Microbial Biofilms Colonizing Plastic Substrates in the Ross Sea (Antarctica)
Abstract
1. Introduction
- To study the prokaryotic and microalgal communities within the microbial biofilms and to assess their spatial patterns in relation to the physico-chemical, trophic (nutrient concentration) and microbiological characteristics of the examined sites
- To analyze variations on a short- and long-term timescale of the abundance, biomass and functional metabolism of microbial biofilm assemblage components
- To investigate whether, and to what extent, a differential response of microbial biofilm community (prokaryotes and microalgae) can be envisaged in the taxonomic structure, prokaryotic abundance and metabolism in sites exposed to natural perturbations (such as salinity gradients due to the proximity to a glacier in Tethys Bay) or anthropogenic disturbance (such as the discharge of treated wastewater in Road Bay) compared to control, unperturbed, sites
- To identify potential associations between a particular typology of pressure (natural/anthropic) and key features (i.e., prokaryotic abundance/metabolic patterns dominant algal species,) and evaluate their role as candidate bioindicators of environmental changes in this extreme environment.
2. Materials and Methods
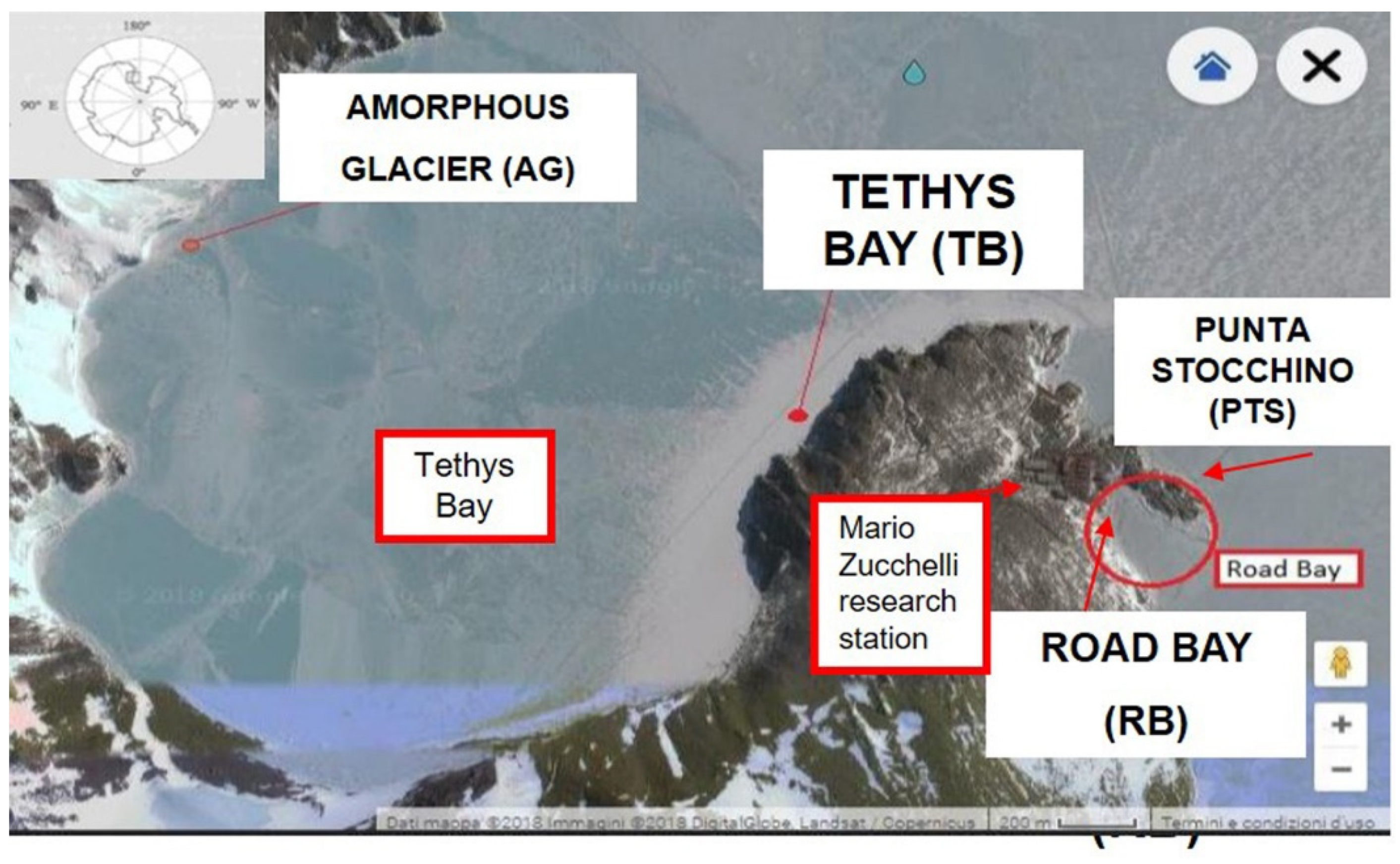
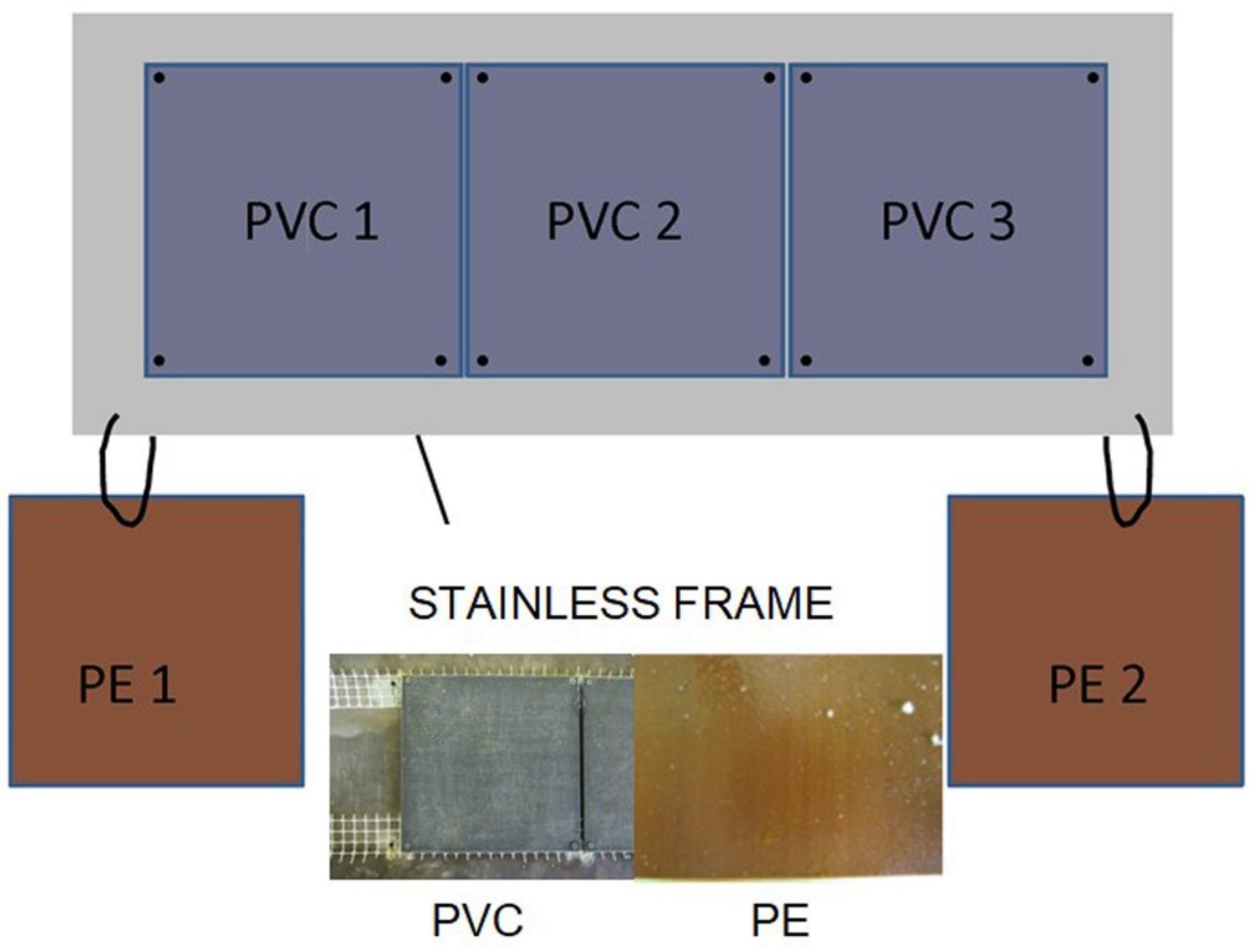
2.1. Experimental Design
- (1)
- Road Bay, a small bay near the Mario Zucchelli station research settlement, was chosen as representative of a human-impacted area since its waters receive the treated wastewater coming from the research station;
- (2)
- Tethys Bay, a large and isolated bay 2 km north of Road Bay, was chosen as representative of a naturally-impacted area, due to the presence of a large glacier (Amorphous Glacier) approaching the shore, as a natural forcing; here, a salinity gradient was expected to occur following climate warming.
