Tarrant, A.M. Heat shock protein expression during stress and diapause in the marine copepod Calanus finmarchicus. J. Insect Physiol. 2011, 57, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Tarrant, A.M.; Baumgartner, M.F.; Verslycke, T.; Johnson, C.L. Differential gene expression in diapausing and active Calanus finmarchicus (Copepoda). Mar. Ecol. Prog. Ser. 2008, 355, 193–207. [Google Scholar] [CrossRef]
- Heggland, E.I.; Tröße, C.; Eichner, C.; Nilsen, F. Heavy and light chain homologs of ferritin are essential for blood-feeding and egg production of the ectoparasitic copepod Lepeophtheirus salmonis. Mol. Biochem. Parasitol. 2019, 232, 111197. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Carotenuto, Y.; Roncalli, V. Glutathione S-transferases in marine copepods. J. Mar. Sci. Eng. 2021, 9, 1025. [Google Scholar] [CrossRef]
- Roncalli, V.; Uttieri, M.; Capua, I.D.; Lauritano, C.; Carotenuto, Y. Chemosensory-Related Genes in Marine Copepods. Mar. Drugs 2022, 20, 681. [Google Scholar] [CrossRef]
- Roncalli, V.; Lauritano, C.; Carotenuto, Y. First report of OvoA gene in marine arthropods: A new candidate stress biomarker in copepods. Mar. Drugs 2021, 19, 647. [Google Scholar] [CrossRef]
- Cieslak, M.C.; Castelfranco, A.M.; Roncalli, V.; Lenz, P.H.; Hartline, D.K. t-Distributed Stochastic Neighbor Embedding (t-SNE): A tool for eco-physiological transcriptomic analysis. Mar. Genom. 2020, 51, 100723. [Google Scholar] [CrossRef]
- Roncalli, V.; Cieslak, M.C.; Lenz, P.H. Transcriptomic responses of the calanoid copepod Calanus finmarchicus to the saxitoxin producing dinoflagellate Alexandrium fundyense. Sci. Rep. 2016, 6, 25708. [Google Scholar] [CrossRef]
- Asai, S.; Sanges, R.; Lauritano, C.; Lindeque, P.K.; Esposito, F.; Ianora, A.; Carotenuto, Y. De novo transcriptome assembly and gene expression profiling of the copepod Calanus helgolandicus feeding on the PUA-producing diatom Skeletonema marinoi. Mar. Drugs 2020, 18, 392. [Google Scholar] [CrossRef]
- Russo, E.; Lauritano, C.; d’Ippolito, G.; Fontana, A.; Sarno, D.; von Elert, E.; Ianora, A.; Carotenuto, Y. RNA-Seq and differential gene expression analysis in Temora stylifera copepod females with contrasting non-feeding nauplii survival rates: An environmental transcriptomics study. Bmc Genom. 2020, 21, 693. [Google Scholar] [CrossRef]
- Russo, E.; d’Ippolito, G.; Fontana, A.; Sarno, D.; D’Alelio, D.; Busseni, G.; Ianora, A.; von Elert, E.; Carotenuto, Y. Density-dependent oxylipin production in natural diatom communities: Possible implications for plankton dynamics. ISME J. 2020, 14, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Christie, A.E.; Fontanilla, T.M.; Nesbit, K.T.; Lenz, P.H. Prediction of the protein components of a putative Calanus finmarchicus (Crustacea, Copepoda) circadian signaling systems using a de novo assembled transcriptome. Comp. Biochem. Physiol. D-Genom. Proteom. 2013, 8, 165–193. [Google Scholar] [CrossRef] [PubMed]
- Roncalli, V.; Cieslak, M.C.; Passamaneck, Y.; Christie, A.E.; Lenz, P.H. Glutathione S-transferase (GST) gene diversity in the crustacean Calanus finmarchicus–contributors to cellular detoxification. PLoS ONE 2015, 10, e0123322. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Roncalli, V.; Ambrosino, L.; Cieslak, M.C.; Ianora, A. First de novo transcriptome of the copepod Rhincalanus gigas from Antarctic waters. Biology 2020, 9, 410. [Google Scholar] [CrossRef] [PubMed]
- Hartline, D.K.; Cieslak, M.C.; Castelfranco, A.M.; Lieberman, B.; Roncalli, V.; Lenz, P.H. De novo transcriptomes of six calanoid copepods (Crustacea): A resource for the discovery of novel genes. Sci. Data 2023, 10, 242. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Barreto, F.S.; Watson, E.T.; Lima, T.G.; Willett, C.S.; Edmands, S.; Li, W.; Burton, R.S. Genomic signatures of mitonuclear coevolution across populations of Tigriopus californicus. Nat. Ecol. Evol. 2018, 2, 1250–1257. [Google Scholar] [CrossRef]
- Andrews, S.C. The Ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2010, 1800, 691–705. [Google Scholar] [CrossRef] [PubMed]
- Uttieri, M. Trends in copepod studies. In Trends in Copepod Studies—Distribution, Biology and Ecology; Nova Science Publishers Inc.: New York, NY, USA, 2018; pp. 1–11. [Google Scholar]
