StrawberryNet: Fast and Precise Recognition of Strawberry Disease Based on Channel and Spatial Information Reconstruction
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
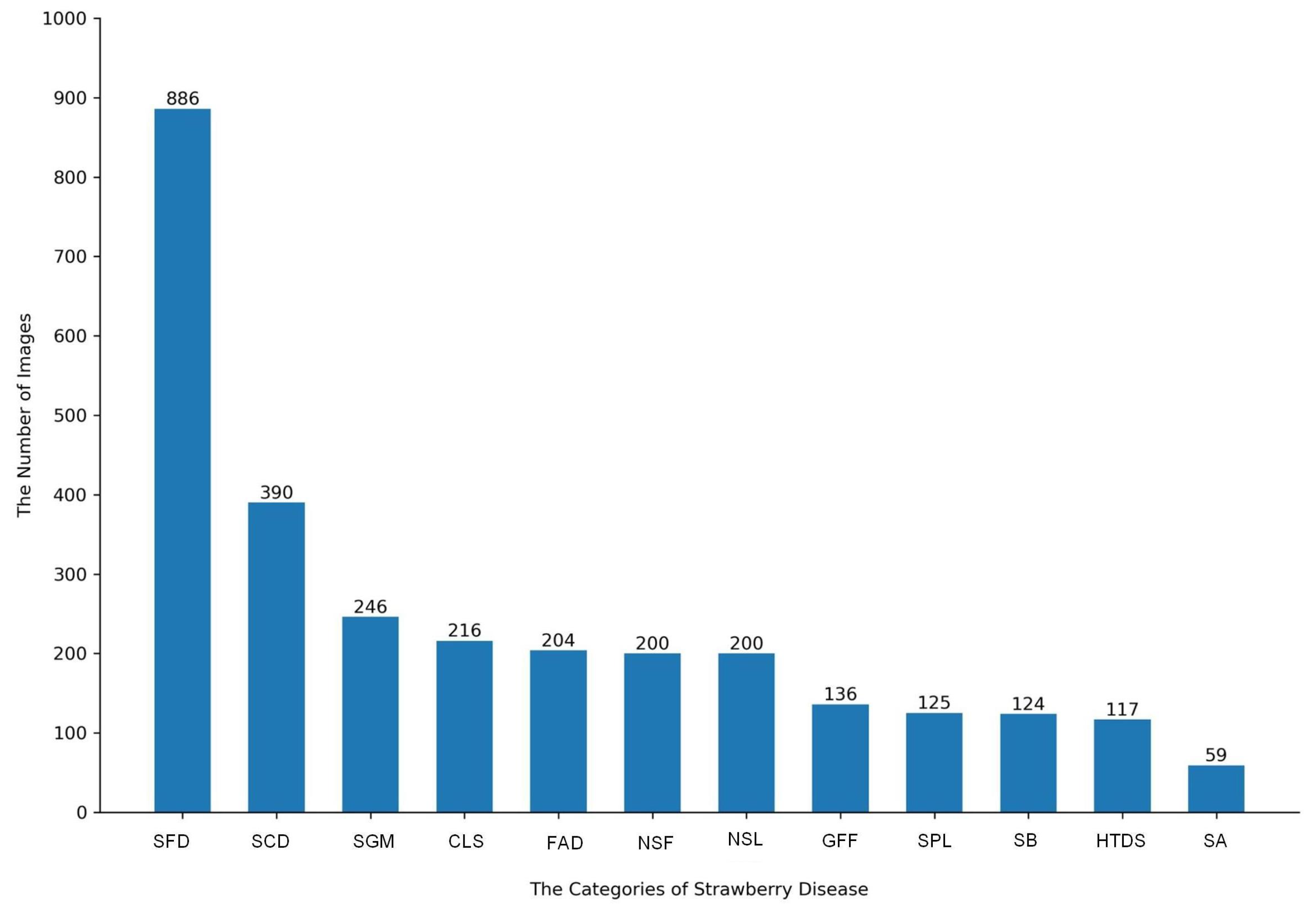
2.1. Strawberry Disease Dataset
2.2. Proposed Method
2.2.1. Overall Architecture of StrawberryNet
2.2.2. Review of Depthwise Separable Convolution and Partial Convolution
2.2.3. FasterNet Stage
2.2.4. Discriminative Feature Extraction Block
2.3. Evaluation Metrics
3. Experimental Results and Analysis
3.1. Experimental Settings
3.2. Experimental Results
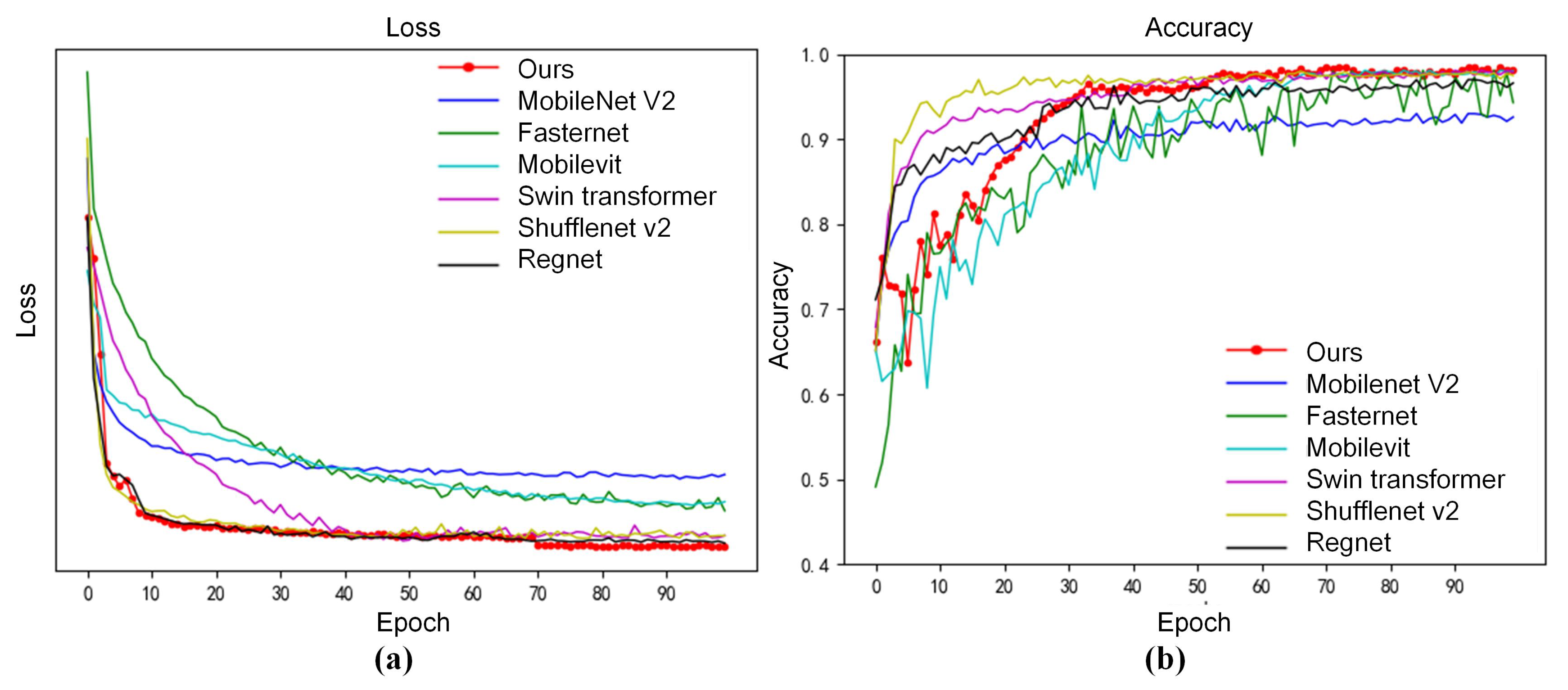
3.2.1. Overall Performance on Strawberry Disease Dataset
3.2.2. Ablation Experiments
3.2.3. Efficiency Analysis of Strawberry Disease
3.2.4. Visualized Analysis of StrawberryNet
4. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, D.; Wang, X.; Chen, Y.; Wu, Y.; Zhang, X. Strawberry ripeness classification method in facility environment based on red color ratio of fruit rind. Comput. Electron. Agric. 2023, 214, 108313. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, S.; Li, N.; Faheem, M.; Li, P. Development and Field Test of an Autonomous Strawberry Plug Seeding Transplanter for Use in Elevated Cultivation. Appl. Eng. Agric. 2019, 35, 1067–1078. [Google Scholar]
- Dong, A.Y.; Wang, Z.; Huang, J.J.; Song, B.A.; Hao, G.F. Bioinformatic tools support decision-making in plant disease management. Trends Plant Sci. 2021, 26, 953–967. [Google Scholar] [PubMed]
- Zhou, Y.; Zhou, H.; Chen, Y. An automated phenotyping method for Chinese Cymbidium seedlings based on 3D point cloud. Plant Methods 2024, 20, 151. [Google Scholar] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Dai, J.; Qi, H.; Xiong, Y.; Li, Y.; Zhang, G.; Hu, H.; Wei, Y. Deformable Convolutional Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 764–773. [Google Scholar] [CrossRef]
