1. Introduction
In modern dairy farming, Jersey cattle are highly valued for their distinct production performance and superior milk quality [
1]. Reproductive traits rank among the most critical functional traits in dairy cattle, directly impacting herd productivity and economic sustainability. However, in recent years, intense selection for milk production traits, coupled with the negative genetic correlation between reproductive and milk production traits, has resulted in a decline in the reproductive performance of Jersey cows [
2]. Adding to the complexity of genetic improvement is the intricate genetic architecture of reproductive traits themselves, characterized by low heritability (typically less than 5%) and high susceptibility to environmental influences [
3]. Furthermore, recording reproductive data is often hampered by subjectivity and missing records, complicating accurate phenotypic and genetic evaluation. Therefore, identifying effective strategies to enhance reproductive capability is crucial. Identifying readily measurable traits with favorable genetic correlations to reproduction offers a crucial indirect strategy for genetic improvement [
4,
5].
Body conformation traits are vital for dairy cattle genetic improvement, serving not only as direct selection criteria but also as tools for evaluating economically important traits like reproduction [
6]. This utility stems from the physiological links between specific conformation traits and key reproductive processes, including estrus expression, ovulation, and calving ease (CE) [
7], supporting their role as supplementary fertility indicators. Substantial evidence supports this link: for instance, rump angle (RA) shows genetic correlations with calving interval (0.32) [
8], CE (−0.28) [
9] and retained placenta incidence (0.38) [
10]. Rump width (RW) is correlated with CE (0.15), commencement of luteal activity (−0.25) [
11], and early ovulation (EO, 0.55) [
9]. Similarly, stature demonstrates correlations with age at puberty (0.28) and pelvic dimensions [
12,
13], and both loin strength (LS) and body depth (BD) have been connected to CE and embryonic loss, respectively [
8,
9,
14]. While these findings highlight the broad relevance of conformation traits, their relationships with reproductive traits in Jersey cattle remain underexplored.
While phenotypic and genetic correlations provide valuable insights into the relationships between conformation and reproductive traits, uncovering the underlying genetic mechanisms requires genome-level investigation. Genome-wide association studies (GWAS) provides a powerful approach for mapping genetic variants underlying complex traits [
15]. Prior GWAS in cattle have identified loci influencing both conformation and reproduction, suggesting shared genetic control. For example, the
PLAG1 gene affects ST and body weight, and is also associated with fertility traits like AFC [
16,
17,
18]. Similarly, variants near
LCORL and
NCAPG impact ST, calf size, and CE [
19]. However, GWAS specifically targeting both reproduction and conformation traits in Jersey cattle are rarely reported.
This study systematically evaluates six conformation traits (LS, RA, BD, RW, ST, bone quality-BQ) and six reproductive traits (Age at First Service—AFS, AFC, GL, Number of Artificial Inseminations per Successful Conception—AIS, Calf Birth Weight—BiW, and CE) in Jersey cattle to achieve three main objectives: to estimate their genetic parameters and correlations, to assess the utility of conformation in predicting reproduction, and to identify associated genomic regions via GWAS. Through this integrated approach, we seek to elucidate the genetic architecture linking conformation to reproduction and explore its application for marker-assisted selection in Jersey cattle.
4. Discussion
Reproductive efficiency directly influences herd productivity and is therefore crucial for farm profitability and sustainable development. Although integrating fertility traits into breeding goals is a well-established strategy, direct selection for reproductive traits remains challenging due to their low heritability and difficulties in data recording. [
35]. Jersey cattle, known for their small size, high milk quality, early sexual maturity, high conception rate, and offspring survival, represent a valuable genetic resource [
36]. Understanding the genetic basis of their favorable reproductive characteristics is essential for breed improvement and may also provide insights applicable to other dairy breeds. Presently, China’s Jersey population remains in the early stages of introduction and local breeding programs, where limited pedigree and reproductive records impede direct genetic selection for fertility traits. Notably, accumulating evidence supports significant genetic correlations between specific conformation traits and reproductive performance [
37]. These objectively measurable conformation traits may offer potential supplementary selection criteria for enhancing genetic evaluations of fertility in data-limited contexts [
38]. Therefore, this study aimed to systematically evaluate the potential of conformation traits as predictors for reproductive performance. We estimated genetic parameters, performed GWAS, and evaluated the practical utility of significant variants via genomic prediction, thereby establishing an integrated genetic foundation for future selection strategies.
