Identification and Characterization of Maize Yellow Mosaic Virus Causing Mosaic Symptoms on Maize in Taiwan
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Isolation, and Preliminary Detection
2.2. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
2.3. Virus Isolation
2.4. Establishment of Non-Viruliferous Aphid Colonies
2.5. Transmission Assay
2.6. Rapid Amplification of cDNA Ends (RACE)
2.7. Whole Genome Amplification
2.8. Sequence and Phylogenetic Analysis
2.9. Sample Collection in Maize Field
3. Results
3.1. Occurrence of MaYMV
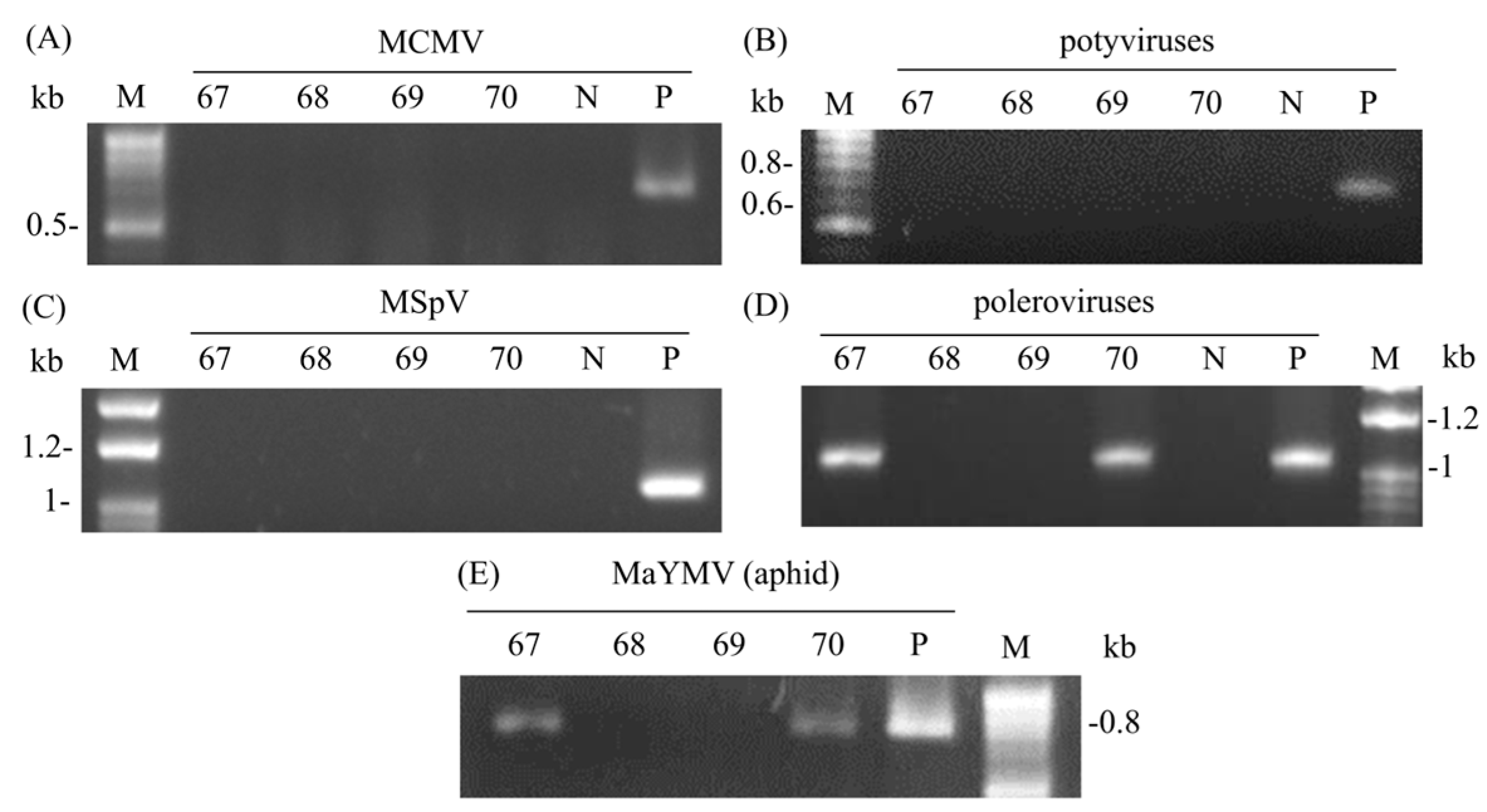
3.2. Isolation of Virus
3.3. Host Range Test
3.4. Whole Viral Genome and Individual Gene Sequences Analysis
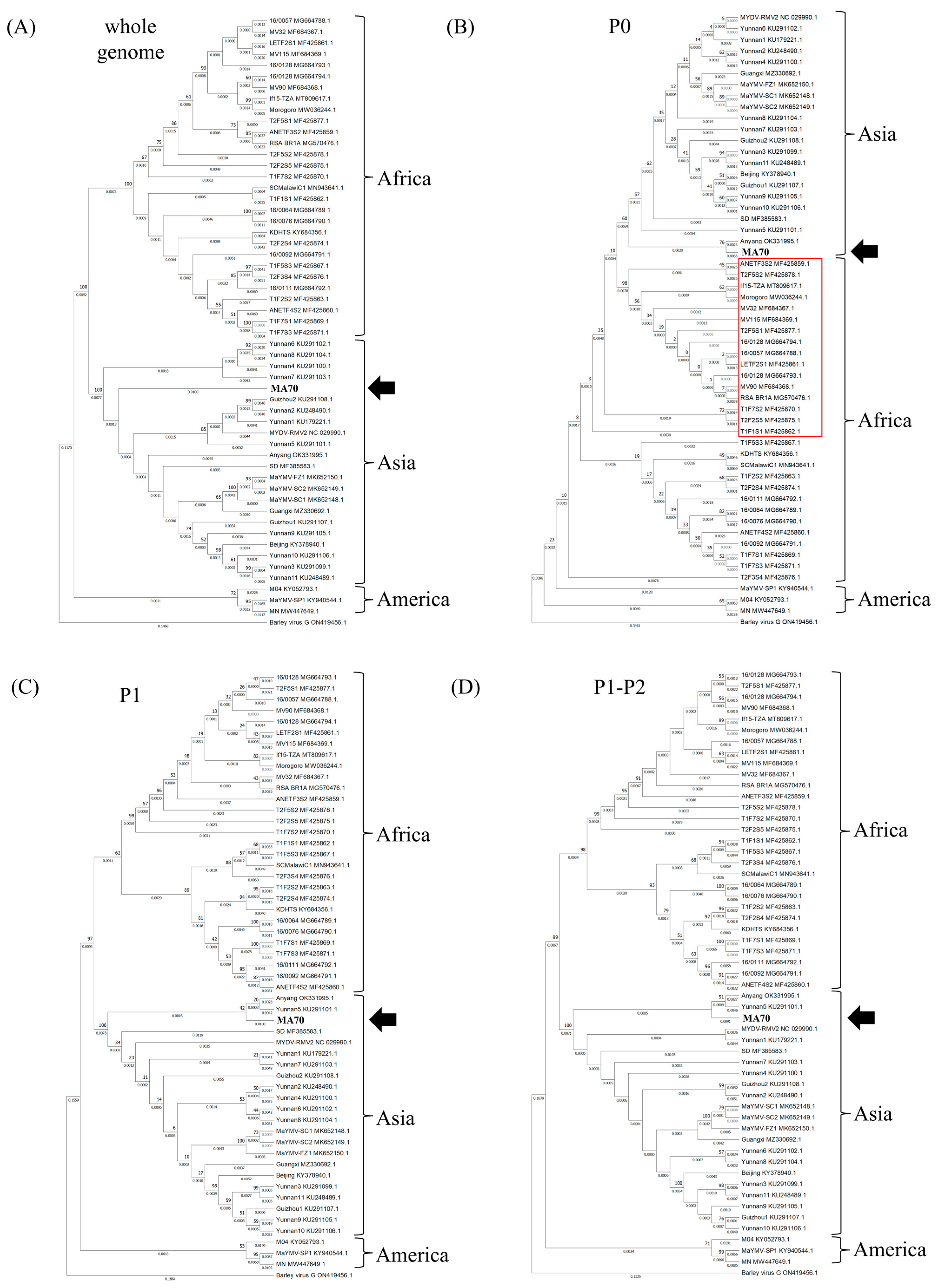
3.5. Phylogenetic Analysis
3.6. Field Survey of MaYMV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. FAOSTAT Statistical Database. Available online: https://www.fao.org/faostat/en/#compare (accessed on 10 February 2024).