2.2. Environmental Parameters
2.3. Prokaryotic Community: Abundance, Biomass, Morphological Types and Metabolism
2.4. Microalgae: Abundance, Biomass and Composition
2.5. Data Elaboration and Statistical Analyses
3. Results
3.1. Physical and Chemical Characterization of the Study Areas
3.2. Microbial Biofilm Community
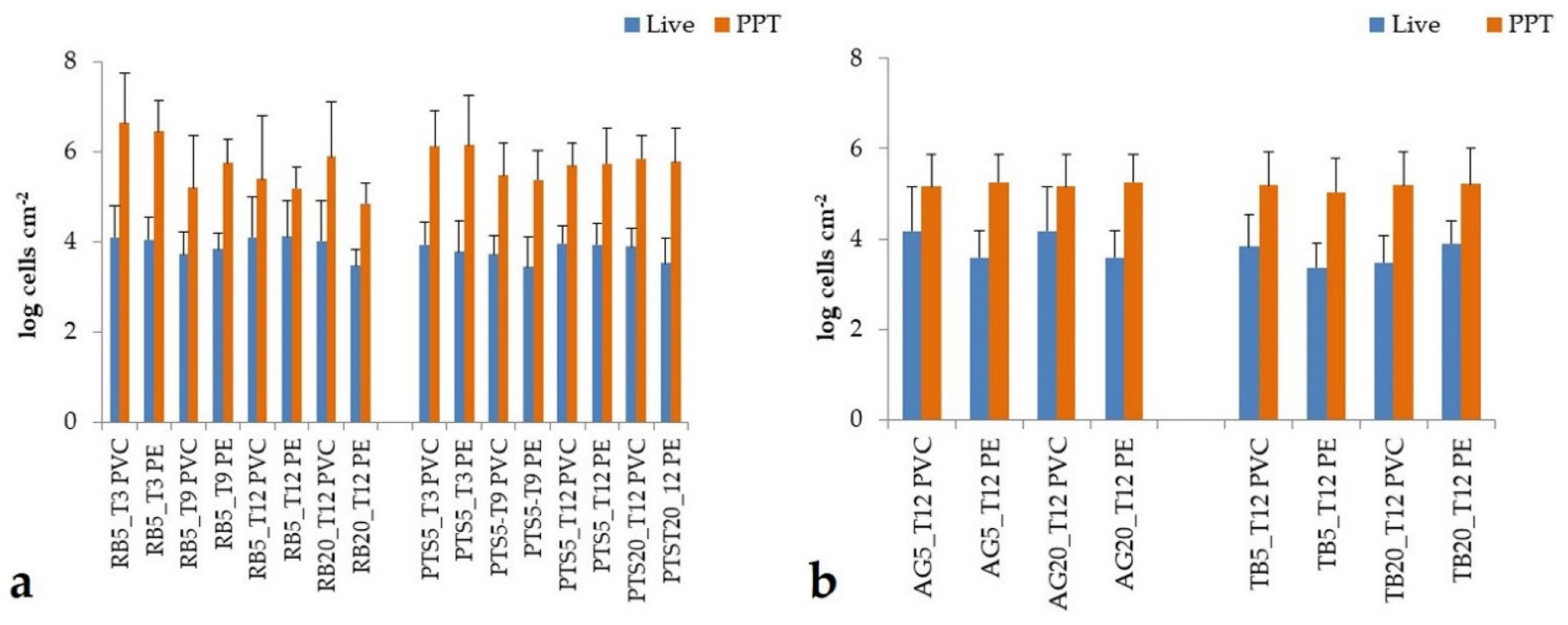
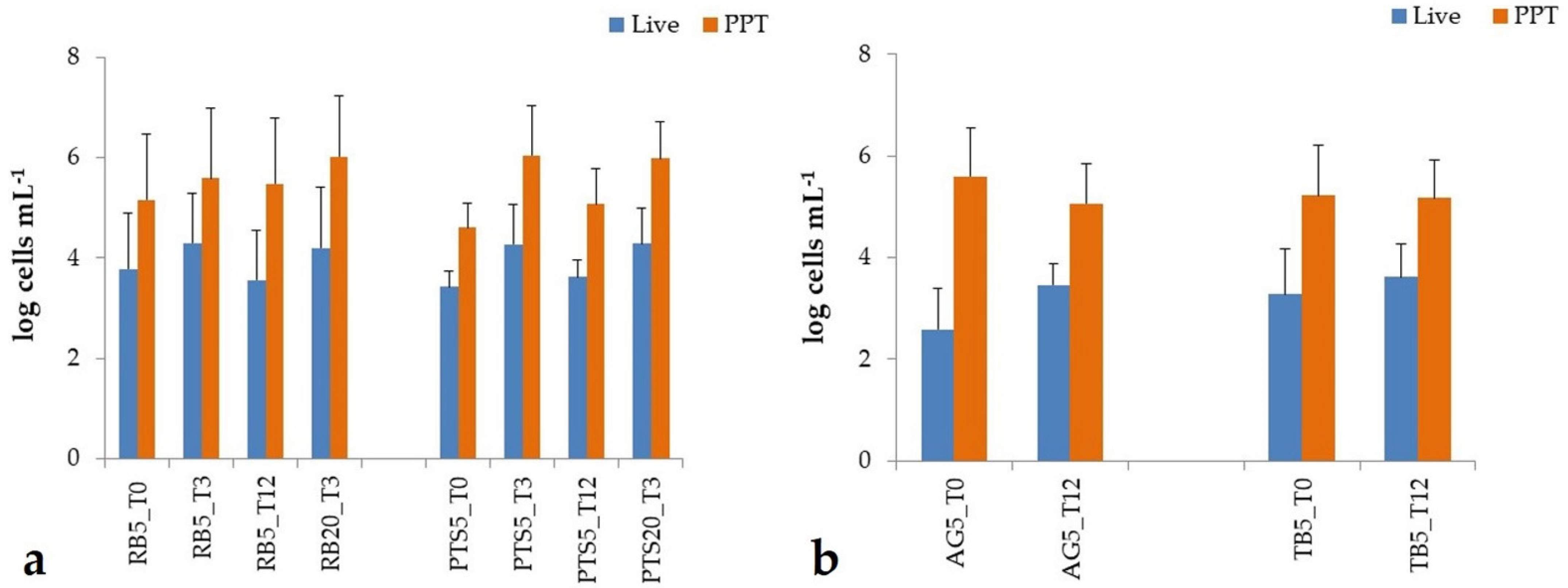
3.2.1. Prokaryotic Biofilm Abundance
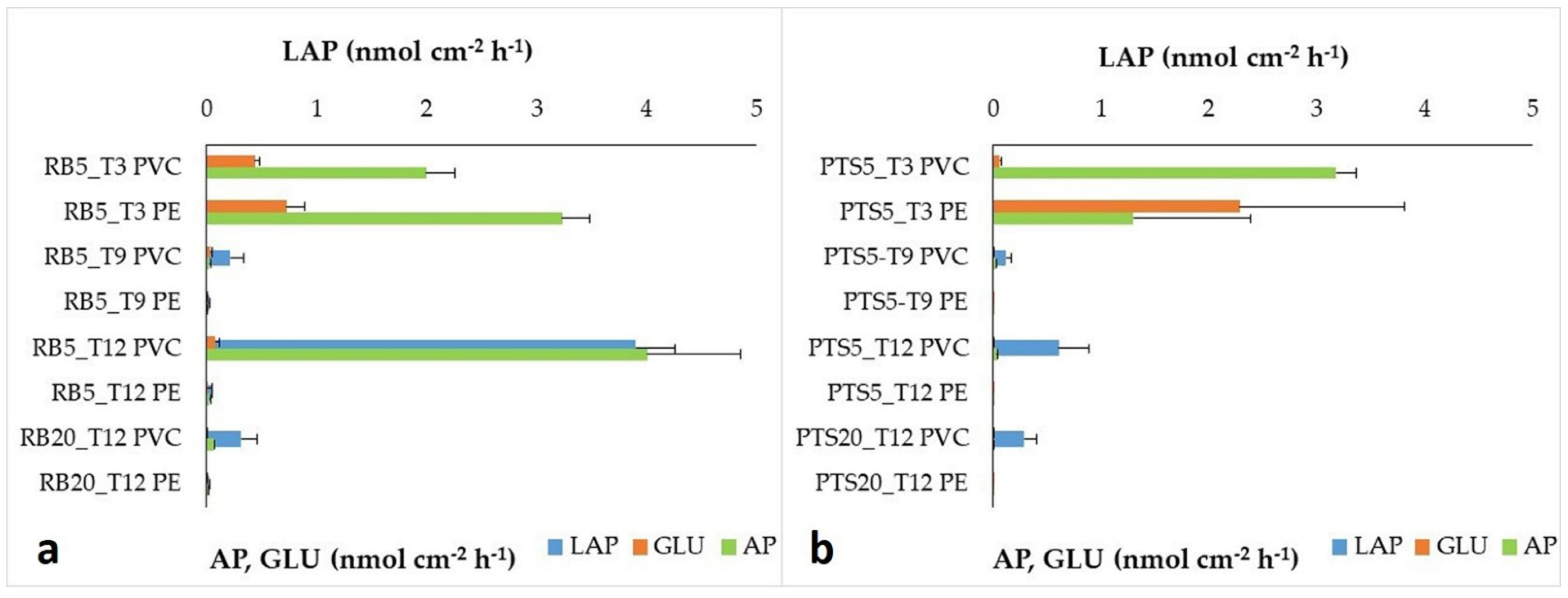
3.2.2. Heterotrophic Microbial Biofilm Metabolism
3.2.3. Microalgal Biofilm Assemblages
3.3. Water Microbial Community
3.3.1. Prokaryotic Abundance
3.3.2. Heterotrophic Microbial Community Metabolism
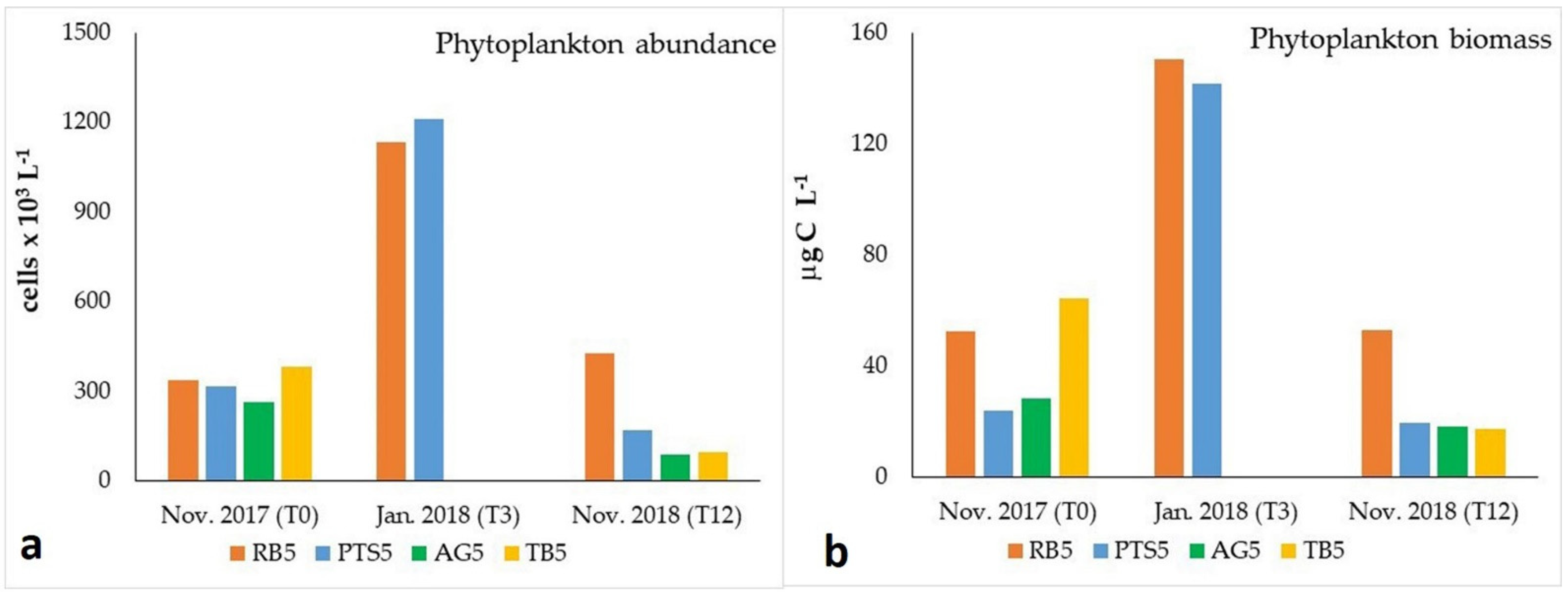
3.3.3. Phytoplankton Abundance and Biomass
3.4. Comparison between the Plastisphere and the Surrounding Planktonic Communities
3.5. Statistical Analysis of Data: Pearson Correlation
4. Discussion
4.1. Spatial and Temporal Patterns of Microbial Biofilms
4.2. Within the Plastisphere, Differences between PVC and PE
4.3. Diversity of the Plastisphere versus Surrounding Water and Factors Affecting Colonization
5. Conclusions
- The abundance and metabolic ability of the prokaryotic biofilm community colonizing the benthic domain of stations of Road and Tethys Bays differed significantly, with high microbial abundance and activity levels recorded at the Road Bay and Amorphous Glacier stations. At Road Bay, this finding underlined that human activities—although sewage waste underwent specific treatments—stimulated microbial growth and metabolism. Also in Tethys Bay, microbial biofilms colonizing PVC at the less haline site, Amorphous Glacier, exhibited the highest abundance of prokaryotic living cells and levels of LAP activity probably favored by the detritus released from ice melting.
