Screening Cultivated Eggplant and Wild Relatives for Resistance to Bacterial Wilt (Ralstonia solanacearum)
Abstract
:1. Introduction
2. Material and Methods
2.1. Seeds and Plant Growth Conditions
2.2. Pathogen and Resistance Assays in First Trial
2.3. Evaluation of Resistant and Moderately Resistant Accessions in Second Trial
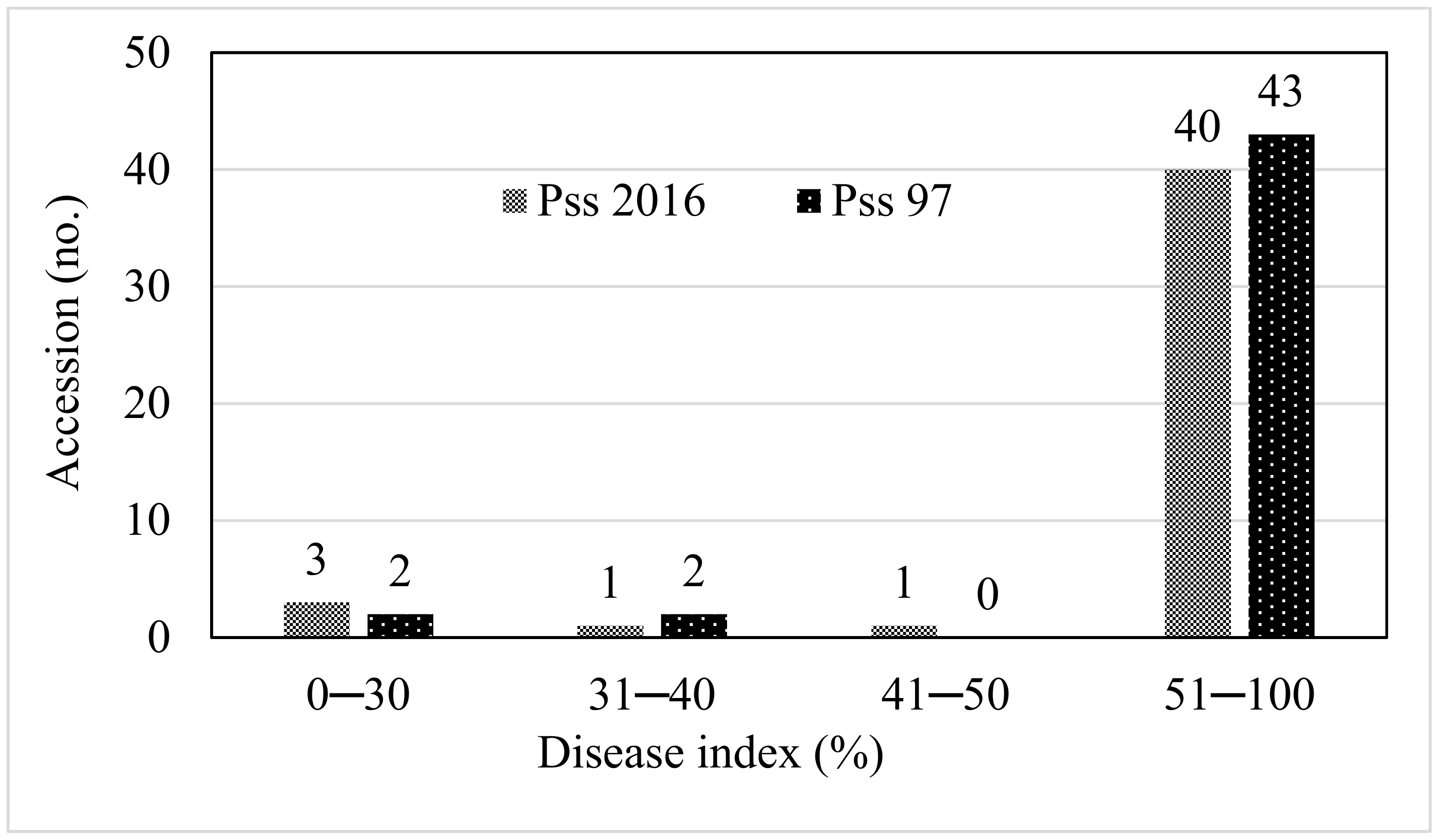
3. Results
Second Trial
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Genin, S.; Denny, T.P. Pathogenomics of the Ralstonia solanacearum Species Complex. Annu. Rev. Phytopathol. 2012, 50, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Huet, G. Breeding for resistances to Ralstonia solanacearum. Front. Plant Sci. 2014, 5, 715. [Google Scholar] [CrossRef] [PubMed]
- Elphinstone, J.G. The current bacterial wilt situation. A global view. In Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex; Allen, C., Prior, P., Hayward, A.C., Eds.; APS Press: Saint Paul, MN, USA, 2005; pp. 9–28. [Google Scholar]
- Peeters, N.; Guidot, A.; Vailleau, F.; Valls, M. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 2013, 14, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Wenneker, M.; Verdel, M.S.W.; Groeneveld, R.M.W.; Kempenaar, C.; Van Beuningen, A.R.; Janse, J.D. Ralstonia (Pseudomonas) solanacearum race 3 (biovar 2) in surface water and natural weed hosts: First report on stinging nettle (Urtica dioica). Eur. J. Plant Pathol. 1999, 105, 307–315. [Google Scholar] [CrossRef]
- Janse, J.D.; Van Den Beld, H.E.; Elphinstone, J.; Simpkins, S.; Tjou-Tam-Sin, N.N.A.; Van Vaerenbergh, J. Introduction to Europe of Ralstonia solanacearum biovar 2, race 3 in Pelargonium zonale cuttings. J. Plant Pathol. 2004, 86, 147–155. [Google Scholar]
- Swanson, J.K.; Yao, J.; Tans-Kersten, J.; Allen, C. Behavior of Ralstonia solanacearum Race 3 Biovar 2 during latent and active infection of geranium. Phytopathology 2005, 95, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Fegan, M.; Prior, P.; Allen, C.; Hayward, A.C. How complex is the Ralstonia solanacearum species complex. In Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex; Allen, C., Prior, P., Hayward, A.C., Eds.; American Phytopathological Society Press: St. Paul, MN, USA, 2005; pp. 449–461. [Google Scholar]
- Wicker, E.; Lefeuvre, P.; De Cambiaire, J.C.; Lemaire, C.; Poussier, S.; Prior, P. Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J. 2012, 6, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, A.; Daunay, M.-C.; Frary, A.; Palloix, A.; Wang, J.