Abstract
Infestation by Varroa destructor is responsible for high mortality rates in Apis mellifera colonies worldwide. This study was designed to develop and test under field conditions a new free software (VarroDetector) based on a deep learning approach for the automated detection and counting of Varroa mites using smartphone images of sticky boards collected in honeybee colonies. A total of 204 sheets were collected, divided into four frames using green strings, and photographed under controlled lighting conditions with different smartphone models at a minimum resolution of 48 megapixels. The Varroa detection algorithm comprises two main steps: First, the region of interest where Varroa mites must be counted is established. From there, a one-stage detector is used, namely YOLO v11 Nano. A final verification was conducted counting the number of Varroa mites present on new sticky sheets both manually through visual inspection and using the VarroDetector software and comparing these measurements with the actual number of mites present on the sheet (control). The results obtained with the VarroDetector software were highly correlated with the control (R2 = 0.98 to 0.99, depending on the smartphone camera used), even when using a smartphone for which the software was not previously trained. When Varroa mite numbers were higher than 50 per sheet, the results of VarroDetector were more reliable than those obtained with visual inspection performed by trained operators, while the processing time was significantly reduced. It is concluded that the VarroDetector software Version 1.0 (v. 1.0) is a reliable and efficient tool for the automated detection and counting of Varroa mites present on sticky boards collected in honeybee colonies.
1. Introduction
Honeybees are among the most essential pollinators, contributing significantly to the reproductive success of both cultivated and wild plant species [1,2]. Their role in agricultural ecosystems is particularly relevant. It has been estimated that nearly 70% of all crop species globally rely on bees to some extent for successful pollination, underscoring their importance in food production [1]. However, in recent years, alarming reports of widespread colony losses have emerged, raising serious concerns among scientists, policymakers, and the general public regarding the long-term viability of honeybee populations [3,4,5]. The causes of this decline include pests and diseases, bee management, including beekeeping practices and breeding, the change in climatic conditions, agricultural practices, and the use of pesticides [6]. The decline of these pollinators could have profound ecological and economic consequences, necessitating urgent research and conservation efforts to mitigate potential impacts.
Among the sanitary causes of colony losses, infestation by the ectoparasitic mite Varroa destructor (hereafter Varroa) represents the most critical pathogenic threat to the western honeybee, Apis mellifera, on a global scale [7,8,9]. Following its worldwide dissemination between the 1950s and 1990s, Varroa has profoundly disrupted apicultural practices and economic viability [8]. The mite is an obligate parasite of honeybee colonies, exhibiting a reproductive cycle tightly synchronized with host brood development. It feeds on the hemolymph and fat body tissue of both developing and adult bees [10]. As a consequence, it reduces honeybee weight, immune responses, and lifespan and alters the flying and orientation abilities of foragers [11,12]. Furthermore, Varroa also acts as an efficient biological vector for multiple honeybee viruses [8,11]. The synergistic effects of viral opportunism and exponential mite proliferation often culminate in lethal viral outbreaks, typically leading to colony collapse within two to three years [8].
Given that the eradication of varroosis does not appear to be a realistic goal, control measures to mitigate its negative impact are essential. In any control strategy, monitoring the level of Varroa infestation is fundamental. One of the most common monitoring methods is analyzing the natural mite fall onto sticky boards placed at the bottom of the hive [9,13,14]. This method offers several advantages over other diagnostic options, because (1) it avoids colony disruption and handling or harming bees, (2) it is widely recognized as the most accurate and consistent approach for estimating the total mite population, as it provides a comprehensive assessment of the entire colony rather than relying on a limited bee sample, (3) it enables a rapid and accurate evaluation of the effectiveness of treatments against Varroa, and (4) it is particularly useful for detecting and selecting Varroa-resistant colonies across a large number of hives [7].
Varroa mite counting on sticky boards is typically performed by visual inspection and demands substantial time and effort. Moreover, the small size of the mites (about 1–1.8 mm long and 1.5–2 mm wide), the presence of debris, and the potentially high mite density further complicate accurate counting, making the method prone to errors and inconsistencies [9]. The challenges associated with Varroa mite detection underscore the need for developing advanced technologies capable of rapidly and accurately estimating mite populations [15]. In this context, artificial intelligence (AI)-based systems are particularly well-suited for image-based diagnostics. These techniques utilize deep learning algorithms trained on large datasets to identify patterns and morphological features that may be challenging to detect with the human eye and with traditional computer vision techniques. However, little research has been carried out aiming to automatize Varroa mite detection and counting on adult honeybees [16,17,18,19], pupae [20], and bottom boards [21,22]. Moreover, in some cases, the results were not satisfactory, some are commercial, and most of them remain at the prototype stage and have not been properly validated in the field. The objective of this study was to develop and test under field conditions a new free software (VarroDetector) based on a deep learning approach for the automated detection and counting of Varroa mites using smartphone images of sticky boards collected in honeybee colonies. To this end, VarroDetector combines image processing techniques and a lightweight neuronal network to detect Varroa mites from sticky sheets, even under the presence of debris and high mite density. This standalone solution delivers precise detection without requiring internet connectivity or cloud computing resources. Designed for efficiency, and being a free tool compatible with widely used commercial smartphones and computers, it can be highly beneficial for beekeepers, technicians, and researchers.
