Abstract
Phytobacter palmae WL65, isolated from the rice rhizosphere, was confirmed as P. palmae through whole-genome analysis. WL65 exhibited key plant growth-promoting (PGP) characteristics, including nitrogen fixation (nifA, nifB, nifD, nifE, nifF, nifH, nifJ, nifK, nifL, nifS, nifU, nifW, and nifX), phosphate solubilization (pstA, pstB, pstC, pstS, phnC, phnD, phnE, and phnV), siderophore production (fhuA, fhuB, fhuC, fhuD, fhuF, feoA, feoB, feoC, acrA, acrB, acrE, acrR, and acrZ), and phytohormone biosynthesis (trpA, trpB, trpC, trpE, trpGD, trpR, and trpS). WL65 also contains an enterobactin biosynthetic gene cluster, essential for iron acquisition and enhancing both bacterial survival and plant growth. This study provides the first genomic insights into the PGP characteristics of P. palmae. The application of WL65 in rice cultivation as a biostimulant resulted in effective root colonization, supported by biofilm formation genes (pgaA, pgaB, pgaC), which enhance bacterial adhesion. The treatment significantly improved rice growth, increasing plant height (5.8%), panicle length (10.2%), and seed yield (34.5%). Soil analysis revealed improved nutrient availability, including increased organic matter (21%), phosphorus (38.4%), potassium (29.8%), and calcium (27%) levels. These findings suggest that WL65 is a promising biofertilizer candidate for improving soil fertility and nutrient uptake in sustainable agriculture.
1. Introduction
The genus Phytobacter (family Enterobacteriaceae, order Enterobacterales) was reclassified from Enterobacter and is recognized as a plant growth-promoting (PGP) bacteria. It currently comprises four accepted species: P. diazotrophicus, P. palmae, P. massiliensis, and P. ursingii. P. diazotrophicus, the type species, was first isolated as a nitrogen-fixing endophyte from Oryza rufipogon in China, highlighting its role in plant growth promotion [1]. P. palmae, isolated from oil palm leaves in Singapore, also exhibits nitrogen-fixing capabilities, further supporting its PGP potential [2]. In contrast, P. ursingii, isolated from clinical samples in Brazil, is distinguished by its ability to utilize L-sorbose and D-serine [3]. Its identification in healthcare-associated infections led to a revision of the genus description. The most recent addition, P. massiliensis, was reclassified from Metakosakonia massiliensis based on genome relatedness and phylogenomic analyses [4]. Together, accumulating the genomic, biochemical, and ecological studies of Phytobacter species highlights their varied habitats, ranging from plant-associated environments to clinical situations, and their importance as PGP and potential biotechnological agents. Traditional bacterial identification methods, including morphology, biochemical tests, and 16S rRNA sequencing, often yield ambiguities, particularly among closely related taxa like Phytobacter species. Whole-genome sequencing (WGS) has emerged as a powerful tool to resolve taxonomic uncertainties, offering high-resolution genomic data, robust phylogenomic analyses, functional gene annotation, and key genetic feature identification.
Recently, Almuzara et al. [5] utilized WGS to investigate three clinical cases associated with Phytobacter spp. in Argentina. Through comparative genomic analyses, the isolates were identified as P. diazotrophicus and P. ursingii. The study highlighted the presence of antibiotic-resistant genes within these genomes, underscoring the clinical relevance of Phytobacter species and their potential role in healthcare-associated infections. This finding also demonstrates the versatility of WGS in linking genomic features to both ecological roles and pathogenic potential. In addition, a Phytobacter sp. isolate, WL65, was recently obtained from the rhizosphere soil of Jasmine rice (Oryza sativa L. cultivar KDML105) in Thailand. This isolate, initially identified by the morphological characters and 16S rRNA gene sequencing, exhibited plant growth-promoting properties and was subsequently used in combination with other plant growth-promoting rhizobacteria as a biostimulant to enhance rice growth [6]. However, the Phytobacter sp. WL65 had not yet been extensively characterized. Therefore, the objectives of this study were to conduct a comprehensive whole-genome sequencing analysis, including digital DNA-DNA hybridization, and to perform an extensive biochemical characterization to accurately identify the species and gain deeper insights into the functional potential of the isolates as plant growth promoters. Additionally, secondary metabolic pathways were predicted through genomic analysis, and the potential application of the isolate WL65 as a biofertilizer for rice cultivation was evaluated.