- Agriculture and Food Agency, Ministry of Agriculture. Agricultural Situation Report Resource Network. Available online: https://agr.afa.gov.tw (accessed on 16 February 2024).
- Signoret, P.-A.; Lapierre, H. Virus diseases of maize. In Viruses and Virus Diseases of Poaceae (Gramineae); Signoret, P.-A., Lapierre, H., Eds.; Mieux Comprendre: Paris, France, 2004; pp. 617–696. [Google Scholar]
- Sastry, K.S.; Mandal, B.; Hammond, J.; Scott, S.W.; Briddon, R.W. Zea mays. In Encyclopedia of Plant Viruses and Viroids; Sastry, K.S., Mandal, B., Hammond, J., Scott, S.W., Briddon, R.W., Eds.; Springer: New Delhi, India, 2020; pp. 2824–2853. [Google Scholar]
- Uyemoto, J.K.; Bockelman, D.L.; Claflin, L.E. Severe outbreak of corn lethal necrosis disease in Kansas. Plant Dis. 1980, 64, 99–100. [Google Scholar] [CrossRef]
- Niblett, C.; Claflin, L. Corn lethal necrosis—A new virus disease of corn in Kansas. Plant Dis. Rep. 1978, 62, 15–19. [Google Scholar]
- Mahuku, G.; Lockhart, B.E.; Wanjala, B.; Jones, M.W.; Kimunye, J.N.; Stewart, L.R.; Cassone, B.J.; Sevgan, S.; Nyasani, J.O.; Kusia, E.; et al. Maize lethal necrosis (MLN), an emerging threat to maize-based food security in sub-Saharan Africa. Phytopathology 2015, 105, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.C.; Chou, C.M.; Chen, C.T.; Tsai, C.H.; Lin, F.C. First report of maize chlorotic mottle virus on sweet corn in Taiwan. Plant Dis. 2014, 98, 1748. [Google Scholar] [CrossRef]
- Lin, S.J. Research of sugarcane mosaic disease. Rep. Taiwan Sugar Exp. Stat. 1968, 47, 139–153. Available online: https://taiwanebook.ncl.edu.tw/zh-tw/book/NCL-9910010577/reader (accessed on 15 December 2025).
- Deng, T.C. Identification of maize dwarf mosaic virus B strain and screening for resistance of corn. J. Agric. Res. China 1985, 34, 195–206. [Google Scholar] [CrossRef]
- Chen, C.; Tsai, J.; Chiu, R.; Chen, M. Purification, characterization, and serological analysis of maize stripe virus in Taiwan. Plant Dis. 1993, 77, 367–372. [Google Scholar] [CrossRef]
- Chou, C.M.; Chen, C.T.; Chien, Y.H.; Tsai, C.H.; Chen, C.C.; Lin, F.C.; Chen, Y.J.; Deng, T.C. Phylogenetic analysis and occurrence of maize chlorotic mottle virus infecting maize in Taiwan. J. Taiwan Agric. Res. 2018, 67, 191–208. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, G.; Wu, J.; Liu, Y.; Qian, Y.; Zhou, X. Characterization of a novel polerovirus infecting maize in China. Viruses 2016, 8, 120. [Google Scholar] [CrossRef]
- Gonçalves, M.; Godinho, M.; Alves-Freitas, D.; Varsani, A.; Ribeiro, S. First report of maize yellow mosaic virus infecting maize in Brazil. Plant Dis. 2017, 101, 2156. [Google Scholar] [CrossRef]
- Guadie, D.; Abraham, A.; Tesfaye, K.; Winter, S.; Menzel, W.; Knierim, D. First report of maize yellow mosaic virus infecting maize (Zea mays) in Ethiopia. Plant Dis. 2018, 102, 1044. [Google Scholar] [CrossRef]
- Read, D.A.; Featherstone, J.; Rees, D.J.G.; Thompson, G.D.; Roberts, R.; Flett, B.C.; Mashingaidze, K.; Berger, D.K.; Welgemoed, T.; Pietersen, G. First report of maize yellow mosaic virus (MaYMV) on maize (Zea mays) in Tanzania. J. Plant Pathol. 2019, 101, 203. [Google Scholar] [CrossRef]
- Bernreiter, A.; Teijeiro, R.G.; Jarrin, D.; Garrido, P.; Ramos, L. First report of maize yellow mosaic virus infecting maize in Ecuador. New Dis. Rep. 2017, 36, 2044–20588. [Google Scholar] [CrossRef]