- Significant differences in the microbial (prokaryotes and microalgae) abundance and composition were recorded at successive sampling times, especially after 3 months of immersion. As the samplings covered different seasonal periods (i.e., 3 months: late summer; 9 months: autumn-winter, 12 months: early summer), differences in microbial abundance, biomass and functional diversity suggested that microbial biofilm community was differently modulated by seasonally changing environmental variables.
- Prokaryotic communities were found to colonize PVC panels with high abundance and metabolic activity rates, while microalgal communities developed more on the PE panels. The good ability of microbes to colonize the plastic substrates suggested that the biofilm lifestyle compared to the behavior of single planktonic organisms provides a protected ecological niche and a strategy functional to overcome the hard environmental conditions of the Antarctic seabed.
- Among the parameters assayed in this study, the enzymes leucine aminopeptidase and alkaline phosphatase, as well as the taxonomic composition of the microalgal communities, were the most responsive variables to environmental changing conditions, suggesting their role as potential candidate sentinels for early detection of environmental natural or anthropic-related disturbances. Moreover, the huge microalgal biomass detected on the plastic panels, quantitatively higher than other coastal ecosystems, demonstrated a better adaptation of the sessile organism communities than the planktonic ones to the cold Antarctic conditions.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Convey, P.; Peck, L.S. Antarctic environmental change and biological responses. Sci. Adv. 2019, 11, eaaz0888. [Google Scholar] [CrossRef]
- Corsi, I.; Bergami, E.; Caruso, G. Plastics in polar regions. Environ. Int. 2021, 149, 106203. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Bergami, E.; Singh, N.; Corsi, I. Plastic Occurrence, Sources, and Impacts in Antarctic Environment and Biota. Water Biol. Secur. 2022, 1, 100034. [Google Scholar] [CrossRef]
- Davey, M.; O’Toole, G. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef] [PubMed]
- Huggett, M.; Nedved, B.; Hadfield, M. Effects of initial surface wettability on biofilm formation and subsequent settlement of Hydroides elegans. Biofoulingg 2009, 25, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Molino, P.J.; Childs, S.; Hubbard, M.R.; Carey, J.M.; Burgman, M.A.; Wetherbee, R. Development of the primary bacterial microfouling layer on antifouling and fouling release coatings in temperate and tropical environments in Eastern Australia. Biofouling 2009, 25, 149–162. [Google Scholar] [CrossRef]
- Dobretsov, S.; Thomason, J.C. The development of marine biofilms on two commercial non-biocidal coatings: A comparison between silicone and fluoropolymer technologies. Biofouling 2011, 27, 869–880. [Google Scholar] [CrossRef]
- Anderson, R.O. Marine and estuarine natural microbial biofilms: Ecological and biogeochemical dimensions. AIMS Microbiol. 2016, 2, 304–331. [Google Scholar] [CrossRef]
- Salta, M.; Wharton, J.A.; Blache, Y.; Stokes, K.R.; Briand, J.F. Marine biofilms on artificial surfaces: Structure and dynamics. Environ. Microbiol. 2013, 15, 2879–2893. [Google Scholar] [CrossRef]
- Zhang, W.; Ding, W.; Li, Y.-X.; Tam, C.; Bougouffa, S.; Wang, R.; Pei, B.; Chiang, H.; Leung, P.; Lu, Y.; et al. Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nat. Commun. 2019, 10, 517. [Google Scholar] [CrossRef]
- Patil, J.; Anil, A. Biofilm diatom community structure: Influence of temporal and substratum variability. Biofouling 2005, 21, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G. Microbial Colonization in Marine Environments: Overview of Current Knowledge and Emerging Research Topics. J. Mar. Sci. Eng. 2020, 8, 78. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Slikas, B.; Boyd, G.D.; Melvin, D.W.; Morrall, C.E.; Proskurowski, G.; Mincer, T.J. The biogeography of the Plastisphere: Implications for policy. Front. Ecol. Environ. 2015, 13, 541–546. [Google Scholar] [CrossRef]
- Bryant, J.A.; Clemente, T.M.; Viviani, D.A.; Fong, A.A.; Thomas, K.A.; Kemp, P.; Karl, D.M.; White, A.E.; DeLong, E.F. Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. mSystems 2016, 1, e00024-16. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Dey, S.; Rout, A.K.; Behera, B.K.; Ghosh, K. Plastisphere community assemblage of aquatic environment: Plastic-microbe interaction, role in degradation and characterization technologies. Environ. Microbiome 2022, 17, 32. [Google Scholar] [CrossRef]
- Barnes, D.K.A.; Conlan, K.E. Disturbance, colonization and development of Antarctic benthic communities. Philos. Trans. R. Soc. B 2007, 362, 11–38. [Google Scholar] [CrossRef]
- Alabiso, G.; Montini, U.; Milillo, M. One year corrosion test of four stainless steel grades at Ny-Alesund (Svalbard Islands): June 2004–June 2005. In CNR-IAMC Technical Report; National Research Council: Taranto, Italy, 2005. [Google Scholar]
- Webster, N.S.; Negri, A.P. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 2006, 8, 1177–1190. [Google Scholar] [CrossRef]
- Lee, Y.M.; Cho, K.H.; Hwang, K.; Kim, E.H.; Kim, M.; Hong, S.G.; Lee, H.K. Succession of bacterial community structure during the early stage of biofilm development in the Antarctic marine environment. Korean J. Microbiol. 2016, 52, 49–58. [Google Scholar] [CrossRef]
- Smith, W.O., Jr.; Ainley, D.G.; Arrigo, K.R.; Dinniman, M.S. The Oceanography and Ecology of the Ross Sea. Ann. Rev. Mar. Sci. 2014, 6, 469–487. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Caruso, G.; Rizzo, C.; Papale, M.; Azzaro, M. Bacterial communities versus anthropogenic disturbances in the Antarctic coastal marine environment. Environ. Sustain. 2019, 2, 297–310. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020, 18, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Zettler, E.R.; Mincer, T.J.; Amaral-Zettler, L.A. Life in the “Plastisphere”: Microbial Communities on Plastic Marine Debris. Environ. Sci. Technol. 2013, 47, 7137–7146. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, A. A roadmap for a Plastisphere. Mar. Pollut. Bull. 2021, 167, 112322. [Google Scholar] [CrossRef]
- Rowlands, E.; Galloway, T.; Manno, C. A Polar outlook: Potential interactions of micro- and nano-plastic with other anthropogenic stressors. Sci. Total Environ. 2021, 754, 142379. [Google Scholar] [CrossRef]
- Schwartz, T.; Hoffmann, S.; Obst, U. Formation and bacterial composition of young, natural biofilms obtained from public bank-filtered drinking water systems. Wat. Res. 1998, 32, 2787–2797. [Google Scholar] [CrossRef]