2. Materials and Methods
The experiment was carried out between October 2024 and March 2025 in two commercial apiaries in Aniés (Huesca, Spain), comprising a total of 70 Langstroth hives equipped with modified bottom boards with mesh floor and with varying levels of Varroa infestation. During the trial, a sticky sheet was placed beneath each beehive for four days to collect naturally fallen Varroa mites. A total of 204 sheets were collected and transported to the laboratory, where they were photographed under controlled lighting conditions.
2.1. Image Acquisition
The sheets were numbered with the colony number and divided into four frames of 23.5 × 18.5 cm using green strings (Figure 1). Each frame was photographed either with an iPhone14 Pro Max (Apple Inc., Cupertino, CA, USA, using iOS 18 software) at 48 megapixels and with a Xiaomi Poco X5 Pro (Xiaomi Inc., Beijing, China, using Android 14 software) at 108 megapixels. The images exhibited a highly variable number of Varroa mites, ranging from 1 to 351.

Figure 1.
Example images of a sticky sheet divided in four frames (photographs).
2.2. Dataset Description
The dataset comprised 357 images containing Varroa obtained from 90 of sticky sheets. These images were randomly divided into approximately 80% for training and 20% for validation, ensuring that the proportion of Varroa mites remained consistent across both sets. More than 10,000 and 3000 Varroa were analyzed for training and validation, respectively. All images were manually annotated by experts, who delineated each Varroa mite using bounding boxes.
2.3. Algorithm Description
The Varroa detection algorithm comprises two main steps: First, the region of interest where Varroa mites must be counted should be established. The images contain green strings that divide the sticky board into frames, and the developed models first detect these strings in the image. This process involves color-based image filtering followed by the application of the Hough transform to identify two perpendicular lines that delineate the counting area. Subsequently, the software identifies Varroa mites within the designated area rather than across the entire image to avoid counting the same Varroa in different images. For this, a one-stage detector is used, namely YOLO v11 Nano, a state-of-the-art lightweight neural network architecture containing approximately 2.6 M parameters that has demonstrated broad applicability across multiple domains. This neural network is employed, since it provides good accuracy with very high speed, which is important, since the goal is to allow inference on standard computers without a graphics processing unit (GPU). In particular, YOLOv11 was launched in September 2024 and was the last available version at the time of research. It follows the principles of the outstanding paper [23], where the well-known YOLO architecture [24] was presented for the first time, but this version introduces novel components, like a computationally efficient implementation of the cross-stage partial (CSP) bottleneck, for improving feature extraction and also an attention mechanism (CSP with spatial attention, named C2PSA) that allows the model to focus more effectively on important regions in the image. All this within a nano architecture [25], which incorporates improvements, such as residual projection–expansion–projection macroarchitectures to significantly reduce the number of parameters and the complexity of the network.
Figure 2 depicts the architecture diagram of the YOLOv11 network. The network comprises three main components: backbone, neck, and head. The backbone essentially functions as a feature extraction network that processes input images to derive hierarchical visual representations. The neck consists of intermediate layers that connect the backbone to the head, enhancing feature fusion across different scales. The head is the final detection layer that takes features from the neck and outputs the final predictions. The backbone incorporates multiple convolutional layers frequently followed by C3K2 blocks—an architectural innovation designed to improve feature aggregation. The architecture also incorporates a new spatial pyramid pooling—fast (SPPF) layer to perform multi-scale feature extraction by applying max-pooling operations at different kernel sizes to capture information at various receptive fields in a more efficient way than the original SPP. The architecture additionally utilizes skip connections (pathways that allow information to bypass one or more layers), which enable the network to preserve and utilize features from earlier layers, combining low-level details with high-level semantic information.
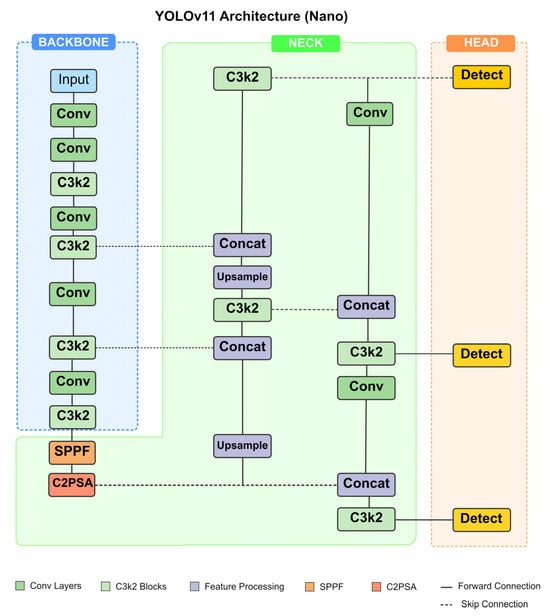
Figure 2.
Architecture diagram of the YOLOv11 network used in this article.
A diagram of the workflow of the application is depicted in Figure 3. A brief summary follows:
- Step 1: Conversion from RGB to HSV colorspace is performed, followed by string color filtering of the image.
- Step 2: Morphological operations (erosion and dilation) are applied to refine the filtered result.