- Welgemoed, T.; Pierneef, R.; Read, D.A.; Schulze, S.E.; Pietersen, G.; Berger, D.K. Next-generation sequencing reveals past and current widespread occurrence of maize yellow mosaic virus in South Africa. Eur. J. Plant Pathol. 2020, 158, 237–249. [Google Scholar] [CrossRef]
- Nithya, K.; VishnuVardhan, J.; Balasaravanan, S.; Vishalakshi, D.; Kaverinathan, K.; Viswanathan, R. First report of maize yellow mosaic virus (MaYMV) infecting sugarcane in India and its molecular characterization. Australas. Plant Pathol. 2021, 50, 633–638. [Google Scholar] [CrossRef]
- Yahaya, A.; Al Rwahnih, M.; Dangora, D.; Gregg, L.; Alegbejo, M.; Lava Kumar, P.; Alabi, O. First report of maize yellow mosaic virus infecting sugarcane (Saccharum spp.) and itch grass (Rottboellia cochinchinensis) in Nigeria. Plant Dis. 2017, 101, 1335. [Google Scholar] [CrossRef]
- Lim, S.; Yoon, Y.; Jang, Y.; Bae, D.; Kim, B.S.; Maharjan, R.; Yi, H.; Bae, S.; Lee, Y.H.; Lee, B. First report of maize yellow mosaic virus infecting Panicum miliaceum and Sorghum bicolor in South Korea. Plant Dis. 2018, 102, 689. [Google Scholar] [CrossRef]
- Palanga, E.; Longué, R.; Koala, M.; Néya, J.; Traoré, O.; Martin, D.P.; Peterschmitt, M.; Filloux, D.; Roumagnac, P. First report of maize yellow mosaic virus infecting maize in Burkina Faso. New Dis. Rep. 2017, 35, 26. [Google Scholar] [CrossRef]
- Tanui, C.; Anami, S.; Wamaitha, J.; Wanjal, B.W. Occurrence of maize yellow mosaic virus and evidence of co-infection with maize lethal necrosis viruses in Bomet County, Kenya. Afr. J. Plant Sci. 2021, 15, 299–308. [Google Scholar]
- Adams, I.; Braidwood, L.; Stomeo, F.; Phiri, N.; Uwumukiza, B.; Feyissa, B.; Mahuku, G.; Wangi, A.; Smith, J.; Mumford, R. Characterising maize viruses associated with maize lethal necrosis symptoms in Sub-Saharan Africa. bioRxiv 2017. bioRxiv:161489. [Google Scholar]
- Delfosse, V.C.; Barrios Barón, M.P.; Distéfano, A.J. What we know about poleroviruses: Advances in understanding the functions of polerovirus proteins. Plant Pathol. 2021, 70, 1047–1061. [Google Scholar] [CrossRef]
- Guo, M.; Yuan, X.; Song, Y.; Liu, Y.; Wang, X. First report of maize yellow mosaic virus (MaYMV) naturally infecting wheat in China. Plant Dis. 2022, 106, 2763. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.; Jiang, X.; Yang, X.; Liu, X.; Wang, Y.; Liu, Y.; Li, H.; Yuan, H.; Shi, Y. First report of maize yellow mosaic virus on weed hosts in China. Plant Dis. 2023, 107, 4036. [Google Scholar] [CrossRef]
- Mlotshwa, S.; Khatri, N.; Todd, J.; Tran, H.H.; Stewart, L.R. First report of cDNA clone-launched infection of maize plants with the polerovirus maize yellow mosaic virus (MaYMV). Virus Res. 2021, 295, 198297. [Google Scholar] [CrossRef] [PubMed]
- Ohlson, E.W.; Khatri, N.; Wilson, J.R. Experimental host and vector ranges of the emerging maize yellow mosaic polerovirus. Plant Dis. 2024, 108, 1246–1251. [Google Scholar] [CrossRef]
- Gonçalves, M.C.; Ramos, A.; Nascimento, T.; Harakava, R.; Duarte, A.P.; Lopes, J.R. Aphid transmission of maize yellow mosaic virus: An emerging polerovirus. Trop. Plant Pathol. 2020, 45, 544–549. [Google Scholar] [CrossRef]
- Castello, J.; Rogers, S.; Bachand, G.; Fillhart, R.; Murray, J.; Weidemann, K.; Bachand, M.; Almond, M. Detection and partial characterization of tenuiviruses from black spruce. Plant Dis. 2000, 84, 143–147. [Google Scholar] [CrossRef]