-F.; Dintinger, J.; Chiroleu, F.; Wicker, E.; Prior, P. Bacterial wilt resistance in tomato, pepper, and eggplant: Genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 2011, 101, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Pradhanang, P.; Ji, P.; Momol, M.; Olson, S.M.; L. Mayfield, J.; Jones, J. Application of acibenzolar-S-Methyl enhances host resistance in tomato against Ralstonia solanacearum. Plant Dis. 2005, 89, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, A.; Fujisawa, M.; Hamasaki, R.; Kawasaki, T.; Fujie, M.; Yamada, T. Biocontrol of Ralstonia solanacearum by treatment with Lytic Bacteriophages. Appl. Environ. Microbiol. 2011, 77, 4155–4162. [Google Scholar] [CrossRef] [PubMed]
- HS, A.; Askora, A.; Kawasaki, T.; Fujie, M.; Yamada, T. Utilization of Filamentous Phage RSM3 to control bacterial wilt caused by Ralstonia solanacearum. Plant Dis. 2012, 96, 1204–1209. [Google Scholar]
- Keatinge, J.D.H.; Lin, L.J.; Ebert, A.W.; Chen, W.Y.; Hughes, J.D.A.; Luther, G.C.; Wang, J.F.; Ravishankar, M. Overcoming biotic and abiotic stresses in the Solanaceae through grafting: Current status and future perspectives. Biol. Agric. Hortic. 2014, 30, 272–287. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.A.X.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. MPP Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimault, V.; Anais, G.; Prior, P. Distribution of Pseudomonas solanacearum in the stem tissues of tomato plants with different levels of resistance to bacterial wilt. Plant Pathol. 1994, 43, 663. [Google Scholar] [CrossRef]
- Hartman, G.L.; Hong, W.F.; Hayward, A.C. Potential of biological and chemical control of bacterial wilt. ACIAR Proc. Aust. Cent. Int. Agric. Res. 1993, 45, 322–326. [Google Scholar]
- Murakoshi, S.; Takahashi, M. Trials of some control of tomato bacterial wilt caused by Pseudomonas solanacearum. Bull. Kanagawa Hortic. Exp. Stn. 1984, 31, 50–56. [Google Scholar]
- Weller, D.M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 1988, 26, 379–407. [Google Scholar] [CrossRef]
- Lemessa, F.; Zeller, W. Screening rhizobacteria for biological control of Ralstonia solanacearum in Ethiopia. YBCON Biol. Control 2007, 42, 336–344. [Google Scholar] [CrossRef]
- Singh, D.; Yadav, D.K.; Sinha, S.; Upadhyay, B.K. Utilization of plant growth promoting Bacillus subtilis isolates for the management of bacterial wilt incidence in tomato caused by Ralstonia solanacearum race 1 biovar 3. Indian Phytopath 2012, 65, 18–24. [Google Scholar]
- Boshou, L. A broad review and perspective on breeding for resistance to bacterial wilt. In Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex; Allen, C., Prior, P., Hayward, A.C., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2005; pp. 225–238. [Google Scholar]
- Denny, T.P. Plant Pathogenic Ralstonia Species. In Plant-Associated Bacteria; Gnanamanickam, S.S., Ed.; Springer: Dordrecht, The Netherlands, 2006; pp. 573–644. [Google Scholar]