- Romaní, A.M.; Borrego, C.M.; Díaz-Villanueva, V.; Freixa, A.; Gich, F.; Ylla, I. Shifts in microbial community structure and function in light- and dark-grown biofilms driven by warming. Environ. Microbiol. 2014, 16, 2550–2567. [Google Scholar] [CrossRef]
- Pastor, A.; Freixa, A.; Skovsholt, L.J.; Wu, N.; Romaní, A.M.; Riis, T. Microbial Organic Matter Utilization in High-Arctic Streams: Key Enzymatic Controls. Microb. Ecol. 2019, 78, 539–554. [Google Scholar] [CrossRef]
- Strickland, J.D.H.; Parsons, T.R. A Practical Handbook of Seawater Analysis, 2nd ed.; Fisheries Research Board of Canada Bulletin 157: Ottawa, ON, Canada, 1972; pp. 1–310. [Google Scholar]
- Aminot, A.; Chaussepied, M. Manuel des Analyses Chimiques en Milieu Marin; Jouve, Ed.; Centre National Pour l’Exploitation des Océans: Brest, France, 1983; pp. 1–395. [Google Scholar]
- Porter, K.G.; Feig, Y.S. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 1980, 25, 943–948. [Google Scholar] [CrossRef]
- La Ferla, R.; Maimone, G.; Azzaro, M.; Conversano, F.; Brunet, C.; Cabral, A.S.; Paranhos, R. Vertical distribution of the prokaryotic cell size in the Mediterranean Sea. Helgol. Mar. Res. 2012, 66, 635–650. [Google Scholar] [CrossRef]
- Cappello, S.; Caruso, G.; Bergami, E.; Macrì, A.; Venuti, V.; Majolino, D.; Corsi, I. New insights into the structure and function of the prokaryotic communities colonizing plastic debris collected in King George Island (Antarctica): Preliminary observations from two plastic fragments. J. Hazard. Mater. 2021, 414, 125586. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Pedà, C.; Cappello, S.; Leonardi, M.; La Ferla, R.; Lo Giudice, A.; Maricchiolo, G.; Rizzo, C.; Maimone, G.; Rappazzo, A.C.; et al. Effects of microplastics on trophic parameters, abundance and metabolic activities of seawater and fish gut bacteria in mesocosm conditions. Environ. Sci. Pollut. Res. 2018, 25, 30067–30083. [Google Scholar] [CrossRef] [PubMed]
- Nedoma, J.; Garcia, J.C.; Comerma, M.; Simek, K.; Armengol, J. Extracellular phosphatases in a Mediterranean reservoir: Seasonal, spatial and kinetic heterogeneity. Freshwater Biol. 2006, 51, 1264–1276. [Google Scholar] [CrossRef]
- Totti, C.; De Stefano, M.; Facca, C.; Ghirardelli, L.A. Microfitobenthos. In Manuale di Metodologie di Campionamento e Studio del Benthos Marino Mediterraneo; Gambi, M.C., Dappiano, M., Eds.; Società Italiana di Biologia Marina: Genova, Italy, 2003; Volume 10, pp. 263–284. [Google Scholar]
- Edler, L.; Elbrächter, M. The Utermöhl method for quantitative phytoplankton analysis. In Microscopic and Methods for Quantitative Phytoplankton Analysis; Karlson, B., Cusack, C., Bresnan, E., Eds.; UNESCO IOC Manuals and Guides: Paris, France, 2010; Volume 55, pp. 13–20. [Google Scholar]
- Andersen, P.; Throndsen, J. Estimating cell numbers. In Manual on Harmful Marine Microalgae. Monographs on Oceanographic Methodology No. 11; Hallegraeff, G.M., Anderson, D.M., Cembella, A.D., Eds.; UNESCO Publishing: Paris, France, 2004; pp. 99–130. [Google Scholar]
- Hillebrand, H.; Durselen, C.D.; Kirschtel, D.; Pollingher, U.; Zohary, T. Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 1999, 35, 403–424. [Google Scholar] [CrossRef]
- Menden-Deuer, S.; Lessard, E.J. Carbon to volume relationships for dinoflagellates, diatoms, and other protist plankton. Limnol. Oceanogr. 2000, 45, 569–579. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software package for education and data analysis. Paleontol. Electron. 2001, 4, 9. [Google Scholar]
- Clarke, K.R.; Gorley, R.N.; Somerfield, P.J.; Warwick, R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, 3rd ed.; PRIMER-E Ltd.: Plymouth, UK, 2014. [Google Scholar]
- Dussud, C.; Hudec, C.; George, M.; Fabre, P.; Higgs, P.; Bruzaud, S.; Delort, A.-M.; Eyheraguibel, B.; Meistertzheim, A.-L.; Jacquin, J.; et al. Colonization of Non-biodegradable and Biodegradable Plastics by Marine Microorganisms. Front. Microbiol. 2018, 9, 1571. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Loeder, M.G.; Gerdts, G.; Osborn, A.M. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol. Ecol. 2014, 90, 478–492. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Osborn, A.M.; Duhaime, M.B. Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS ONE 2016, 11, e0159289. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Kreikemeyer, B.; Labrenz, M. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 2018, 8, 2709. [Google Scholar] [CrossRef]
- Anderson, M.J. Variations in biofilms colonizing artificial surfaces: Seasonal effects and effects of grazers. J. Mar. Biol. Assoc. 1995, 75, 705–714. [Google Scholar] [CrossRef]
- Nagarkar, S.; Williams, G.A.; Subramanian, G.; Saha, S.K. Cyanobacteria-dominated biofilms: A high quality food resource for intertidal grazers. Hydrobiologia 2004, 512, 89–95. [Google Scholar] [CrossRef]
- Bulleri, F. Experimental evaluation of early patterns of colonisation of space on rocky shores and seawalls. Mar. Environ. Res. 2005, 60, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Leite, L.G.; Ciotti, A.M.; Christofoletti, R.A. Abundance of biofilm on intertidal rocky shores: Can trampling by humans be a negative influence? Mar. Environ. Res. 2012, 79, 111–115. [Google Scholar] [CrossRef]
- Orvain, F.; de Crignis, M.; Guizien, K.; Lefebvre, S.; Mallet, C.; Takahashi, E.; Dupuy, C. Tidal and seasonal effects on the short-term temporal patterns of bacteria, microphytobenthos and exopolymers in natural intertidal biofilms (Brouage, France). J. Sea Res. 2014, 92, 6–18. [Google Scholar] [CrossRef]
- Morelle, J.; Maire, O.; Richard, A.; Slimani, A.; Orvain, F. Contrasted impact of two macrofaunal species (Hediste diversicolor and Scrobicularia plana) on microphytobenthos spatial distribution and photosynthetic activity at microscale. Mar. Environ. Res. 2021, 163, 105228. [Google Scholar] [CrossRef]
- Tolker-Nielsen, T.; Molin, S. Spatial organization of microbial biofilm communities. Microb. Ecol. 2000, 40, 75–84. [Google Scholar] [CrossRef]
- Huang, R.; Boney, A.D. Growth interactions between littoral diatoms and juvenile marine algae. J. Exp. Mar. Biol. Ecol. 1984, 81, 21–45. [Google Scholar] [CrossRef]
- Hung, O.S.; Thiyagarajan, V.; Zhang, R.; Wu, R.S.S.; Qian, P.Y. Attachment of Balanus amphitrite larvae to biofilms originating from contrasting environments. Mar. Ecol. Prog. Ser. 2007, 333, 229–242. [Google Scholar] [CrossRef][Green Version]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef]
- Wilson, C.; Lukowicz, R.; Merchant, S.; Valquier-Flynn, H.; Caballero, J.; Sandoval, J.; Okuom, M.; Huber, C.; Brooks, T.D.; Wilson, E.; et al. Quantitative and Qualitative Assessment Methods for Biofilm Growth: A Mini-review. Res. Rev. J. Eng. Technol. 2017, 6. Available online: https://www.rroij.com/open-access/quantitative-and-qualitative-assessment-methods-for-biofilm-growth-a-minireview-.pdf (accessed on 22 February 2021).