- Knierim, D.; Deng, T.; Tsai, W.; Green, S.; Kenyon, L. Molecular identification of three distinct Polerovirus species and a recombinant cucurbit aphid-borne yellows virus strain infecting cucurbit crops in Taiwan. Plant Pathol. 2010, 59, 991–1002. [Google Scholar] [CrossRef]
- Pappu, S.; Pappu, H.; Chang, C.; Culbreath, A.; Todd, J. Differentiation of biologically distinct peanut stripe potyvirus strains by a nucleotide polymorphism-based assay. Plant Dis. 1998, 82, 1121–1125. [Google Scholar] [CrossRef]
- Colinet, D.; Kummert, J. Identification of a sweet potato feathery mottle virus isolate from China (SPFMV-CH) by PCR with degenerate primers. J. Virol. Methods 1993, 45, 149–159. [Google Scholar] [CrossRef]
- Stewart, L.R.; Todd, J.; Willie, K.; Massawe, D.; Khatri, N. A recently discovered maize polerovirus causes leaf reddening symptoms in several maize genotypes and is transmitted by both the corn leaf aphid (Rhopalosiphum maidis) and the bird cherry-oat aphid (Rhopalosiphum padi). Plant Dis. 2020, 104, 1589–1592. [Google Scholar] [CrossRef]
- Bhat, A.I.; Rao, G.P. Transmission of viruses by aphids. In Characterization of Plant Viruses: Methods and Protocols; Bhat, A.I., Rao, G.P., Eds.; Humana: New York, NY, USA, 2020; pp. 69–75. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Delp, B.; Stowell, L.; Marois, J. Evaluation of field sampling techniques for estimation of disease incidence. Phytopathology 1986, 76, 1299–1305. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Mann, K.S.; Johnson, K.N. Plant virus–insect vector interactions: Current and potential future research directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.R.; Chen, J.S.; Yang, J.; Huang, X.C.; Huang, M.T.; Gao, S.J. First report of maize yellow mosaic virus infecting sugarcane in China. Plant Dis. 2019, 103, 2482. [Google Scholar] [CrossRef]
- Sun, S.R.; Chen, J.S.; He, E.Q.; Huang, M.T.; Fu, H.Y.; Lu, J.J.; Gao, S.J. Genetic variability and molecular evolution of maize yellow mosaic virus populations from different geographic origins. Plant Dis. 2021, 105, 896–903. [Google Scholar] [CrossRef]
- Chang, L.Z.; Huang, S.C. Achievement and perspectives of sorghum variety improvement in Taiwan. Improv. Prod. Technol. Upl. Crops Taiwan 1995, 49, 125–133. [Google Scholar]
- Hull, R. Plant-to-plant movement. In Plant Virology; Hull, R., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 669–751. [Google Scholar]
- Loxdale, H.D.; Hardie, J.; Halbert, S.; Foottit, R.; Kidd, N.A.; Carter, C.I. The relative importance of short- and long-range movement of flying aphids. Biol. Rev. 1993, 68, 291–311. [Google Scholar] [CrossRef]
- Chung, B.N.; Koh, S.W.; Choi, K.S.; Joa, J.H.; Kim, C.H.; Selvakumar, G. Temperature and CO2 level influence potato leafroll virus infection in Solanum tuberosum. Plant Pathol. J. 2017, 33, 522. [Google Scholar] [CrossRef]
- Kaakeh, W.; Dutcher, J.D. Effect of rainfall on population abundance of aphids (Homoptera: Aphididae) on pecan. J. Entomol. Sci. 1993, 28, 283. [Google Scholar] [CrossRef]
- Marabi, R.; Das, S.; Bhowmick, A.; Pachori, R.; Sharma, H. Seasonal population dynamics of whitefly (Bemisia tabaci Gennadius) in soybean. J. Entomol. Zool. Stud. 2017, 5, 169–173. [Google Scholar]
- Central Weather Administration. Daily Rainfall. Available online: https://www.cwa.gov.tw/V8/C/D/DailyPrecipitation.html (accessed on 1 June 2024).