- Step 3: Edge detection is applied via the Canny algorithm.
- Step 4: Hough transform is utilized for straight line detection.
- Step 5: The resulting segments are sorted by length, with the longest segments selected. These segments are extended to the image boundaries. Intersection points are calculated, and segments forming approximately 90-degree angles are preserved and drawn as complete lines to the edges in a binary image.
- Step 6: Contour detection is performed, resulting in the delimitation of four distinct regions.
- Step 7: The largest region is isolated and converted to a binary mask (white pixels on black background).
- Step 8: The generated mask is applied to the original image for selecting the region of interest within the strings.
- Step 9: Predictions are made with the YOLO deep learning model.
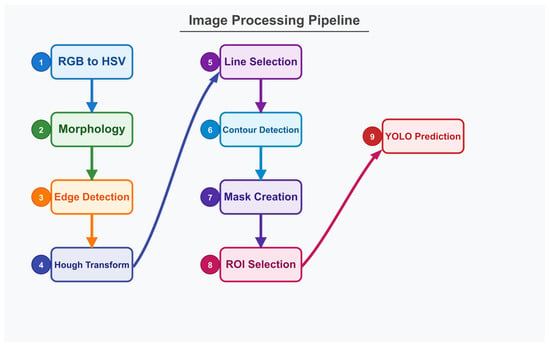
Figure 3.
Diagram of the workflow of the VarroDetector software.
Figure 4 illustrates the processing workflow applied to a representative image, demonstrating each transformation stage.
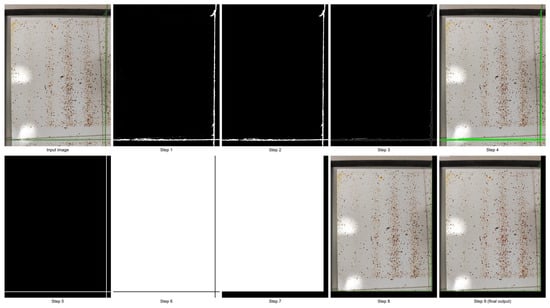
Figure 4.
Workflow of the VarroDetector software applied to one image.
The neuronal network was trained on the training set for 500 epochs, with an early stopping criterion if there was no improvement in 50 epochs. The input image size of the network was chosen to be 8000, instead of the default 640 value. This choice allowed the network to detect small objects as the Varroa mites, though with significant computational demands that were mitigated through the use of a nano network and single-image batch processing. Data augmentation (standard changes in scale, saturation, color, brightness, and flips) and regularization techniques (dropout = 0.05 and weight_decay = 0.001) were employed to enhance model robustness. Validation images were used during the model training process to tune the hyperparameters and choose the best model. The trained model obtained a precision of 0.925, a recall of 0.921, and a mean average precision (mAP) score of 0.956 on the validation set [26].
The code is open-source and is available at https://github.com/jodivaso/varrodetector (accessed on 25 April 2025). The code has been developed using the Python language. The aforementioned repository also contains stand-alone executables for Windows and Linux. The software needs neither installation nor internet connection to be used. A representative sample of the sticky sheet images are also provided in the aforementioned webpage.
2.4. Software Interface
The software features an intuitive user interface, as illustrated in Figure 5, and can be used by any user without the need of computer or AI skills. Based on Figure 5, the main parts of the interface are enumerated:
- (1)
- Select input folder: A button to allow users to select a folder containing the images to be analyzed. If the folder includes subfolders, the program recursively also processes all images within the directory. This functionality is particularly useful for analyzing multiple sticky sheets simultaneously, especially when each sheet’s four images are stored in separate subfolders.
- (2)
- Save button: The program allows to export statistical data in CSV format. Additionally, annotated images with detected mites and their corresponding labels are saved in YOLO format.
- (3)
- Help button: This button displays information about the application and a guide about the basic controls and functionality available.
- (4)
- Image viewer: The program employs the YOLO neural network to detect Varroa mites and displays the processed image with detected bounding boxes marked on the right panel. Users can zoom and pan within the image and manually adjust detections by adding, modifying, or removing identified Varroa mites.
- (5)
- Threshold slider: The detection sensitivity can be adjusted using a confidence slider, which controls the neural network’s prediction threshold confidence. This threshold can be modified per image or applied uniformly across all images. Lower confidence levels increase detections but may also introduce more false positives.
- (6)
- Region of interest (ROI): The software allows the user to restrict the area to count mites. This may be used, for example, in rare instances where string recognition fails.
- (7)
- Statistics panel: This panel provides key information, including the total number of Varroa mites detected across all images, the number of mites within the same subfolder as the selected image (representing a single sheet), and the count of mites in the currently selected image.
- (8)
- List of images: This panel provides the names of the images that have been analyzed and allows the user to select which image to display in the image viewer. The selected image is marked on a dark blue background, whereas those images contained in the same subfolder are marked in light blue.