- Rosenberg, M.; Azevedo, N.F.; Ivask, A. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep. 2019, 9, 6483. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Numerical dominance and phylotype diversity of marine Rhodobacter species during early colonization of submerged surfaces in coastal marine waters as determined by 16S ribosomal DNA sequence analysis and fluorescence in situ hybridization. Appl. Environ. Microbiol. 2002, 68, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Romaní, A.; Sabater, S. Influence of Algal Biomass on Extracellular Enzyme Activity in River Biofilms. Microb. Ecol. 2000, 40, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Romaní, A.M.; Giorgi, A.; Acuña, V.; Sabater, S. The influence of substratum type and nutrient supply on biofilm organic matter utilization in streams. Limnol. Oceanogr. 2004, 49, 1713–1721. [Google Scholar] [CrossRef]
- Misic, C.; Covazzi Harriague, A. Development of marine biofilm on plastic: Ecological features in different seasons, temperatures, and light regimes. Hydrobiologia 2019, 835, 129–145. [Google Scholar] [CrossRef]
- Bruni, V.; Maugeri, T.L.; Monticelli, L. Faecal Pollution Indicators in the Terra Nova Bay (Ross Sea, Antarctica). Mar. Pollut. Bull. 1997, 34, 908–912. [Google Scholar] [CrossRef]
- Lobelle, D.; Cunliffe, M. Early microbial biofilm formation on marine plastic debris. Mar. Pollut. Bull. 2011, 62, 197–200. [Google Scholar] [CrossRef]
- Hillebrand, H.; Worm, B.; Lotze, H.K. Marine microbenthic community structure regulated by nitrogen loading and grazing pressure. Mar. Ecol. Prog. Ser. 2000, 204, 27–38. [Google Scholar] [CrossRef]
- Jenkins, S.R.; Arenas, F.; Arrontes, J.; Bussell, J.; Castro, J.; Coleman, R.A.; Hawkins, S.J.; Kay, S.; Martinez, B.; Oliveros, J.; et al. European-scale analysis of seasonal variability in limpet grazing activity and microalgal abundance. Mar. Ecol. Prog. Ser. 2001, 211, 193–203. [Google Scholar] [CrossRef]
- Thompson, R.C.; Norton, T.A.; Hawkins, S.J. Physical stress and biological control regulate the producer–consumer balance in intertidal biofilms. Ecology 2004, 85, 1372–1382. [Google Scholar] [CrossRef]
- Thompson, R.C.; Moschella, P.S.; Jenkins, S.R.; Norton, T.A.; Hawkins, S.J. Differences in photosynthetic marine biofilms between sheltered and moderately exposed rocky shores. Mar. Ecol. Prog. Ser. 2005, 296, 53–63. [Google Scholar] [CrossRef]
- Xu, X.; Wang, S.; Gao, F.; Li, J.; Zheng, L.; Sun, C.; He, C.; Wang, Z.; Qu, L. Marine microplastic-associated bacterial community succession in response to geography, exposure time, and plastic type in China’s coastal seawaters. Mar. Pollut. Bull. 2019, 145, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Vivier, B.; Claquin, P.; Lelong, C.; Lesage, Q.; Peccate, M.; Hamel, B.; Georges, M.; Bourguiba, A.; Sebaibi, N.; Boutouil, M.; et al. Influence of infrastructure material composition and microtopography on marine biofilm growth and photobiology. Biofouling 2021, 37, 740–756. [Google Scholar] [CrossRef]
- Miao, L.; Wang, P.; Hou, J.; Yao, Y.; Liu, Z.; Liu, S.; Li, T. Distinct community structure and microbial functions of biofilms colonizing microplastics. Sci. Total Environ. 2019, 650, 2395–2402. [Google Scholar] [CrossRef]
- Odobel, C.; Dussud, C.; Philip, L.; Derippe, G.; Lauters, M.; Eyheraguibel, B.; Burgaud, G.; Ter Halle, A.; Meistertzheim, A.-L.; Bruzaud, S.; et al. Bacterial Abundance, Diversity and Activity During Long-Term Colonization of Non-biodegradable and Biodegradable Plastics in Seawater. Front. Microbiol. 2021, 12, 734782. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef]
- Maugeri, T.L.; Gugliandolo, C.; Bruni, V. Heterotrophic bacteria in the Ross Sea (Terra Nova Bay, Antarctica). New Microbiol. 1996, 19, 67–76. [Google Scholar]
- Caruso, G.; Dell’Acqua, O.; Caruso, R.; Azzaro, M. Phenotypic characterization of bacterial isolates from marine waters and plastisphere communities of the Ross Sea (Antarctica). J. Clin. Microbiol. Biochem. Technol. 2022, 8, 001–009. [Google Scholar] [CrossRef]
- Casabianca, S.; Bellingeri, A.; Capellacci, S.; Sbrana, A.; Russo, T.; Corsi, I.; Penna, A. Ecological implications beyond the ecotoxicity of plastic debris on marine phytoplankton assemblage structure and functioning. Environ. Pollut. 2021, 290, 118101. [Google Scholar] [CrossRef]
- Zhao, S.; Zettler, E.R.; Amaral-Zettler, L.A.; Mincer, T.J. Microbial carrying capacity and carbon biomass of plastic marine debris. ISME J. 2021, 15, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.; Langer, T.M.; Hüer, T.; Hofmann, T.; Herndl, G.J. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS ONE 2019, 14, e0217165. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.M.Y.; Thiyagarajan, V.; Tsoi, M.M.Y.; Qian, P.Y. Qualitative and quantitative changes in marine biofilms as a function of temperature and salinity in summer and winter. Biofilms 2005, 2, 183–195. [Google Scholar] [CrossRef]
- Mejdandžić, M.; Ivanković, T.; Pfannkuchen, M.; Godrijan, J.M.; Pfannkuchen, D.M.; Hrenović, J.; Ljubešić, Z. Colonization of diatoms and bacteria on artificial substrates in the northeastern coastal Adriatic Sea. Acta Bot. Croat. 2015, 74, 407–422. [Google Scholar] [CrossRef]
- Briand, J.-F.; Barani, A.; Garnieri, C.; Réhel, K.; Urvois, F.; LePoupon, C.; Bouchez, A.; Debroas, D.; Bressy, C. Spatio-temporal variations of marine biofilm communities colonizing artificial substrata including antifouling coatings in contrasted French coastal environments. Microb. Ecol. 2017, 74, 585–598. [Google Scholar] [CrossRef]