Currently, among conformation traits, the rump traitsexert the most significant influence on reproductive performance, including lumbar strength, RA, and RW [
37]. In China’s dairy cattle classification system, rump traits constitute 10% of the total score, while LS accounts for 8% within the body capacity category (18% weighting) [
39]. This study estimated heritability of LS in Jersey cattle at 0.20, which aligns with Wiggans et al.’s reported estimate (0.21) [
40] for Jersey cattle but is lower than Chinese Holstein values (0.25–0.38) [
37,
41]. LS evaluates structural soundness of the loin through vertebral depression or arching, which affects visceral suspension and birth canal stability, thereby influencing CE and calf survival. However, we detected no significant genetic correlation between LS and CE in Jersey cattle, contrasting with the moderate negative correlation (approximately −0.11) observed in Holsteins [
41]. This discrepancy may stem from Jersey cattle’s inherently compact conformation and well-developed hindquarter structure, where LS scores may not fully reflect functional support capacity. Moreover, Jersey cattle’s generally favorable CE likely the contribute to nonsignificant correlations. Notably, LS showed a genetic correlation of −0.24 with AIS, indicating Jersey cattle with stronger LS require fewer inseminations. A robust loin structure also facilitates body condition maintenance in higher lactations, enhancing durability and longevity [
42]. RA and RW reflect pelvic architecture, which plays a critical role in calving ease and calf viability [
43]. Optimal pelvic conformation (broad with moderate slope) promotes unassisted calving, reduces dystocia risk, and facilitates postpartum fluid expulsion, thereby decreasing reproductive disorders and metritis incidence [
44]. In this study, heritability estimates for RA (0.16) and RW (0.33) in Jersey cattle were moderate to low, consistent with ranges reported in Holsteins [
39], though slightly differing from some previous Jersey-specific estimates [
40]. Genetically, both RW and RA were negatively correlated with CE, while RA showed no significant association with BiW. Notably, RW and RA were also correlated with AFC and AFS, and RW exhibited a negative genetic correlation with GL.These findings align with Royal et al. [
11] and Carthy et al. [
9], who reported genetic correlations between RW and both the onset of luteal activity (−0.25) and early ovulation (0.55). Together, these results demonstrate intrinsic links between pelvic development and reproductive maturation processes in Jersey cattle.
Beyond rump traits, this study revealed genetic correlations between ST, BD, BQ and reproductive performance, particularly with AFC. Multiple regression further confirmed these conformation traits as significant predictors of reproductive outcomes, with RW consistently influencing multiple traits. Phenotypically, Jersey cattle with greater stature, deeper body depth, and stouter skeletal structure exhibited advanced development, leading to earlier insemination and higher calf birth weights. These observations aligned well with the phenotypic associations identified for AFC, AFS, and BiW. Heritability estimates for ST (0.62) and BQ (0.31) were higher than typical dairy cattle ranges (ST: 0.32–0.53; BQ: 0.05–0.30), whereas BD (0.05) was lower (0.10–0.17) [
39]. Most of the six conformation traits analyzed exhibited moderate-to-strong genetic correlations and highly significant phenotypic correlations, with largely consistent directional trends. Given that the conformation traits were assessed primarily in primiparous Jersey cattle, ST, BD, and BQ are considered particularly relevant for selecting well-developed individuals with early breeding potential [
45]. In contrast, in mature cows, body development should be moderated, particularly in BD and BQ, to prevent oversized conformation and associated health risks such as metabolic and reproductive disorders [
46].