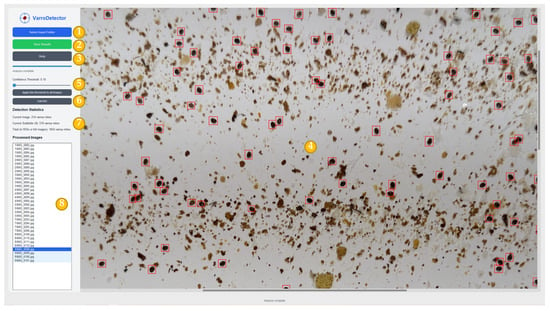
Figure 5.
Interface of the VarroDetector software. (1) Select imput folder button, (2) Save button, (3) Help button, (4) Image viewer, (5) Threshold slider, (6) Region of interest, (7) Statistics panel, (8) List of images. Varroa mites detected by VarroDetector are shown in red.
2.5. Verification (Testing)
The reliability of the VarroDetector software was tested capturing new images of 114 sticky sheets, since new images unseen by the underlying model are needed for a fair testing. To assess the repeatability of the results generated by the VarroDetector software, each of the 114 sheets captured using the Xiaomi smartphone was analyzed in the following three distinct orientations: the original orientation, rotated 90°, and rotated 180°.
A final verification was conducted aiming to evaluate the accuracy of the VarroDetector software in counting Varroa mites compared to visual inspection conducted by human observers. To this end, new images of 114 sticky sheets were captured, since new images unseen by the underlying model are needed for a fair testing. The number of Varroa mites present on new sticky sheets across different beehives and sampling dates was counted both manually through visual inspection and using the VarroDetector software, using the default threshold. The images were captured with the two smartphone cameras previously used for neural network training (iPhone 14 Pro Max and Xiaomi Poco X5 Pro) and an additional smartphone (Samsung Galaxy S24 Ultra, Suwon, Republic of Korea; at 50 megapixels), for which the VarroDetector had not been previously trained. Additionally, the time required for visual counting and for the automatic analysis with the VarroDetector was recorded.
Visual inspection was carried out by three experienced operators to evaluate the reliability of the results and determine whether the accuracy of visual inspection was influenced by the operator. The accuracy of both the VarroDetector and visual inspection was assessed by comparing the number of Varroa mites detected by each method to the actual number of mites present on the sheet (control). This reference value was established through a cross-check between the VarroDetector output and visual observation. After the analysis with the VarroDetector, each image was selected by a trained operator, who magnified the image on the screen, and then added, removed, or confirmed the identified mites using the edition capacities of the software. In cases of uncertainty, a direct visual inspection of the sheet was performed using magnifying glasses.
2.6. Statistical Analysis
Statistical analyses were performed using the SPSS package, version 24.0 (IBM SPSS Statistics, Chicago, IL, USA). In the verification (testing), various parameters were analyzed to assess the efficiency and accuracy of the VarroDetector software in comparison to human visual inspection and the actual count (control). Normality distributions and the variance homogeneity of the median value score for each set were checked using the Kolmogorov–Smirnov [27] and Levene tests [28], respectively. Table 1 summarizes the statistical analyses applied to each of the parameters examined.

Table 1.
Summary of the statistical analyses applied in relation to each evaluated parameter.
3. Results
3.1. Detection Time Efficiency
Figure 6 illustrates the time trends associated with visual and VarroDetector counting. The time required for visual inspection (blue dots and line) exhibited high variability (35–845 s) depending on the number of Varroa mites in the sheet. As the mite count increased, the detection time increased exponentially. In contrast, the processing time per sheet using the VarroDetector method (orange dots and line, only time for iPhone images is represented) remained nearly constant, independent of the number of mites present. For all observations, the processing time with the VarroDetector remained below 50 s. Time was greater when using the iPhone, since their images are raw files (DNG format). These images need more space (usually 10× or 20× compared to a JPG) and also require some processing steps performed by VarroDetector (adjust of white balance, color profiles, etc.), which are not needed for JPG files. For the Xiaomi and Samsung images, the analysis time per sheet was reduced to 24–40 s, depending on the image resolution and computer used. This processing time included the processing for border detection and the Varroa detection in the four images corresponding to each sticky sheet. The time required for image acquisition and classification was not considered.
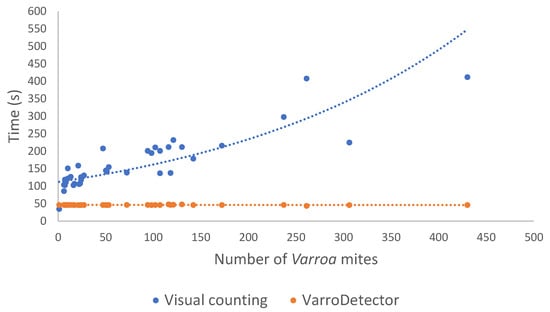
Figure 6.
Comparison of the efficiency of the two methods (visual inspection and VarroDetector) in the analysis time of each sticky sheet in relation to the number of Varroa mites that they contained.
3.2. Repeatability of the VarroDetector as a Function of Sheet Orientations
Figure 7 shows the Pearson correlation between the replicates as a function of sheet orientations. A strong correlation among replicates regardless of sheet orientation was obtained (R2 > 0.99), highlighting good repeatability of measurements for the VarroDetector software.
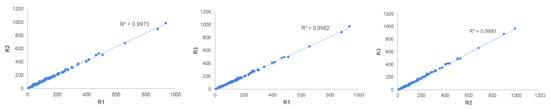
Figure 7.