- Cardoso, S.C.; Soares, A.C.F.; Brito, A.D.S.; Santos, A.P.D.; Laranjeira, F.F.; Carvalho, F.F. Evaluation of tomato rootstocks and its use to control bacterial wilt disease avaliação de porta-enxertos de tomateiro e o uso da enxertia no controle da murcha bacteriana. Semin. Agrar. 2012, 33, 595–604. [Google Scholar] [CrossRef]
- Kumar, B.A.; Raja, P.; Pandey, A.K.; Rabindro, P. Evaluation of wilt resistance of wild Solanum species through grafting in brinjal. Int. J. Curr. Microbiol. 2017, 6, 3464–3469. [Google Scholar]
- Bittner, R.J.; Arellano, C.; Mila, A.L. Effect of temperature and resistance of tobacco cultivars to the progression of bacterial wilt, caused by Ralstonia solanacearum. Plant Soil 2016, 408, 299–310. [Google Scholar] [CrossRef]
- Ranil, R.; Niran, H.M.L.; Plazas, M.; Fonseka, R.; Fonseka, H.H.; Vilanova, S.; Andújar, I.; Gramazio, P.; Fita, A.; Prohens, J. Improving seed germination of the eggplant rootstock Solanum torvum by testing multiple factors using an orthogonal array design. Sci. Hort. 2015, 193, 174–181. [Google Scholar] [CrossRef]
- Buddenhagen, I.W. Designations of races in Pseudomonas solanacearum. Phytopathology 1962, 52, 726. [Google Scholar]
- Hayward, A.C. Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 1964, 27, 265–277. [Google Scholar] [CrossRef]
- Cook, D. Genetic diversity of Pseudomonas solanacearum: Detection of restriction fragment length polymorphisms with DNA probes that specify virulence and the hypersensitive response. Mol. Plant Microbe Interact. 1989, 2, 113. [Google Scholar] [CrossRef]
- He, L.Y. Characteristics of strains of Pseudomonas solanacearum from China. Plant Dis. 1983, 67, 1357–1361. [Google Scholar] [CrossRef]
- Kelman, A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology 1954, 44, 693–695. [Google Scholar]
- Kado, C.; Heskett, M. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology 1970, 60, 969–976. [Google Scholar] [CrossRef]
- Hanson, P.M.; Wang, J.-F.; Licardo, O.; Hanudin; Mah, S.Y.; Hartman, G.L.; Lin, Y.-C.; Chen, J. Variable reaction of tomato lines to bacterial wilt evaluated at several locations in southeast Asia. HortScience 1996, 31, 143. [Google Scholar] [CrossRef]
- Winstead, N.N.; Kelman, A. Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathology 1952, 42, 623–634. [Google Scholar]
- Aslam, M.N.; Hussain, M.A.; Raheel, M. Assessment of resistance to bacterial wilt incited by Ralstonia solanacearum in tomato germplasm. J. Plant Dis. Prot. 2017, 124, 585–590. [Google Scholar] [CrossRef]
- Scott, J.W.; Wang, J.-F.; Hanson, P.M. Breeding tomatoes for resistance to bacterial wilt, a global view. In I International Symposium on Tomato Diseases; ISHS Acta Horticulturae: Orlando, FL, USA, 2004; pp. 161–172. [Google Scholar]