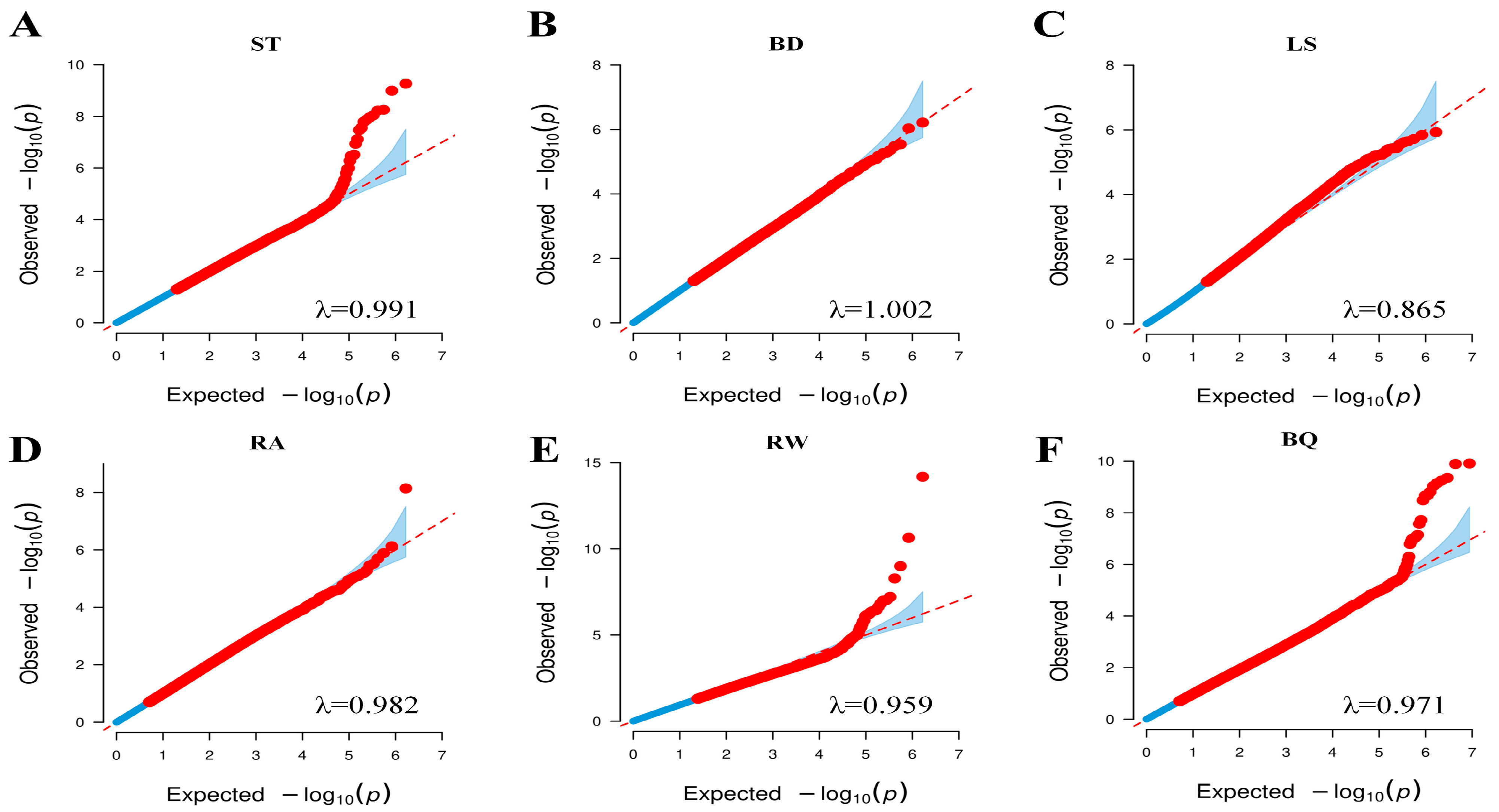
The accuracy of GWAS is influenced by adequate control of population structure and characterization of LD patterns [
47]. Our analysis identified three genetic subpopulations reflecting the diverse origins of the cattle, and the first three principal components (collectively explaining 15.27% of genetic variation) were included as covariates to control for stratification. LD decayed rapidly in this Jersey population, with mean r
2 dropping to approximately 0.08 at 200 kb—a faster rate than reported in Holstein cattle [
48] and Iranian water buffalo [
49], suggesting greater genetic diversity or distinct demographic history [
50]. This rapid decay highlights the value of using high-density SNP data, such as the 10.5 million imputed markers analyzed here. We employed the FarmCPU model, which iteratively combines fixed and random effect models to improve detection power and control false positives in structured populations [
51]. Its efficacy in handling complex architectures has been demonstrated in crops and other species [
52,
53], supporting its use in this study.