Linear correlation between VarroDetector replicates as a function of image orientation (R1 long side of the sheet in the same orientation as in the colony seen from behind, R2 90° rotation from the initial position, and R3 180° rotation from the initial position.
3.3. Accuracy of Varroa Counting Methods (Visual Inspection and VarroDetector) Compared to the Real Value
Table 2 presents the accuracy of the two counting methods, visual and VarroDetector, based on the calculation of their standard deviations relative to the real data. The data were divided into five categories based on the Varroa count ranges per sheet. The results of the Friedman test for paired samples showed that, in the ranges between 51 and 200 Varroa mites per sheet, no significant differences were observed between the VarroDetector with respect to the real data. However, high significant differences in the standard deviation of the error in the visual count compared to the real data were obtained in all categories but also in the results obtained with VarroDetector in the range N ≤ 50 and N > 200.

Table 2.
Differences in standard deviation of the errors made with the two counting methods (visual vs. VarroDetector) compared to the real data. ** Denotes the presence of significant difference (p < 0.01).
Figure 8 shows the correlations between the replicates of the visual observations and of the VarroDetector from images captured with different Smartphones with respect to the real data (control). The higher reliability was obtained with the VarroDetector software combined with the iPhone (R2 = 0.991) followed by the Xiaomi (R2 = 0.985) and Observer 3 (R2 = 0.985). The results obtained from images captured with the Samsung smartphone, for which VarroDetector had not been previously trained, were slightly lower than those of the other two smartphones but still acceptable (R2 = 0.978). When comparing to the reference line, all the methods, except the VarroDetector using the Xiaomi smartphone (that captured the images at higher resolution), underestimated the number of Varroa, with higher deviations in high-density regions.
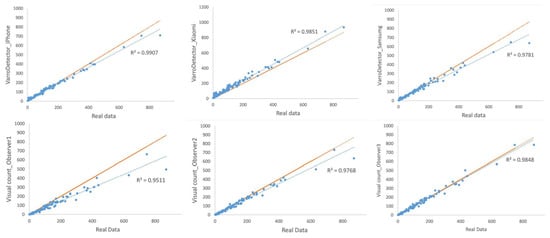
Figure 8.
Linear correlation for Varroa mite counting methods compared to the real data (blue circles and line) and comparison with the 1:1 reference line (orange line).
Figure 9 presents an example in which both the ground truth Varroa mites and the detections produced by VarroDetector are shown, including instances of missed detections and false positives.
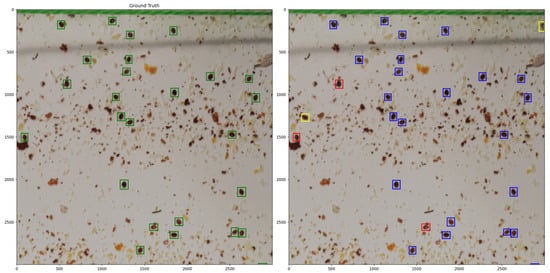
Figure 9.
Crop of an image in which ground truth Varroa are highlighted in green on the left, while on the right, Varroa mites detected by VarroDetector are shown in blue, false negatives in red, and false positives in yellow.
Table 3 shows the cumulative percentage error of visual counting (average of the three observers) and VarroDetector-based counting when compared with real data. The data were divided in the same categories as in Table 1. For the sheets containing between 0 and 50 Varroa mites, the VarroDetector method was less reliable than visual counting. In subsequent mite count ranges, the percentage error was lower for the VarroDetector compared to the visual inspection.

Table 3.
Cumulative percentage error for visual inspection and with the VarroDetector method compared to the real data.
4. Discussion
In this study, a new open-source and automated AI tool for detecting and counting natural Varroa mite fall was developed. The study validated its performance, efficacy, and reliability by comparing it with human visual counts and real data. Previous studies have attempted to use AI models for the analysis of Varroa destructor infestation on adult honeybees [16,17,18,19], and on honeybee pupae [20]. Although promising, some of them obtained unsatisfactory results [16,17,19,20], remain at the prototype stage [16,17,19,20], and/or have not been properly validated in the field [16,17,18,19,20].
In Italy, a commercial digital portable scanner coupled with an AI algorithm (BeeVS) focusing on the analysis Varroa mites on sticky sheets has been developed [22]. This system has been recently validated in the field by Scutaru et al. [22]. Although offering reliable results, it requires special equipment (digital scanner) and subscription, the models are not publicly available, and the algorithm relies on two-stage neural networks that runs on a central server.
In a recent work, we introduced a precise and accurate open-source AI algorithm for locating and counting Varroa mites using images of the sticky sheets taken by smartphone cameras [21]. The procedure was based on the use of lower resolution images than in the present study (12 megapixels) that required higher magnifications to adequately discriminate the Varroa mites. Consequently, it was necessary to increase the number of photographs per sheet to eight. Furthermore, the algorithm relies on two-stage neural networks that demand substantial computational resources. Consequently, the analysis must be conducted on a centralized server. Both aspects that restrict its practical applicability have been overcome with the new VarroDetector software. The use of higher resolution smartphone photographs allowed us to reduce the images per sheet to only four, and the application of one-stage YOLO v11 nano neuronal network made the analysis in low-end computers possible without GPUs. New features have also been introduced that facilitated the analysis, such as the automatic selection of the region of interest where Varroa must be counted, and the analysis of the images from same directory (sheet) together. Moreover, VarroDetector is the only available tool that allows user interaction during the analysis process, enabling the adjustment of the most appropriate threshold and facilitating quick, intuitive corrections after analysis. This feature makes it a robust and adaptable tool, suitable both for fast and precise automatic determinations and for scenarios where maximum accuracy is required.