2. Materials and Methods
2.1. Bacterial Strains
Phytobacter sp. isolate WL65 was obtained from the Department of Microbiology, Faculty of Science, Srinakharinwirot University, Thailand. WL65 was originally isolated from the rhizosphere soil of organic Thai Jasmine rice (Oryza sativa L. cultivar KDML105) grown in Sakon Nakhon province, Thailand. WL65 was cultured on Luria–Bertani (LB) agar at 30 °C and preserved as a stock culture in 15% (v/v) glycerol at −80 °C for long-term storage.
2.2. Physiological and Biochemical Characteristics
The isolate WL65 was observed for phenotypic characteristics of bacterial colonies on a nitrogen-free medium, nutrient agar, and MacConkey agar after incubation at 30 °C and 37 °C for 24 h. Gram staining, endospore formation, and cellular shape were assessed. Catalase and oxidase activities were tested according to Aslanzadeh et al. [7]. Motility was determined using a semi-solid medium containing 0.4% (w/v) soft agar. The biochemical characteristics were analyzed using the API 20E test strip (bioMérieux, Marcy-l’Étoile, France) following the instructions of the manufacturer.
2.3. 16S rRNA Gene Sequences and Phylogenetic Analysis
The genomic DNA of isolate WL65 was extracted using the FavorprepTM Tissue genomic DNA extraction mini kit following the instructions of the manufacturer. The 16S rRNA gene was amplified using universal primers 27F and 1492R [8]. The purified PCR amplicon was sequenced by Macrogen Inc. (Seoul, Republic of Korea). The nucleotide sequence was submitted to the GenBank database under accession number PP326909.
The 16S rRNA gene sequence was carefully verified using the BioEdit software v7.2 [9] and aligned with sequences of the Phytobacter species and related genera obtained from the GenBank database using ClustalX [10]. A phylogenetic tree was constructed using the maximum likelihood (ML) method with a bootstrap analysis based on 1000 replications, implemented in MEGA X [11].
2.4. Genome Sequencing and Genome Annotation
Genomic DNA of the isolate WL65 from Section 2.3 was used to prepare the library using the Nextera XT DNA library preparation kit, and the extracted DNA was sequenced using the Illumina system through commercial services of Macrogen Inc. (Seoul, Republic of Korea). The quality of the obtained raw reads was checked using FastQC v0.11.9, followed by adapter and quality trimming with Trimmomatic v0.39 [12]. De novo assembly was performed using SPAdes v3.15.4 [13], and the assembly quality was evaluated with QUAST v5.2.0 [14]. Genome annotation and comprehensive genome analysis for the isolate WL65 were performed using SEED and Rapid Annotation using Subsystem Technology, RAST (https://rast.nmpdr.org) (accessed on 9 January 2025); Pathosystems Resource Integration Center, PATRIC (https://www.bv-brc.org) (accessed on 13 January 2025); and the Functional Annotation and Classification of Proteins of Prokaryotes, FACoP (http://facop.molgenrug.nl) (accessed on 18 January 2025) [15,16]. The circular genome map was constructed using the Proksee server (https://proksee.ca) (accessed on 18 January 2025), which generated a map such as coding sequences (CDS), transfer RNA (tRNAs), ribosomal RNA (rRNAs), and guanine-cytosine (GC) skew content. These data were integrated with the results from Prokka (Rapid Prokaryotic Genome Annotation) [17,18].
2.5. Genome Mining and Phylogenomic Analysis
To identify and classify secondary metabolite biosynthesis gene clusters and gene functions, the genomes of isolate WL65 were analyzed using AntiSMASH v8 (Antibiotics & Secondary Metabolite Analysis Shell) [19] (https://antismash.secondarymetabolites.org) (accessed on 9 January 2025). To identify the genes involved in antibiotic resistance and plasmids, the genomes data were analyzed using ResFinder v4.5.0 [20] (http://genepi.food.dtu.dk/resfinder) (accessed on 28 January 2025) and PlasmidFinder v2.1 [21] (https://cge.food.dtu.dk/services/PlasmidFinder) (accessed on 28 January 2025).
To compare the prokaryotic genome sequences, the average nucleotide identity (ANI) values between the genome of isolate WL65 and the reference strains from the GenomesDB database were calculated using JSpeciesWS [22] (https://jspecies.ribohost.com/jspeciesws/#home) (accessed on 8 January 2025). To assess bacterial strain identification at the genus or species level, pairwise genome sequence comparisons and phylogenomic tree construction were conducted using the Genome BLAST Distance Phylogeny (GBDP) approach and GGDC v4.0 (the Genome-to-Genome Distance Calculator) via the TYGS (Type Strain Genome Server) platform [23,24] (https://tygs.dsmz.de) (accessed on 29 January 2025). The genome was aligned against reference sequences using the progressive Mauve algorithm via Geneious Prime v.2025.0.3 [25].