- Carson, H.S.; Nerheim, M.S.; Carroll, K.A.; Eriksen, M. The plastic-associated microorganisms of the North Pacific Gyre. Mar. Pollut. Bull. 2013, 75, 126–132. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Labrenz, M. Marine Microbial Assemblages on Microplastics: Diversity, Adaptation, and Role in Degradation. Ann. Rev. Mar. Sci. 2020, 12, 209–232. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Seasonal dynamics of particle-associated and free-living marine Proteobacteria in a salt marsh tidal creek as determined using fluorescence in situ hybridization. Environ. Microbiol. 2002, 4, 287–295. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2000, 66, 467–475. [Google Scholar] [CrossRef]
- Dang, H.; Li, T.; Chen, M.; Huang, G. Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 2008, 74, 52–60. [Google Scholar] [CrossRef]
- Dang, H.; Chen, R.; Wang, L.; Shao, S.; Dai, L.; Ye, Y.; Guo, L.; Huang, G.; Klotz, M.G. Molecular characterization of putative biocorroding microbiota with a novel niche detection of Epsilon- and Zetaproteobacteria in Pacific Ocean coastal seawaters. Environ. Microbiol. 2011, 13, 3059–3074. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wuertz, S. Bacteria and archaea on earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Silvadon, P.; Barnier, C.; Urlos, L.; Grimaud, R. Biofilm formation as a microbial strategy to assimilate particulate substrates. Environ. Microbiol. Rep. 2019, 11, 749–764. [Google Scholar] [CrossRef]
- Catão, C.P.E.; Pollet, T.; Garnier, C.; Barry-Martinet, R.; Rehel, K.; Linossier, I.; Tunin-Ley, A.; Turquet, J.; Briand, J.F. Temperate and tropical coastal waters share relatively similar microbial biofilm communities while free-living or particle-attached communities are distinct. Mol. Ecol. 2021, 30, 2891–2904. [Google Scholar] [CrossRef]
- Kirstein, I.V.; Wichels, A.; Krohne, G.; Gerdts, G. Mature biofilm communities on synthetic polymers in seawater—Specific or general? Mar. Environ. Res. 2018, 142, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Catão, E.C.P.; Pollet, T.; Misson, B.; Garnier, C.; Ghiglione, J.-F.; Barry-Martinet, R.; Maintenay, M.; Bressy, C.; Briand, J.-F. Shear stress as a major driver of marine biofilm communities in the NW Mediterranean Sea. Front. Microbiol. 2019, 10, 1768. [Google Scholar] [CrossRef] [PubMed]
- Laganà, P.; Caruso, G.; Corsi, I.; Bergami, E.; Venuti, V.; Majolino, D.; La Ferla, R.; Azzaro, M.; Cappello, S. Do plastics serve as a possible vector for the spread of antibiotic resistance? First insights from bacteria associated to a polystyrene piece from King George Island (Antarctica). Int. J. Hyg. Environ. Health 2019, 222, 89–100. [Google Scholar] [CrossRef]
- Mammo, F.K.; Amoah, I.D.; Gani, K.M.; Pillay, L.; Ratha, S.K.; Bux, F.; Kumari, S. Microplastics in the environment: Interactions with microbes and chemical contaminants. Sci. Total Environ. 2020, 743, 140518. [Google Scholar] [CrossRef]
- Harrison, J.P.; Hoellein, T.J.; Sapp, M.; Tagg, A.S.; Ju-Nam, Y.; Ojeda, J.J. Microplastic-Associated Biofilms: A Comparison of Freshwater and Marine Environments. In Handbook of Environmental Chemistry; Barcelò, D., Kostianoy, A.G., Eds.; Springer: Cham, Switzerland, 2018; Volume 58, pp. 181–201. [Google Scholar] [CrossRef]
- Saggiomo, M.; Escalera, L.; Bolinesi, F.; Rivaro, P.; Saggiomo, V.; Mangoni, O. Diatom diversity during two austral summers in the Ross Sea (Antarctica). Mar. Micropaleontol. 2021, 165, 101993. [Google Scholar] [CrossRef]
- Armand, L.K.; Cornet-Barthaux, V.; Mosseri, J.; Quéguiner, B. Late summer diatom biomass and community structure on and around the naturally iron-fertilised Kerguelen Plateau in the Southern Ocean. Deep-Sea Res. II Top. Stud. Oceanogr. 2008, 55, 653–676. [Google Scholar] [CrossRef]
- Arrigo, K.R.; Robinson, D.H.; Worthen, D.L.; Dunbar, R.B.; DiTullio, G.R.; VanWoert, M.; Lizotte, M.P. Phytoplankton community structure and the drawdown of nutrients and CO2 in the Southern Ocean. Science 1999, 283, 365–367. [Google Scholar] [CrossRef]
- Arrigo, K.R.; van Dijken, G.L.; Bushinsky, S. Primary production in the Southern Ocean, 1997–2006. J. Geophys. Res. 2008, 113, C08004. [Google Scholar] [CrossRef]
- Tréguer, P.; Nelson, D.M.; van Bennekom, A.J.; DeMaster, D.J.; Leynaert, A.; Quéguiner, B. The silica balance in the world ocean: A reestimate. Science 1995, 268, 375–379. [Google Scholar] [CrossRef]
- Smith, W.O.; Ainley, D.G.; Cattaneo-Vietti, R. Trophic interactions within the Ross Sea continental shelf ecosystem. Philos. Trans. R. Soc. Ser. B Biol. Sci. 2007, 362, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, O.; Saggiomo, M.; Bolinesi, F.; Castellano, M.; Povero, P.; Saggiomo, V.; DiTullio, G.D. Phaeocystis antarctica unusual summer bloom in stratified antarctic coastal waters (Terra Nova Bay, Ross Sea). Mar. Environ. Res. 2019, 151, 104733. [Google Scholar] [CrossRef]
- Bolinesi, F.; Saggiomo, M.; Ardini, F.; Castagno, P.; Cordone, A.; Fusco, G.; Rivaro, P.; Saggiomo, V.; Mangoni, O. Spatial-related community structure and dynamics in phytoplankton of the Ross Sea, Antarctica. Front. Mar. Sci. 2020, 7, 1092. [Google Scholar] [CrossRef]
- Saggiomo, M.; Escalera, L.; Saggiomo, V.; Bolinesi, F.; Mangoni, O. Phytoplankton blooms below the Antarctic landfast ice during the melt season between late spring and early summer. J. Phycol. 2021, 57, 541–550. [Google Scholar] [CrossRef]
- Tittel, J.; Bissinger, V.; Zippel, B.; Gaedke, U.; Bell, E.; Lorke, A.; Kamjunke, N. Mixotrophs combine resource use to outcompete specialists: Implications for aquatic food webs. Proc. Natl. Acad. Sci. USA 2003, 100, 12776–12781. [Google Scholar] [CrossRef]