- Jiang, J.-L.; Wang, G.-Z.; Mao, M.-G.; Wang, K.-J.; Li, S.-J.; Zeng, C.-S. Differential gene expression profile of the calanoid copepod, Pseudodiaptomus annandalei, in response to nickel exposure. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2013, 157, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Hafez, T.; Bilbao, D.; Etxebarria, N.; Duran, R.; Ortiz-Zarragoitia, M. Application of a biological multilevel response approach in the copepod Acartia tonsa for toxicity testing of three oil Water Accommodated Fractions. Mar. Environ. Res. 2021, 169, 105378. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, B.; Jakobsen, H.H.; Stief, P.; Drillet, G.; Hansen, B.W. Copepod swimming behavior, respiration, and expression of stress-related genes in response to high stocking densities. Aquac. Rep. 2017, 6, 35–42. [Google Scholar] [CrossRef]
- Schminke, H.K. Entomology for the copepodologist. J. Plankton Res. 2007, 29, i149–i162. [Google Scholar] [CrossRef]
- Pham, D.Q.; Winzerling, J.J. Insect ferritins: Typical or atypical? Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2010, 1800, 824–833. [Google Scholar] [CrossRef]
- Whiten, S.R.; Eggleston, H.; Adelman, Z.N. Ironing out the details: Exploring the role of iron and heme in blood-sucking arthropods. Front. Physiol. 2018, 8, 1134. [Google Scholar] [CrossRef]
- Dunkov, B.C.; Georgieva, T.; Yoshiga, T.; Hall, M.; Law, J.H. Aedes aegypti ferritin heavy chain homologue: Feeding of iron or blood influences message levels, lengths and subunit abundance. J. Insect Sci. 2002, 2, 7. [Google Scholar] [CrossRef]
- Geiser, D.L.; Chavez, C.A.; Flores-Munguia, R.; Winzerling, J.J.; Pham, D.Q.D. Aedes aegypti ferritin: A cytotoxic protector against iron and oxidative challenge? Eur. J. Biochem. 2003, 270, 3667–3674. [Google Scholar] [CrossRef]
- Chen, T.; Amons, R.; Clegg, J.S.; Warner, A.H.; MacRae, T.H. Molecular characterization of artemin and ferritin from Artemia franciscana. Eur. J. Biochem. 2003, 270, 137–145. [Google Scholar] [CrossRef]
- Skottene, E.; Tarrant, A.M.; Olsen, A.J.; Altin, D.; Østensen, M.-A.; Hansen, B.H.; Choquet, M.; Jenssen, B.M.; Olsen, R.E. The β-oxidation pathway is downregulated during diapause termination in Calanus copepods. Sci. Rep. 2019, 9, 16686. [Google Scholar] [CrossRef] [PubMed]
- Kučerová, L.; Kubrak, O.I.; Bengtsson, J.M.; Strnad, H.; Nylin, S.; Theopold, U.; Nässel, D.R. Slowed aging during reproductive dormancy is reflected in genome-wide transcriptome changes in Drosophila melanogaster. Bmc Genom. 2016, 17, 50. [Google Scholar] [CrossRef] [PubMed]
- Lenz, P.H.; Roncalli, V. Diapause within the Context of Life-History Strategies in Calanid Copepods (Calanoida: Crustacea). Biol. Bull 2019, 237, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Lenz, P.H.; Roncalli, V.; Cieslak, M.C.; Tarrant, A.M.; Castelfranco, A.M.; Hartline, D.K. Diapause vs. reproductive programs: Transcriptional phenotypes in a keystone copepod. Commun. Biol. 2021, 4, 426. [Google Scholar] [CrossRef]
- Roncalli, V.; Cieslak, M.C.; Castelfranco, A.M.; Hopcroft, R.R.; Hartline, D.K.; Lenz, P.H. Post-diapause transcriptomic restarts: Insight from a high-latitude copepod. BMC Genom. 2021, 22, 409. [Google Scholar]
- Barroeta, Z.; Villate, F.; Uriarte, I.; Iriarte, A. Impact of Colonizer Copepods on Zooplankton Structure and Diversity in Contrasting Estuaries. Estuaries Coasts 2022, 45, 2592–2609. [Google Scholar] [CrossRef]
- Camatti, E.; Pansera, M.; Bergamasco, A. The copepod Acartia tonsa dana in a microtidal Mediterranean lagoon: History of a successful invasion. Water 2019, 11, 1200. [Google Scholar] [CrossRef]
- Carotenuto, Y.; Vitiello, V.; Gallo, A.; Libralato, G.; Trifuoggi, M.; Toscanesi, M.; Lofrano, G.; Esposito, F.; Buttino, I. Assessment of the relative sensitivity of the copepods Acartia tonsa and Acartia clausi exposed to sediment-derived elutriates from the Bagnoli-Coroglio industrial area. Mar. Environ. Res. 2020, 155, 104878. [Google Scholar] [CrossRef]
- Støttrup, J.G.; Richardson, K.; Kirkegaard, E.; Pihl, N.J. The cultivation of Acartia tonsa Dana for use as a live food source for marine fish larvae. Aquaculture 1986, 52, 87–96. [Google Scholar] [CrossRef]
- Gonzàlez-Porta, M.; Frankish, A.; Rung, J.; Harrow, J.; Brazma, A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013, 14, R70. [Google Scholar] [CrossRef]
- Taneri, B.; Snyder, B.; Gaasterland, T. Distribution of alternatively spliced transcript isoforms within human and mouse transcriptomes. J. Omics Res. 2011, 1, 1–5. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).