- Wang, J.-F.; Hanson, P.; Barnes, J.A. Worldwide evaluation of an international set of resistance sources to bacterial wilt in tomato. In Bacterial Wilt Disease: Molecular and Ecological Aspects; Prior, P., Allen, C., Elphinstone, J., Eds.; Springer: Berlin, Germany, 1998; pp. 269–275. [Google Scholar]
- Rotino, G.L.; Sala, T.; Toppino, L. Eggplant in Alien Gene Transfer in Crop Plants; Pratap, A., Kumar, J., Eds.; Springer: New York, NY, USA, 2014; pp. 381–409. [Google Scholar]
- Rahman, M.A.; Rashid, M.A.; Hossain, M.M.; Salam, M.A.; Masum, A.S.M.H. Grafting Compatibility of Cultivated Eggplant Varieties with Wild Solanum Species. Pak. J. Biol. Sci. 2002, 5, 755–757. [Google Scholar]
- Ramesh, R.; Achari, G.; Asolkar, T.; D’Souza, M.; Singh, N. Management of bacterial wilt of brinjal using wild brinjal (Solanum torvum Sw) as root stock. Indian Phytopathol. 2016, 69, 260–265. [Google Scholar]
- Yamakawa, K.; Mochizuki, H. Nature and inheritance of Fusarium-wilt resistance in eggplant cultivars and related wild Solanum species. Bull. Veg. Ornam. Crop. Res. Station. Ser. A 1979, 6, 19–27. [Google Scholar]
- Plazas, M.; Vilanova, S.; Gramazio, P.; Rodríguez-Burruezo, A.; Fita, A.; Herraiza, F.J.; Ranil, R.; Fonseka, R.; Niran, L.; Fonseka, H.; et al. Interspecific hybridization between eggplant and wild relatives from different genepools. J. Am. Soc. Hortic. Sci. 2016, 141, 34–44. [Google Scholar] [CrossRef]
- Gramazio, P.; Prohens, J.; Plazas, M.; Mangino, G.; Herraiz, F.J.; García-Fortea, E.; Vilanova, S. Genomic tools for the enhancement of vegetable crops: A case in eggplant. Not. Bot. Horti Agrobot. Cluj Napoca 2018, 46, 1–13. [Google Scholar] [CrossRef]
- Prior, P.; Bart, S.; Leclercq, S.; Darrasse, A.; Anais, G. Resistance to bacterial wilt in tomato as discerned by spread of Pseudomonas (Burholderia) solanacearum in the stem tissues. Plant Pathol. 1996, 45, 720–726. [Google Scholar] [CrossRef]
- Santhosha, H.M.; Indiresh, K.M.; Gopalakrishnan, C.; Singh, T.H. Evaluation of brinjal genotypes against bacterial wilt caused by Ralstonia solanacearum. Hortl. Sci. 2015, 10, 74–78. [Google Scholar]
- Vasse, J.; Danoun, S.; Trigalet, A. Microscopic studies of root infection in resistant tomato cultivar Hawaii. In Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex; Allen, C., Prior, P., Hayward, A.C., Eds.; APS Press: St. Paul, MN, USA, 2005; pp. 285–291. [Google Scholar]
- Hoque, M.O.; HO, M.L.; Chowdhury, B.C. Screening tomato varieties for resistance to bacterial wilt. Bangladesh J. Agril. Res. 1981, 6, 55. [Google Scholar]

| Taxa and Accession Code a | Country of Origin | Genepool | Accession Code in |