- Mnih, V.; Heess, N.; Graves, A.; Kavukcuoglu, K. Recurrent models of visual attention. Adv. Neural Inf. Process. Syst. 2014, 27. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Chen, L.; Zhang, H.; Xiao, J.; Nie, L.; Shao, J.; Liu, W.; Chua, T.S. SCA-CNN: Spatial and Channel-Wise Attention in Convolutional Networks for Image Captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Yang, J.; Li, C.; Zhang, P.; Dai, X.; Xiao, B.; Yuan, L.; Gao, J. Focal Self-attention for Local-Global Interactions in Vision Transformers. arXiv 2021, arXiv:2107.00641. [Google Scholar]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7794–7803. [Google Scholar]
- Liu, R.; Deng, H.; Huang, Y.; Shi, X.; Lu, L.; Sun, W.; Wang, X.; Dai, J.; Li, H. Fuseformer: Fusing fine-grained information in transformers for video inpainting. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 14040–14049. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar] [CrossRef]
- Han, K.; Wang, Y.; Tian, Q.; Guo, J.; Xu, C.; Xu, C. GhostNet: More Features From Cheap Operations. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 1577–1586. [Google Scholar] [CrossRef]
- Chen, J.; Kao, S.h.; He, H.; Zhuo, W.; Wen, S.; Lee, C.H.; Chan, S.H.G. Run, Don’t Walk: Chasing Higher FLOPS for Faster Neural Networks. In Proceedings of the 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, 17–24 June 2023; pp. 12021–12031. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Hou, Q.; Lu, C.Z.; Cheng, M.M.; Feng, J. Conv2Former: A Simple Transformer-Style ConvNet for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 8274–8283. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, J.; Wu, S. Multiple disease detection method for greenhouse-cultivated strawberry based on multiscale feature fusion Faster R_CNN. Comput. Electron. Agric. 2022, 199, 107176. [Google Scholar] [CrossRef]
- Attallah, O. Tomato leaf disease classification via compact convolutional neural networks with transfer learning and feature selection. Horticulturae 2023, 9, 149. [Google Scholar] [CrossRef]
- Faisal, M.; Leu, J.S.; Avian, C.; Prakosa, S.W.; Köppen, M. DFNet: Dense fusion convolution neural network for plant leaf disease classification. Agron. J. 2024, 116, 826–838. [Google Scholar]