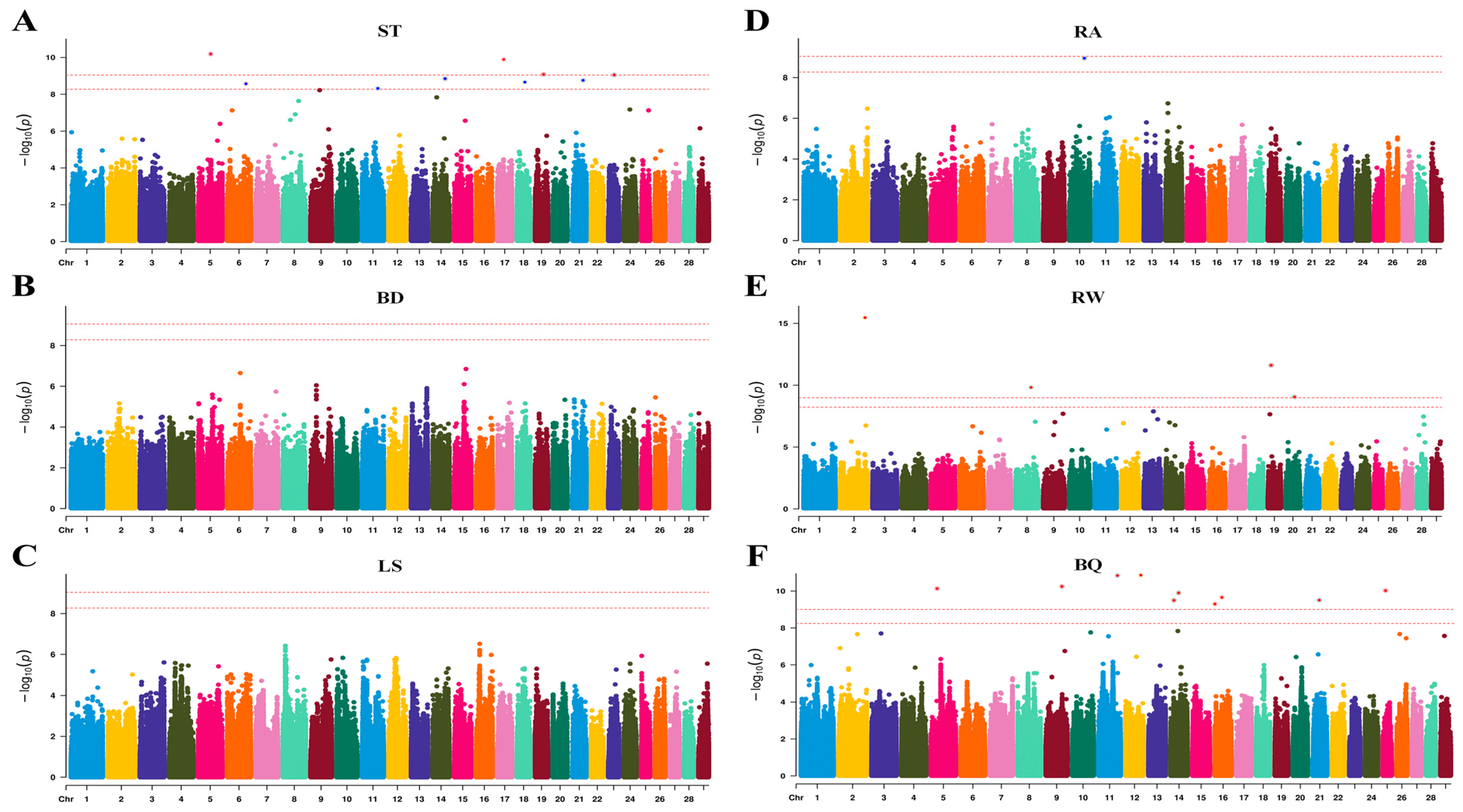
With these methodological foundations laid, the GWAS identified 24 significant SNPs associated with four conformation traits. Functional candidate genes were subsequently screened within 200 kb upstream and downstream of these SNPs, leading to the identification of 14 candidate genes in proximity to the significant loci. Functional characterization revealed that the identified genes predominantly cluster into two categories: those implicated in reproductive processes and those associated with metabolic regulation. Among these, genes with reproductive functions are of particular interest. The reproductive candidate genes associated with ST include
AZIN1, which influences bovine endometrial microenvironments during ovulation via polyamine metabolism pathways [
54,
55]. Also linked to ST is
KRT19, which is highly expressed in the epididymis and sperm of fertile bulls and is associated with sperm acrosomal integrity and membrane function [
56,
57]; it is also implicated in placental development post-fertilization [
56,
58]. In females, it serves as a marker for key gonadal development cells (e.g., GREL cells and ovarian surface epithelia) [
59], participating in ovarian development, folliculogenesis [
60], reproductive tract formation [
61], and endometrial function [
62], with its expression levels varying across reproductive stages [
60]. It is also downregulated in bovine blastocysts derived from vitrified morulae [
63].