The VarroDetector software is based on a deep learning approach, a type of machine learning that uses artificial neuronal networks to learn from data [29]. Deep learning studies usually require hundreds or thousands of images [30,31]. The dataset used for training contained 285 images, but the total number of Varroa in the images was 11917. Being that it is a relatively simple object, the number of images can be considered sufficient for the neural networks to learn to distinguish the Varroa bodies [21].
The algorithm developed is based on the one-stage YOLO neural network architecture. Specific methodologies for small object detection were also studied, such as the slicing aided hyper inference (SAHI) [32] with its image tiling approach, but they were ultimately not implemented, since the performance improvement was minor compared to the very significant increase in inference time. The use of two-stage detectors, like the faster R-CNN family [33], and other deeper architectures were also discarded for the same reasons. Thanks to this choice, the processing time using this neuronal network is very fast, despite the fact of the high-resolution images involved (48 Mpx and greater); when executed on modern processors, the analysis requires only milliseconds per image, while processors from old generations complete the task in a few seconds. All computations are performed locally on the device, eliminating the need for cloud computing infrastructure or dedicated GPUs. The use of high-resolution images was not arbitrarily chosen. Preliminary experiments with lower-resolution cameras proved inadequate, as they produced images too blurry to reliably distinguish mites from environmental debris, such as dirt and soil particles. The current market availability of affordable smartphones equipped with high-resolution cameras makes this requirement readily achievable. For the same reasons, four photographs per sampling sheet are recommended. Currently, the sticky sheet images belonging to the dataset were photographed by smartphones in a controlled environment. Over time, the advances in smartphone camera technologies will make it possible to just take a single picture of the sticky sheet in the field.
The results highlighted the high repeatability of results generated by the VarroDetector software across different image orientations, supporting its reliability. These findings are consistent with those reported in the aforementioned study employing a commercial system [22]. Moreover, a high accuracy of the measurements performed with the VarroDetector software when compared to the real data (R2 = 0.98 to 0.99, depending on the smartphone camera), even when using the Samsung smartphone for which the software was not previously trained. The results were more reliable than those obtained with visual inspection performed by different trained operators. After 50 Varroa mites per sheet, the cumulative percentage error was much lower for the VarroDetector than for the visual counting method when compared with real data. Additionally, a statistically significant difference was found between the Varroa mites determined by visual inspection and the control, while the VarroDetector results differed to the real data only when ≤ 50 and > 200 Varroa mites per sheet were counted. These findings underscore the limitations of visual inspection and the inherent variability of the human eye in performing such assessments, as also reported in previous studies [22,34].
In terms of counting efficiency, the VarroDetector method demonstrated a stable processing time, unaffected by the number of Varroa mites on the sticky sheets. In contrast, visual inspection became increasingly time-consuming as mite numbers grew, following an exponential pattern. Similar results were also obtained when testing the commercial BeeVS scanner [22]. Moreover, the VarroDetector offers the possibility of analysis of an unlimited number of images in one step, ensuring that the processing time is not a constraint, as the operator can attend to other tasks, while the system handles the analysis.
The main limitations of the VarroDetector software are the worse performance than human inspection at low infestation levels, in agreement with the results obtained by others using more complex neuronal networks [22] and its dependency on high-quality images. The software was trained and tested using high-resolution images captured under controlled lighting conditions. The use of lower resolution images would much likely reduce the accuracy, limiting usability. The software was neither tested using field captured images with variable lighting conditions. Moreover, in the rare instances when the green frame strings are not properly detected, a manual ROI selection is needed, which may reduce efficiency. Also, as can be seen in Figure 6, VarroDetector may fail to detect some Varroa mites, especially when they are blurred or surrounded by a lot of similarly colored dirt. Another limitation of our study is the use of two smartphones for training; if a model is trained with the same smartphone that is later tested, the results surely will improve. This is due to the so-called domain shift, a fundamental challenge in machine learning where the distribution of training data differs from testing data. Although, in this case, techniques such as data augmentation or regularization have been used to improve generalization across different smartphones, a switch to a different smartphone camera introduces a small domain shift. However, our results show that the neural network has managed to learn and generalize enough to obtain good results, but it would certainly improve even more if it were trained with the same smartphone used for testing.
VarroDetector is specialized for sticky board images only, excluding other mite detection methods like the inspection of brood cells or live bees. We decided to focus on this approach, because previous attempts to analyze Varroa on brood [20], and live bees [16,17,19] provided unsatisfactory results. However, unlike the study of Sevin et al. on adult bees [18], the current version of VarroDetector software does not integrate with smart hives or IoT devices, limiting its automation potential.