- Liu, B.; Huang, X.; Sun, L.; Wei, X.; Ji, Z.; Zhang, H. MCDCNet: Multi-scale constrained deformable convolution network for apple leaf disease detection. Comput. Electron. Agric. 2024, 222, 109028. [Google Scholar]
- Liu, Y.; Wang, Z.; Wang, R.; Chen, J.; Gao, H. Flooding-based MobileNet to identify cucumber diseases from leaf images in natural scenes. Comput. Electron. Agric. 2023, 213, 108166. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Tran, T.D.; Nguyen, T.T.; Pham, N.M.; Nguyen Ly, P.H.; Luong, H.H. Strawberry disease identification with vision transformer-based models. Multimed. Tools Appl. 2024, 83, 73101–73126. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, G.; Li, X. Swin-MLP: A strawberry appearance quality identification method by Swin Transformer and multi-layer perceptron. J. Food Meas. Charact. 2022, 16, 2789–2800. [Google Scholar] [CrossRef]
- Li, G.; Jiao, L.; Chen, P.; Liu, K.; Wang, R.; Dong, S.; Kang, C. Spatial convolutional self-attention-based transformer module for strawberry disease identification under complex background. Comput. Electron. Agric. 2023, 212, 108121. [Google Scholar] [CrossRef]
- DeVries, T.; Taylor, G.W. Improved regularization of convolutional neural networks with cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Radosavovic, I.; Kosaraju, R.P.; Girshick, R.; He, K.; Dollár, P. Designing network design spaces. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville TN, USA, 11–15 June 2020; pp. 10428–10436. [Google Scholar]






| Methods | Accuracy (%) | Recall (%) | Precision (%) | Specificity (%) | F1-Score | Params (M) |
|---|---|---|---|---|---|---|
| MobileNet | 93.02 | 95.21 | 95.12 | 96.88 | 95.44 | 15.4 |
| RegNet | 97.04 | 96.33 | 96.44 | 98.1 | 96.54 | 15.7 |
| ShuffleNet v2 | 98.02 | 96.11 | 96.52 | 98.77 | 96.7 | 15.6 |
| MobileVit | 98.1 | 96.33 | 95.89 | 98.65 | 96.7 | 5.6 |
| Swin Transformer | 98.34 | 96.54 | 96.77 | 98.99 | 96.8 | 40 |
| L-GhostNet | 98.33 | 96.6 | 96.74 | 98.86 | 96.67 | 5.14 |
| SCSA-Transformer | 99.1 | 98.47 | 97.77 | 99.37 | 97.75 | 24.2 |