- Svanella-Dumas, L.; Vitry, C.; Valade, R.; Robin, N.; Thibord, J.B.; Marais, A.; Candresse, T. First report of barley virus G infecting winter barley (Hordeum vulgare) in France. Plant Dis. 2023, 107, 591. [Google Scholar] [CrossRef]
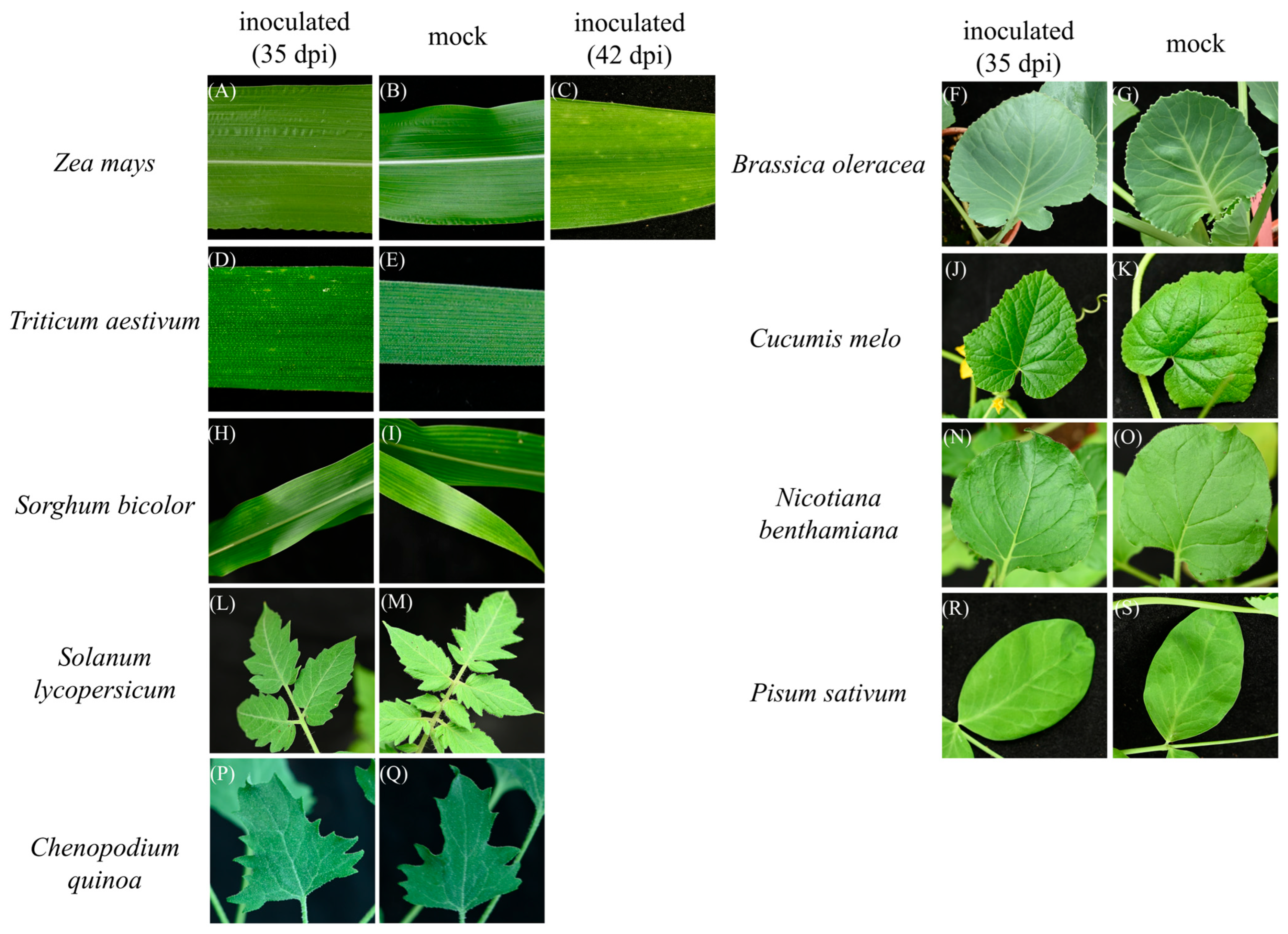

| Species | Cultivar | Infection Ratio (1st) | Infection Ratio (2nd) | Infection Ratio (3rd) |
|---|---|---|---|---|
| Zea mays | Red autumn | 3 a/3 b | 10/10 | 17/20 |
| Sorghum bicolor | Taichung No. 5 | 0/3 | 0/10 | 0/18 |
| Triticum aestivum | Taichung Sel. 2 | 3/3 | 10/10 | 12/20 |
| Solanum lycopersicum | 301 | 0/3 | 0/10 | 0/20 |
| Nicotiana benthamiana | 0/3 | 0/10 | 0/20 | |
| Chenopodium quinoa | 0/3 | 0/10 | 0/20 | |
| Pisum sativum | 0/3 | 0/10 | 0/20 | |
| Brassica oleracea | 0/3 | 0/10 | 0/20 | |
| Cucumis melo | Silver light | 0/3 | 0/10 | 0/20 |
| Whole Genome | P0 | P1 | P1–P2 | P3a | P3 | P4 | P3–P5 | |
|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence | ||||||||
| MaYMV (54 isolate a) | 94.8–96.8% b | 96.7–99.1% | 95.3–98.7% | 95.7–98.7% | 97–100% | 97.8–99.2% | 97.9–99.1% | 93.2–98.5% |
| BVG (ON419456.1) | 77.8% | 60% | 74.6% | 80% | 88.1% | 83.8% | 84.2% | 76.1% |
| Amino acid sequence | ||||||||
| MaYMV (54 isolate) | - | 95.5–98.9% | 95.2–98.7% | 97.3–99.1% | 97.8–100% | 96.4–98.5% | 95.8–99% | 93.9–98.2% |
| BVG (ON419456.1) | - | 48.9% | 67.7% | 78% | 91.1% | 79.8% | 71.5% | 75.6% |