5. Conclusions
In conclusion, VarroDetector is a free and open-source tool for the efficient analysis of Varroa mite fall that simplifies the monitorization and control of this relevant parasite. The software is designed for continuous improvement through the integration of new images from different devices, illuminations, and sticky boards, etc., allowing it to adapt to user needs. These adaptations, together with the development of a mobile app for direct analysis or even a cloud service would facilitate in situ data collection. Users interested in contributing to this feedback process should contact the software developers. Future research may also be oriented to the automatic detection of emerging diseases, such as Tropilaelapsosis, and to the ability to collect data (images, annotations, geolocation), which could be applied by governments for the early identification of pests.
Author Contributions
Conceptualization, J.Y., J.D. and P.S.; data curation, M.C.; formal analysis, J.Y., J.D. and F.J.M.-d.-P.; funding acquisition, J.Y., P.S., B.C. and J.D.; investigation, J.Y., J.D. and F.J.M.-d.-P.; methodology, J.Y., J.D., M.C., F.J.M.-d.-P., M.A.S. and P.S.; resources, M.A.S., P.S. and J.Y.; software, J.D. and F.J.M.-d.-P.; supervision, P.S. and B.C.; writing—original draft, J.Y.; writing—review and editing, F.J.M.-d.-P., M.A.S., P.S., J.D. and J.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by MCIU/AEI/10.13039/501100011033 (grant: PID2023-148475OB-I00), the EU Horizon Europe (grant: 101082073), the DGA-FSE (grant: A07_23R), and La Rioja Government (grant: INICIA2023/02).
Data Availability Statement
The executable files, the full code of VarroDetector, and the trained YOLO model can be found in the following repository: https://github.com/jodivaso/varrodetector (accessed on 25 April 2025).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| DWV | Deformed Wing Virus |
| AI | Artificial Intelligence |
| GPU | Graphics Processing Unit |
| mAP | Mean Average Precision |
| ROI | Region of Interest |
References
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Hung, K.L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172140. [Google Scholar] [CrossRef]
- Soroker, V.; Hetzroni, A.; Yakobson, B.; David, D.; David, A.; Voet, H.; Slabezki, Y.; Efrat, H.; Levski, S.; Kamer, Y.; et al. Evaluation of colony losses in Israel in relation to the incidence of pathogens and pests. Apidologie 2011, 42, 192–199. [Google Scholar] [CrossRef]
- Clermont, A.; Eickermann, M.; Kraus, F.; Georges, C.; Hoffmann, L.; Beyer, M. A survey on some factors potentially affecting losses of managed honey bee colonies in Luxembourg over the winters 2010/2011 and 2011/2012. J. Apicult. Res. 2014, 53, 43–56. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Predictive markers of honey bee colony collapse. PLoS ONE 2012, 7, e32151. [Google Scholar] [CrossRef]
- Hristov, P.; Shumkova, R.; Palova, N.; Neov, B. Factors associated with honey bee colony losses: A mini-review. Vet. Sci. 2020, 7, 166. [Google Scholar] [CrossRef]
- De la Mora, A.; Goodwin, P.H.; Emsen, B.; Kelly, P.G.; Petukhova, T.; Guzman-Novoa, E. Selection of Honey Bee (Apis mellifera) Genotypes for Three Generations of Low and High Population Growth of the Mite Varroa destructor. Animals 2024, 14, 3537. [Google Scholar] [CrossRef]
- Mondet, F.; Beaurepaire, A.; McAfee, A.; Locke, B.; Alaux, C.; Blanchard, S.; Danka, B.; Le Conte, Y. Honey bee survival mechanisms against the parasite Varroa destructor: A systematic review of phenotypic and genomic research efforts. Int. J. Parasitol. 2020, 50, 433–447. [Google Scholar] [CrossRef]
- Dietemann, V.; Nazzi, F.; Martin, S.J.; Anderson, D.L.; Locke, B.; Delaplane, K.S.; Wauquiez, Q.; Tannahill, C.; Frey, E.; Ziegelmann, B.; et al. Standard methods for varroa research. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef]
- Noël, A.; Le Conte, Y.; Mondet, F. Varroa destructor: How does it harm Apis mellifera honey bees and what can be done about it? Emerg. Top. Life Sci. 2020, 4, 45–57. [Google Scholar] [CrossRef] [PubMed]
- DeGrandi-Hoffman, G.; Curry, R. A mathematical model of varroa mite (Varroa destructor anderson and trueman) and honeybee (Apis mellifera L.) population dynamics. Int. J. Acarol. 2004, 30, 259–274. [Google Scholar] [CrossRef]
- Ostiguy, N.; Sammataro, D. A simplified technique for counting Varroa jacobsoni Oud. on sticky boards. Apidologie 2000, 31, 707–716. [Google Scholar] [CrossRef]