|---|---|---|---|
| Germplasm Collection | |||
| S. anguivi | |||
| ANG1 | Ivory coast | Secondary | BBS119 |
| ANG2 | Ivory coast | Secondary | BBS125/B |
| VI048764 | Thailand | Secondary | VI048764 |
| VI050346 | Unknown | Secondary | VI050346 |
| VI050392 | Unknown | Secondary | VI050392 |
| S. campylacanthum | |||
| CAM5 | Tanzania | Secondary | MM680 |
| CAM6 | Tanzania | Secondary | MM700 |
| CAM8 | Kenya | Secondary | MM1426 |
| S. dasyphyllum | |||
| DAS1 | Uganda | Secondary | MM1153 |
| S. elaeagnifolium | |||
| ELE1 | Senegal | Tertiary | MM1627 |
| ELE2 | Greece | Tertiary | ELE2 |
| S. incanum | |||
| MM577 | Israel | Primary | MM577 |
| INC1 | Israel | Primary | MM664 |
| S. insanum | |||
| INS1 | Sri lanka | Primary | SLKINS-1 |
| INS2 | Sri lanka | Primary | SLKINS-2 |
| INS3 | Japan | primary | MM498 |
| VI034853 | Malaysia | Primary | VI034853 |
| VI037989 | Thailand | Primary | VI037989 |
| VI040123 | Thailand | Primary | VI040123 |
| VI040350 | Thailand | Primary | VI040350 |
| VI041106 | Thailand | Primary | VI041106 |
| VI041189 | Thailand | Primary | VI041189 |
| VI054957 | Lao People’s Democratic Republic | Primary | VI054957 |
| VI054962 | Lao People’s Democratic Republic | Primary | VI054962 |
| VI054964 | Lao People’s Democratic Republic | Primary | VI054964 |
| VI054967 | Lao People’s Democratic Republic | Primary | VI054967 |
| VI046583 | Vietnam | Primary | VI046583 |
| S. lichtensteinii | |||
| LIC1 | South Africa | Secondary | MM674 |
| LIC2 | Iran | Secondary | MM677 |
| S. linnaeanum | |||
| LIN1 | Spain | Secondary | JPT0028 |
| LIN3 | Tunisia | Secondary | MM195 |
| VI042691 | Italy | Secondary | VI042691 |
| VI042692 | Italy | Secondary | VI042692 |
| VI042740 | Colombia | Secondary | VI042740 |
| S. melongena | |||
| MEL1 | Ivory coast | Cultivated | BBS-118/B |
| MEL2 | Ivory coast | Cultivated | BBS-146 |
| MEL3 | Ivory coast | Cultivated | BBS-175 |
| MEL4 | Sri Lanka | Cultivated | 7145 |
| MEL5 | Sri Lanka | Cultivated | 8104 |
| MEL6 | Sri Lanka | Cultivated | Ampara |
| ANS26 | Spain | Cultivated | ANS26 |
| S. pyracanthos | |||
| PYR1 | Unknown | Secondary | SOLN-66 |
| S. sisymbriifolium | |||
| SIS1 | Unknown | Tertiary | SOLN-78 |
| SIS2 | Unknown | Tertiary | 1180 |
| S. tomentosum | |||
| TOM1 | South Africa | Secondary | MM992 |
| S. torvum | |||
| TOR2 | Sri Lanka | Tertiary | SLKTOR-2 |
| TOR3 | Unknown | Tertiary | 55953 |
| S. melongena (Checks) | |||
| EG048 | Denmark | Cultivated | VI046095 |
| EG203 | India | Cultivated | VI045276 |
| Taxa and Accession Code a | Pss97 | Pss2016 | ||||
|---|---|---|---|---|---|---|
| W% ± SE b | DI ± SE c | Resistance Category d | W% ± SE | DI ± SE | Resistance Category | |