| Location/No. | Date | MaYMV ratio |
|---|---|---|
| Daya No.1 | 20 October 2023 | 8 a/137 b/5.8% c |
| Houli No.1 | 31 October 2023 | 7/111/6.3% |
| Fengyuan No.1 | 1 November 2023 | 4/113/3.5% |
| Waipu No.1 | 22 November 2023 | 19/151/12.6% |
| Daya No.2 | 15 December 2023 | 32/81/39.5% |
| Wuri No.1 | 3 January 2024 | 60/150/40% |
| Total | 130/743/17.5% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Chen, J.-H.; Ku, H.-M.; Chang, H.-H.; Chang, C.-J.; Jan, F.-J. Identification and Characterization of Maize Yellow Mosaic Virus Causing Mosaic Symptoms on Maize in Taiwan. Agriculture 2026, 16, 27. https://doi.org/10.3390/agriculture16010027
Chen J-H, Ku H-M, Chang H-H, Chang C-J, Jan F-J. Identification and Characterization of Maize Yellow Mosaic Virus Causing Mosaic Symptoms on Maize in Taiwan. Agriculture. 2026; 16(1):27. https://doi.org/10.3390/agriculture16010027
Chicago/Turabian StyleChen, Jing-Han, Hsin-Mei Ku, Ho-Hsiung Chang, Chung-Jan Chang, and Fuh-Jyh Jan. 2026. "Identification and Characterization of Maize Yellow Mosaic Virus Causing Mosaic Symptoms on Maize in Taiwan" Agriculture 16, no. 1: 27. https://doi.org/10.3390/agriculture16010027
APA StyleChen, J.-H., Ku, H.-M., Chang, H.-H., Chang, C.-J., & Jan, F.-J. (2026). Identification and Characterization of Maize Yellow Mosaic Virus Causing Mosaic Symptoms on Maize in Taiwan. Agriculture, 16(1), 27. https://doi.org/10.3390/agriculture16010027