| FasterNet | 98.27 | 97 | 96.81 | 98.8 | 96.9 | 3.58 |
| StrawberryNet | 99.01 | 97.66 | 96.88 | 99.22 | 97.27 | 3.6 |
| Stage I | Stage II | Stage III | Stage IV | Accuracy (%) | FLOPs (G) | Image/s | Params (M) |
|---|---|---|---|---|---|---|---|
| 97.25 | 0.34 | 4.3 | 3.58 | ||||
| ✓ | 98.27 | 0.37 | 4 | 3.6 | |||
| ✓ | 98.15 | 0.36 | 4.1 | 3.6 | |||
| ✓ | 98.34 | 0.37 | 4.2 | 3.6 | |||
| ✓ | 99.01 | 0.42 | 4.3 | 3.6 | |||
| ✓ | ✓ | 98.64 | 0.4 | 4 | 3.73 | ||
| ✓ | ✓ | ✓ | 98.74 | 0.4 | 3.9 | 3.84 | |
| ✓ | ✓ | ✓ | ✓ | 99.01 | 0.42 | 3.7 | 4 |
| Methods | Image/s | Params (M) | FLOPs (G) | Accuracy (%) |
|---|---|---|---|---|
| MobileNet v2 | 3.3 | 15.4 | 0.3 | 97.25 |
| Regnet | 4 | 15.7 | 0.33 | 98.27 |
| ShuffleNet v2 | 4.1 | 15.6 | 0.3 | 98.15 |
| MobileVit | 4.2 | 5.6 | 0.37 | 98.34 |
| Swin Transformer | 4.3 | 40 | 0.41 | 99.01 |
| L-GhostNet | 3.9 | 5.14 | 0.36 | 98.64 |
| SCSA-Transformer | 4.2 | 24.2 | 0.41 | 99.1 |
| FasterNet | 4.3 | 3.58 | 0.34 | 98.74 |
| Our methods | 4.3 | 3.6 | 0.42 | 99.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Jiao, L.; Liu, K.; Liu, Q.; Wang, Z. StrawberryNet: Fast and Precise Recognition of Strawberry Disease Based on Channel and Spatial Information Reconstruction. Agriculture 2025, 15, 779. https://doi.org/10.3390/agriculture15070779
Li X, Jiao L, Liu K, Liu Q, Wang Z. StrawberryNet: Fast and Precise Recognition of Strawberry Disease Based on Channel and Spatial Information Reconstruction. Agriculture. 2025; 15(7):779. https://doi.org/10.3390/agriculture15070779
Chicago/Turabian StyleLi, Xiang, Lin Jiao, Kang Liu, Qihuang Liu, and Ziyan Wang. 2025. "StrawberryNet: Fast and Precise Recognition of Strawberry Disease Based on Channel and Spatial Information Reconstruction" Agriculture 15, no. 7: 779. https://doi.org/10.3390/agriculture15070779
APA StyleLi, X., Jiao, L., Liu, K., Liu, Q., & Wang, Z. (2025). StrawberryNet: Fast and Precise Recognition of Strawberry Disease Based on Channel and Spatial Information Reconstruction. Agriculture, 15(7), 779. https://doi.org/10.3390/agriculture15070779