| Solanum anguivi | ||||||
| VI050346 | 100 ± 0 | 100 ± 0 | S | 41.7 ± 11.0 | 23.3 ± 9.8 | R |
| ANG1 | 100 ± 0 | 96.7 ± 3.3 | S | 95.8 ± 4.2 | 45.8 ± 14.0 | MS |
| ANG2 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 59.2 ± 6.0 | S |
| VI048764 | 100 ± 0 | 92.5 ± 3.8 | S | 100 ± 0 | 91.7 ± 4.2 | S |
| VI050392 | 100 ± 0 | 100 ± 0 | S | 95.8 ± 4.2 | 66.7 ± 9.8 | S |
| S. campylacanthum | ||||||
| CAM5 | 95.8 ± 4.2 | 95.8 ± 4.2 | S | 95.8 ± 4.2 | 51.7 ± 3.6 | S |
| CAM6 | 100 ± 0 | 97.5 ± 2.5 | S | ND | ND | |
| CAM8 | 90.5 ± 9.5 | 83.6 ± 9.9 | S | 100 ± 0 | 70 ± 1.4 | S |
| S. dasyphyllum | ||||||
| DAS1 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 100 ± 0 | S |
| S. elaeagnifolium | ||||||
| ELE1 | 100 ± 0 | 98.7 ± 1.3 | S | 83.3 ± 8.3 | 52.5 ± 3.8 | S |
| ELE2 | 100 ± 0 | 100 ± 0 | S | 66.7 ± 6.7 | 61.3 ± 9.6 | S |
| S. incanum | ||||||
| MM577 | 100 ± 0 | 95 ± 2.5 | S | 66.7 ± 8.3 | 23.3 ± 7.4 | R |
| INC1 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 76.7 ± 5.7 | S |
| S. insanum | ||||||
| INS1 | 100 ± 0 | 98.3 ± 1.7 | S | 100 ± 0 | 96.7 ± 3.3 | S |
| INS2 | 95.8 ± 4.2 | 82.5 ± 6.6 | S | 100 ± 0 | 74.2 ± 2.2 | S |
| INS3 | 100 ± 0 | 100 ± 0 | S | ND | ND | |
| VI034853 | 100 ± 0 | 93.3 ± 4.4 | S | 95.8 ± 4.2 | 89.2 ± 1.7 | S |
| VI037989 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 89.2 ± 1.7 | S |
| VI040123 | 100 ± 0 | 97.5 ± 2.5 | S | 95.8 ± 4.2 | 82.5 ± 10.1 | S |
| VI040350 | 100 ± 0 | 100 ± 0 | S | 95.8 ± 4.2 | 86.7 ± 3 | S |
| VI041106 | 100 ± 0 | 93.3 ± 4.4 | S | 100 ± 0 | 90 ± 3.8 | S |
| VI041189 | 100 ± 0 | 91.7 ± 8.3 | S | 100 ± 0 | 75.8 ± 5.5 | S |
| VI054957 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 92.5 ± 5.2 | S |
| VI054962 | 100 ± 0 | 95 ± 3.8 | S | 100 ± 0 | 93.3 ± 3 | S |
| VI054964 | 95.8 ± 4.2 | 90.8 ± 6.8 | S | 100 ± 0 | 82.5 ± 6.6 | S |
| VI054967 | 100 ± 0 | 97.5 ± 1.4 | S | 62.5 ± 31.5 | 89.2 ± 6.5 | S |
| VI046583 | 100 ± 0 | 97.5 ± 2.5 | S | 91.7 ± 8.3 | 50.3 ± 2.6 | S |
| S. lichtensteinii | ||||||
| LIC1 | 100 ± 0 | 100 ± 0 | S | 95.8 ± 4.2 | 90 ± 6.6 | S |
| LIC2 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 55 ± 7.6 | S |
| S. linnaeanum | ||||||
| LIN1 | 95.8 ± 4.2 | 95.8 ± 4.2 | S | 100 ± 0 | 86.7 ± 4.4 | S |
| LIN3 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 91.7 ± 5.1 | S |
| VI042691 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 76.7 ± 7.1 | S |
| VI042692 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 77.5 ± 2.9 | S |
| VI042740 | 100 ± 0 | 100 ± 0 | S | 95.8 ± 4.2 | 79.2 ± 3.6 | S |
| S. melongena | ||||||
| MEL1 | 100 ± 0 | 97.5 ± 2.5 | S | 100 ± 0 | 100 ± 0 | S |
| MEL2 | 75 ± 12.5 | 100 ± 0 | S | 100 ± 0 | 81.1 ± 3.9 | S |
| MEL3 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 89.2 ± 4.2 | S |