- Pietropaoli, M.; Gajger, I.T.; Costa, C.; Gerula, D.; Wilde, J.; Adjlane, N.; Sánchez, P.A.; Škerl, M.I.S.; Bubnič, J.; Formato, G. Evaluation of two commonly used field tests to assess varroa destructor infestation on honey bee (Apis mellifera) colonies. Appl. Sci. 2021, 11, 4458. [Google Scholar] [CrossRef]
- Roth, M.A.; Wilson, J.M.; Tignor, K.R.; Gross, A.D. Biology and Management of Varroa destructor (Mesostigmata: Varroidae) in Apis mellifera (Hymenoptera: Apidae) Colonies. J. Integr. Pest Manag. 2020, 11, 1. [Google Scholar] [CrossRef]
- Lee, H.G.; Kim, M.J.; Kim, S.B.; Lee, S.; Lee, H.; Sin, J.Y.; Mo, C. Identifying an Image-Processing Method for Detection of Bee Mite in Honey Bee Based on Keypoint Analysis. Agriculture 2023, 13, 1511. [Google Scholar] [CrossRef]
- Bilik, S.; Kratochvila, L.; Ligocki, A.; Bostik, O.; Zemcik, T.; Hybl, M.; Horak, K.; Zalud, L. Visual Diagnosis of the Varroa Destructor Parasitic Mite in Honeybees Using Object Detector Techniques. Sensors 2021, 21, 2764. [Google Scholar] [CrossRef]
- Sevin, S.; Tutun, H.; Mutlu, S. Detection of Varroa mites from honey bee hives by smart technology Var-Gor: A hive monitoring and image processing device. Turk. J. Vet. Anim. Sci. 2021, 45, 487–491. [Google Scholar] [CrossRef]
- Voudiotis, G.; Moraiti, A.; Kontogiannis, S. Deep Learning Beehive Monitoring System for Early Detection of the Varroa Mite. Signals 2022, 3, 506–523. [Google Scholar] [CrossRef]
- Divasón, J.; Martinez-de-Pison, F.J.; Romero, A.; Santolaria, P.; Yániz, J.L. Varroa Mite Detection Using Deep Learning Techniques. In Hybrid Artificial Intelligent Systems; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2023; pp. 326–337. [Google Scholar] [CrossRef]
- Divason, J.; Romero, A.; Martinez-de-Pison, F.J.; Casalongue, M.; Silvestre, M.A.; Santolaria, P.; Yaniz, J.L. Analysis of varroa mite colony infestation level using new open software based on deep learning techniques. Sensors 2024, 24, 3828. [Google Scholar] [CrossRef]
- Scutaru, D.; Bergonzoli, S.; Costa, C.; Violino, S.; Costa, C.; Albertazzi, S.; Capano, V.; Kostić, M.M.; Scarfone, A. An AI-Based Digital Scanner for Varroa destructor Detection in Beekeeping. Insects 2025, 16, 75. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 777–778. [Google Scholar]
- Terven, J.; Córdova-Esparza, D.M.; Romero-González, J.A. A Comprehensive Review of YOLO Architectures in Computer Vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Wong, A.; Famuori, M.; Shafiee, M.J.; Li, F.; Chwyl, B.; Chung, J. YOLO Nano: A Highly Compact You only Look Once Convolutional Neural Network for Object Detection. In Proceedings of the Proceedings 5th Workshop on Energy Efficient Machine Learning and Cognitive Computing, EMC2-NIPS 2019, Vancouver, BC, Canada, 13 December 2019; pp. 22–25. [Google Scholar]
- Padilla, R.; Netto, S.L.; Da Silva, E.A.B. A Survey on Performance Metrics for Object-Detection Algorithms. In Proceedings of the International Conference on Systems, Signals, and Image Processing, Niterói, Brazil, 1–3 July 2020; pp. 237–242. [Google Scholar]
- Smirnov, N.V. Table for estimating the goodness of fit of empirical distributions. Ann. Math. Stat. 1948, 19, 279–281. [Google Scholar] [CrossRef]
- Levene, H. Robust tests for equality of variances. In Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling; Olkin, I., Ghurye, S.G., Hoeffding, W., Madow, W.G., Mann, H.B., Eds.; Stanford University Press: Redwood City, CA, USA, 1960; pp. 278–292. [Google Scholar]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Bates, K.; Le, K.; Lu, H. Deep learning for robust and flexible tracking in behavioral studies for C. elegans. PLoS Comput. Biol. 2022, 18, e1009942. [Google Scholar] [CrossRef]
- Geldenhuys, D.S.; Josias, S.; Brink, W.; Makhubele, M.; Hui, C.; Landi, P.; Bingham, J.; Hargrove, J.; Hazelbag, M.C. Deep learning approaches to landmark detection in tsetse wing images. PLoS Comput. Biol. 2023, 19, e1011194. [Google Scholar] [CrossRef]
- Akyon, F.C.; Altinuc, S.O.; Temizel, A. Slicing aided hyper inference and fine-tuning for small object detection. In Proceedings of the IEEE International Conference on Image Processing (ICIP), Bordeaux, France, 16–19 October 2022; pp. 966–970. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, Canada, 7–12 December 2015; pp. 91–99. [Google Scholar]
- Liu, M.; Cui, M.; Xu, B.; Liu, Z.; Li, Z.; Chu, Z.; Zhang, X.; Liu, G.; Xu, X.; Yan, Y. Detection of Varroa destructor Infestation of Honeybees Based on Segmentation and Object Detection Convolutional Neural Networks. Agriengineering 2023, 5, 1644–1662. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).