KRT35, reported to be upregulated in non-obstructive azoospermia [
64], also functions in estrogen-mediated hair follicle development [
65] and is downregulated in the bovine endometrium during the mid-luteal phase [
66]. The association of these reproductive genes with ST may point to pleiotropic effects where genetic variants influencing skeletal frame size also co-regulate reproductive tissue development or function. Regarding RW,
bta-mir-2285br was identified and is known to associate with early follicular development in the bovine estrous cycle [
67], suggesting a possible genetic link between pelvic width and ovarian physiology. For RA, the significant SNP lay within
KCNH5, a gene specifically expressed in human placenta [
68] whose protein family participates in multiple physiological processes including cell cycle regulation, proliferation control and hormone release from endocrine cells [
69]; this provides a plausible molecular basis for how pelvic angle might influence calving ease. Finally, multiple genes related to BQ have direct reproductive roles.
OR2H1, enriched in testis-derived cells, mediates sperm chemotaxis [
70].
HS6ST3 shows significant correlations with reproductive traits including mating-to-conception intervals and AFS [
71]; its biological functions in heparan sulfate biosynthesis [
71] are essential for embryogenesis, growth factor signaling [
72] and mesoderm formation.
ERCC4 maintains genomic stability in early embryos [
73] and exhibits reduced expression in infertile human sperm [
74]. Together, these genes influence key reproductive processes including sperm quality, ovarian function, endometrial receptivity, and embryonic development.
Another group of genes is associated with lipid metabolism, adipogenesis, and related physiological processes. several metabolism-related genes were identified for ST.
ANXA3 regulates adipogenesis and metabolic processes [
75,
76,
77], directly influencing body weight dynamics [
78].
PKD1L3 participates in glucose and lipid metabolism, where missense variants alter triglyceride levels [
79].
TDRD15 modulates adipose function and spermatogenesis [
80,
81], with documented upregulation in mature equine testes [
82].
KRT33A, a keratin gene, influences claw disorders in Holsteins [
83,
84]. This cluster of associations may reflect a herd-specific phenomenon where individuals with high ST were more prone to over-conditioning under the prevailing management. For RW,
TMEM132E may associate with metabolic disorders [
85]. Genes associated with BQ include the putative obesity gene
PLD5 [
86] and
RCN2, which contributes to osteogenesis [
87] and suppresses appetite-mediated obesity through circadian rhythm modulation [
88]. The detection of these metabolic genes, particularly for ST and BQ, aligns with the documented susceptibility of Jersey cattle to energy imbalance and metabolic disorders. This suggests that these conformation traits also capture meaningful genetic variation in energy balance and fat deposition within this breed.
Lower MAF thresholds may help capture low-frequency variants with potentially larger effects [
89], and previous studies have reported additional loci when applying less stringent MAF filters. To evaluate whether such signals could also be informative in our Jersey population, we compared GWAS results using MAF > 0.01 and MAF > 0.05. The MAF > 0.01 analysis identified several additional candidate genes, including
NTRK2 [
90] and
CEP152 [
91], both of which have plausible links to reproductive biology. However, the overlap of key genes between the two thresholds was limited, and the lower cutoff did not increase the number of robust, consistently interpretable signals across traits. Given that our primary objective is to identify stable and biologically meaningful loci that connect conformation with reproduction, we therefore retained MAF > 0.05 as the main threshold and present the MAF > 0.01 results as supportive evidence highlighting potential low-frequency candidates that warrant further validation.
This study systematically demonstrates, through both genetic correlation analysis and regression analyses, that conformation traits possess significant predictive value for reproductive performance. More importantly, by utilizing these objectively measurable conformation traits as a bridge, we successfully identified multiple reproduction-related SNPs and candidate genes via GWAS, highlighting the pivotal role of conformation traits in elucidating the genetic basis of reproduction. However, several limitations must be acknowledged: subjectivity in scoring and moderate-to-low genetic correlations introduce noise and constrain predictive accuracy [
92], and SNPs identified primarily affect conformation with limited direct impact on reproduction. While these considerations temper expectations and warrant cautious implementation, they do not diminish the important status of conformation traits in breeding practice. Therefore, conformation traits should be regarded as valuable supplementary indicators in genetic evaluations of reproductive performance, particularly offering unique value in populations with incomplete reproductive records [
37]. In summary, this research not only establishes reliable associations between conformation traits and reproductive efficiency, but more significantly, through this strategy, identifies several genomic regions and candidate genes of considerable biological importance, providing new targets for marker-assisted selection in Jersey cattle and laying the groundwork for balanced breeding strategies that integrate high productivity with superior fertility.