| MEL4 | 100 ± 0 | 98.3 ± 1.7 | S | 100 ± 0 | 100 ± 0 | S |
| MEL5 | 91.7 ± 8.3 | 75.8 ± 8.7 | S | 100 ± 0 | 89.2 ± 0.8 | S |
| MEL6 | 100 ± 0 | 96.7 ± 3.3 | S | 100 ± 0 | 100 ± 0 | S |
| ANS26 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 100 ± 0 | S |
| S. pyracanthos | ||||||
| PYR1 | 100 ± 0 | 100 ± 0 | S | 100 ± 0 | 84.2 ± 5.1 | S |
| S. sisymbriifolium | ||||||
| SIS1 | 33.3 ± 22 | 33.3 ± 22 | MR | 41.7 ± 4.2 | 12.5 ± 0 | R |
| SIS2 | 37.5 ± 0 | 37.5 ± 0 | MR | 33.3 ± 4.2 | 9.2 ± 2.2 | R |
| S. tomentosum | ||||||
| TOM1 | 75 ± 0 | 71.7 ± 0.8 | S | 100 ± 0 | 95.8 ± 4.2 | S |
| S. torvum | ||||||
| TOR2 | 16.7 ± 11 | 5.8 ± 3.0 | R | 91.7 ± 8.3 | 64.2 ± 5.5 | S |
| TOR3 | 12.5 ± 0 | 10.8 ± 1.7 | R | 38.9 ± 14.7 | 32.2 ± 16.4 | MR |
| S. melongena (Checks) | ||||||
| EG048 | 100 ± 0 | 98.3 ± 1.7 | S | 100 ± 0 | 88.3 ± 3.0 | S |
| EG203 | 0 ± 0 | 0 ± 0 | R | 50 ± 9.6 | 31.1 ± 8.7 | MR |
| Taxa and Accession Code a | Pss97 | Pss2016 | ||||
|---|---|---|---|---|---|---|
| W% b | DI c | Resistance Category d | W% | DI | Resistance Category | |
| Solanum anguivi | ||||||
| VI050346 | 100.0 | 99.2 | S | 95.8 | 81.7 | S |
| S. incanum | ||||||
| MM577 | 100 | 100 | S | ND | ND | ND |
| S. sisymbriifolium | ||||||
| SIS1 | 20.8 | 17.5 | R | ND | ND | ND |
| SIS2 | 62.5 | 51.7 | S | ND | ND | ND |
| S. torvum | ||||||
| TOR2 | 54.2 | 36.7 | MR | ND | ND | ND |
| TOR3 | 44.4 | 33.3 | MR | ND | ND | ND |
| S. melongena (checks) | ||||||
| EG048 | 100.0 | 100.0 | S | 100.0 | 100.0 | S |
| EG203 | 8.3 | 2.5 | R | 62.5 | 48.8 | MS |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Namisy, A.; Chen, J.-R.; Prohens, J.; Metwally, E.; Elmahrouk, M.; Rakha, M. Screening Cultivated Eggplant and Wild Relatives for Resistance to Bacterial Wilt (Ralstonia solanacearum). Agriculture 2019, 9, 157. https://doi.org/10.3390/agriculture9070157
Namisy A, Chen J-R, Prohens J, Metwally E, Elmahrouk M, Rakha M. Screening Cultivated Eggplant and Wild Relatives for Resistance to Bacterial Wilt (Ralstonia solanacearum). Agriculture. 2019; 9(7):157. https://doi.org/10.3390/agriculture9070157
Chicago/Turabian StyleNamisy, Ahmed, Jaw-Rong Chen, Jaime Prohens, Elmahdy Metwally, Mohammed Elmahrouk, and Mohamed Rakha. 2019. "Screening Cultivated Eggplant and Wild Relatives for Resistance to Bacterial Wilt (Ralstonia solanacearum)" Agriculture 9, no. 7: 157. https://doi.org/10.3390/agriculture9070157
APA StyleNamisy, A., Chen, J.-R., Prohens, J., Metwally, E., Elmahrouk, M., & Rakha, M. (2019). Screening Cultivated Eggplant and Wild Relatives for Resistance to Bacterial Wilt (Ralstonia solanacearum). Agriculture, 9(7), 157. https://doi.org/10.3390/agriculture9070157