- Eddie, B.; Krembs, C.; Neuer, S. Characterization and growth response to temperature and salinity of psychrophilic, halotolerant Chlamydomonas sp. ARC isolated from Chukchi Sea ice. Mar. Ecol. Prog. Ser. 2008, 354, 107–117. [Google Scholar] [CrossRef]
- Jacobs, S.; Giulivi, C.; Mele, P. Freshening of the Ross Sea during the late 20th century. Science 2002, 297, 386–389. [Google Scholar] [CrossRef]
- Andreoli, C.; Tolomio, C.; Moro, I.; Radice, M.; Moschin, E.; Bellato, S. Diatoms and dinoflagellates in Terra Nova Bay (Ross Sea-Antarctica) during austral summer 1990. Polar Biol. 1995, 15, 465–475. [Google Scholar] [CrossRef]
- Mangoni, O.; Saggiomo, V.; Bolinesi, F.; Margiotta, F.; Budillon, G.; Cotroneo, Y.; Misic, C.; Rivaro, P.; Saggiomo, M. Phytoplankton blooms during austral summer in the Ross Sea, Antarctica: Driving factors and trophic implications. PLoS ONE 2017, 12, e0176033. [Google Scholar] [CrossRef]
- Nuccio, C.; Innamorati, M.; Lazzara, L.; Mori, G.; Massi, L. Spatial and temporal distribution of phytoplankton assemblages in the Ross Sea. In Ross Sea Ecology; Guglielmo, L., Ianora, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2000; pp. 231–245. [Google Scholar]




| Station Typology | Latitude | Longitude | Immersion Time (Months) | Station Acronym |
|---|---|---|---|---|
| ROAD BAY | ||||
| IMPACT, −5 m | 74°41′743″ S | 164°07′125″ E | 3, 9, 12 | RB5_(3, 9, 12) |
| IMPACT, −20 m | 74°41′784″ S | 164°07′219″ E | 3, 9, 12 | RB20_(3, 9, 12) |
| CONTROL, −5 m | 74°41′651″ S | 164°07′303″ E | 3, 9, 12 | PTS5_(3, 9, 12) |
| CONTROL, −20 m | 74°41′623″ S | 164°07′343″ E | 3, 9, 12 | PTS20_(3, 9, 12) |
| TETHYS BAY | ||||
| IMPACT, −5 m | 74°41′234″ S | 164°02′135″ E | 12 | AG5_12 |
| IMPACT, −20 m | 74°41′242″ S | 164°02′186″ E | 12 | AG20_12 |
| CONTROL, −5 m | 74°41′417″ S | 164°06′303″ E | 12 | TB5_12 |
| CONTROL, −20 m | 74°41′407″ S | 164°06′311″ E | 12 | TB20_12 |
| Date | Months | T | S | O2 | Chl a | pH | NH4 | NO2 | NO3 | PO4 | N/P | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| °C | Psu | mL L−1 | RFU | µmol L−1 | µmol L−1 | µmol L−1 | µmol L−1 | |||||
| ROAD BAY | ||||||||||||
| RB 5 | 22 November 2017 | 0 | −1.83 | 35.19 | 8.0 | 0.62 | 8.02 | 1.96 | 0.07 | 23.74 | 1.85 | 13.95 |
| RB 20 | 22 November 2017 | 0 | −1.83 | 35.20 | 8.8 | 0.58 | 7.74 | 1.81 | 0.13 | 15.55 | 1.86 | 9.40 |
| RB 5 | 26 January 2018 | 3 | 0.90 | 34.79 | 7.9 | 0.99 | 8.11 | 1.33 | 0.09 | 6.85 | 0.52 | 16.00 |
| RB 20 | 26 January 2018 | 3 | 0.86 | 34.85 | 8.4 | 1.05 | 8.19 | 0.86 | 0.18 | 8.48 | 0.53 | 17.81 |
| RB 5 | 17 November 2018 | 12 | −1.73 | 35.24 | n.d. | 0.66 | n.d. | 1.52 | 0.06 | 26.05 | 1.39 | 19.87 |
| RB 20 | 17 November 2018 | 12 | −1.74 | 35.25 | n.d. | 0.61 | n.d. | 3.52 | 0.12 | 26.33 | 1.45 | 20.67 |
| PTS-5 | 23 November 2017 | 0 | −1.82 | 35.22 | 7.2 | 0.53 | 7.93 | 4.61 | 0.46 | 15.51 | 1.68 | 12.28 |
| PTS 20 | 23 November 2017 | 0 | −1.80 | 35.22 | 6.9 | 0.58 | 7.83 | 3.53 | 0.60 | 20.12 | 1.67 | 14.52 |
| PTS 5 | 26 January 182018 | 3 | 0.91 | 34.84 | 8.4 | 0.72 | 8.10 | 1.66 | 0.08 | 10.76 | 0.75 | 16.72 |
| PTS 20 | 26 January 18 | 3 | 0.87 | 34.84 | 8.4 | 1.05 | 8.09 | 1.22 | 0.10 | 10.44 | 0.63 | 18.64 |
| PTS 5 | 25 November 2018 | 12 | −1.86 | 35.27 | n.d. | 0.66 | n.d. | 0.94 | 0.08 | 25.77 | 1.68 | 15.94 |
| PTS 20 | 25 November 2018 | 12 | −1.73 | 35.25 | n.d. | 0.63 | n.d. | 6.93 | 0.06 | 26.63 | 1.49 | 22.56 |
| TETHYS BAY | ||||||||||||
| AG 5 | 30 November 2017 | 0 | −1.68 | 35.21 | 8.0 | 0.92 | 7.70 | 4.05 | 0.10 | 20.67 | 2.41 | 10.32 |
| AG 20 | 30 November 2017 | 0 | −1.77 | 35.20 | 7.5 | 0.92 | 7.68 | 1.70 | 0.09 | 21.30 | 1.97 | 11.72 |
| AG 5 | 13 November 2018 | 12 | −1.82 | 35.26 | 7.5 | 0.64 | n.d. | 0.94 | 0.10 | 21.87 | 1.52 | 15.07 |
| AG 20 | 13 November 2018 | 12 | −1.82 | 35.26 | n.d. | 0.61 | n.d. | 1.10 | 0.04 | 21.86 | 1.52 | 15.13 |
| TB 5 | 26 November 2017 | 0 | −1.72 | 35.14 | 8.1 | 1.00 | 8.12 | 1.63 | 0.11 | 13.44 | 1.35 | 11.24 |
| TB 20 | 26 November 2017 | 0 | −1.72 | 35.19 | 8.5 | 1.00 | 7.75 | 2.05 | 0.12 | 26.22 | 1.76 | 16.13 |
| TB 5 | 9 November 2018 | 12 | −1.75 | 34.96 | n.d. | 0.63 | n.d. | 1.43 | 0.04 | 24.03 | 1.44 | 17.71 |
| TB 20 | 9 November 2018 | 12 | −1.74 | 35.24 | 9.730 | 0.62 | n.d. | 2.67 | 0.07 | 26.53 | 2.5 | 11.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caroppo, C.; Azzaro, M.; Dell’Acqua, O.; Azzaro, F.; Maimone, G.; Rappazzo, A.C.; Raffa, F.; Caruso, G. Microbial Biofilms Colonizing Plastic Substrates in the Ross Sea (Antarctica). J. Mar. Sci. Eng. 2022, 10, 1714. https://doi.org/10.3390/jmse10111714
Caroppo C, Azzaro M, Dell’Acqua O, Azzaro F, Maimone G, Rappazzo AC, Raffa F, Caruso G. Microbial Biofilms Colonizing Plastic Substrates in the Ross Sea (Antarctica). Journal of Marine Science and Engineering. 2022; 10(11):1714. https://doi.org/10.3390/jmse10111714
Chicago/Turabian StyleCaroppo, Carmela, Maurizio Azzaro, Ombretta Dell’Acqua, Filippo Azzaro, Giovanna Maimone, Alessandro Ciro Rappazzo, Francesco Raffa, and Gabriella Caruso. 2022. "Microbial Biofilms Colonizing Plastic Substrates in the Ross Sea (Antarctica)" Journal of Marine Science and Engineering 10, no. 11: 1714. https://doi.org/10.3390/jmse10111714
APA StyleCaroppo, C., Azzaro, M., Dell’Acqua, O., Azzaro, F., Maimone, G., Rappazzo, A. C., Raffa, F., & Caruso, G. (2022). Microbial Biofilms Colonizing Plastic Substrates in the Ross Sea (Antarctica). Journal of Marine Science and Engineering, 10(11), 1714. https://doi.org/10.3390/jmse10111714