2.6. Phytobacter Palmae WL65 as a Plant Growth-Promoting Rhizobacterium for Rice Cultivation in Pot Experiments
2.6.1. Bacterial Colonization on Rice Roots
The rice (Oryza sativa L.) seeds cultivar RD79 was obtained from the Department of Agriculture, Thailand. The seeds were surface-sterilized by soaking them in 75% ethanol, followed by 0.6% sodium hypochlorite, and were then rinsed with sterile distilled water. The isolate WL65 was cultured in nutrient broth for 48 h at 30 °C with shaking at 150 rpm, and the bacterial suspension was adjusted to 1.0 × 108 CFU/mL in 0.85% (v/v) saline solution. Fifty rice seeds were immersed in 50 mL of bacterial suspension at 30 °C with shaking at 150 rpm for 24 h. The seeds were germinated in Petri dishes for 14 days at 30 °C in the dark. After that, four healthy seedlings were chosen, and the roots were immersed in 50 mL of WL65 suspension (1.0 × 108 CFU/mL) for 1 h before transplanting to a minirhizotron containing 400 g of sterile soil. Four additional healthy seedlings treated with sterile distilled water were used as a negative control. The rice roots were collected at 4 weeks after transplanting (DAT) and carefully washed in sterile water to remove the soils. Then, the roots were air-dried and kept at 4 °C until processing. The root samples were prepared for bacterial colonization analysis by JEOL JSM-IT500HR scanning electron microscope (SEM) observation at the Scientific and Technological Research Equipment Centre, Chulalongkorn University (Thailand).
2.6.2. Effects of P. palmae WL65 on Rice Cultivar RD79 in Pot Experiment
The pot experiment consisted of two treatments: WL65 and a negative control. The experiment was conducted in a greenhouse (4 × 3 × 2 m) at the Faculty of Science, Nakhon Phanom University, Nakhon Phanom province. The planting process and measurements followed the method described by Thamvithayakorn et al. [6]. The soil used for planting was collected from paddy fields in Nong Yart District, Nakhon Phanom province, sieved through a 5 mm mesh to remove stones, and mixed with organic fertilizer (Ngok-Ngam™, Ubon Ratchathani, Thailand) in a ratio of 1000:3 (w/w) before use.
Fifty RD79 rice seeds were surface-sterilized and germinated in soil for 21 days in the greenhouse. Eight seedlings with the same root and shoot length were selected, soaked in 100 mL of the treatment solutions (WL65 and control) for 1 h, and then transplanted into 10-inch pots containing 3 kg of the soil mixture. One seedling was placed in each pot. The pots were irrigated with tap water every two days.
Rice growth parameters, including the rice height, chlorophyll content index, number of panicles per tiller, panicle length, and number of seeds per panicle, were measured. Soil samples were collected and analyzed for physicochemical properties, including the pH, organic matter, total carbon, nitrogen, available phosphorus, potassium, calcium, and magnesium at Kasetsart University, Thailand.
2.7. Statistical Analysis
The experiments were performed in four replications and analyzed using the independent samples t-test in IBM SPSS Statistics v29.0.2. Data visualization was performed using RStudio v2024.12.0 + 467.
3. Results
3.1. Biochemical Characteristics and 16S rRNA Gene Sequence Analysis
Isolate WL65 was previously identified as a member of the genus Phytobacter based on the 16S rRNA gene sequence analysis in our previous study [6]. Phytobacter sp. WL65 is a Gram-negative, rod-shaped, motile bacterium that does not form spores. Its cell dimensions range from 0.62–0.82 μm in width to 1.62–2.96 μm in length. On a nitrogen-free medium and nutrient agar, the colonies appeared smooth, white, and circular with entire edges and measured 2–3 mm in diameter after 24 h of incubation at 30 °C. On the MacConkey agar, the colonies developed a pink color after incubation at 37 °C for 24 h. Biochemical characterization of isolate WL65 was performed using the API 20E system and compared to four reference Phytobacter species: P. palmae S29T, P. ursingii ATCC 27989T, P. diazotrophicus DSM 17806T, and P. massiliensis JC163T (Table 1). The biochemical profile of isolate WL65 showed positive catalase activity but negative oxidase activity. It was capable of fermenting glucose, mannitol, sorbitol, rhamnose, saccharose, amygdalin, and arabinose but was unable to ferment inositol and melibiose. Additionally, it tested positive for citrate utilization and indole production.

Table 1.
Biochemical characteristics of Phytobacter sp. WL65 and another four Phytobacter species based on the API20E analysis.
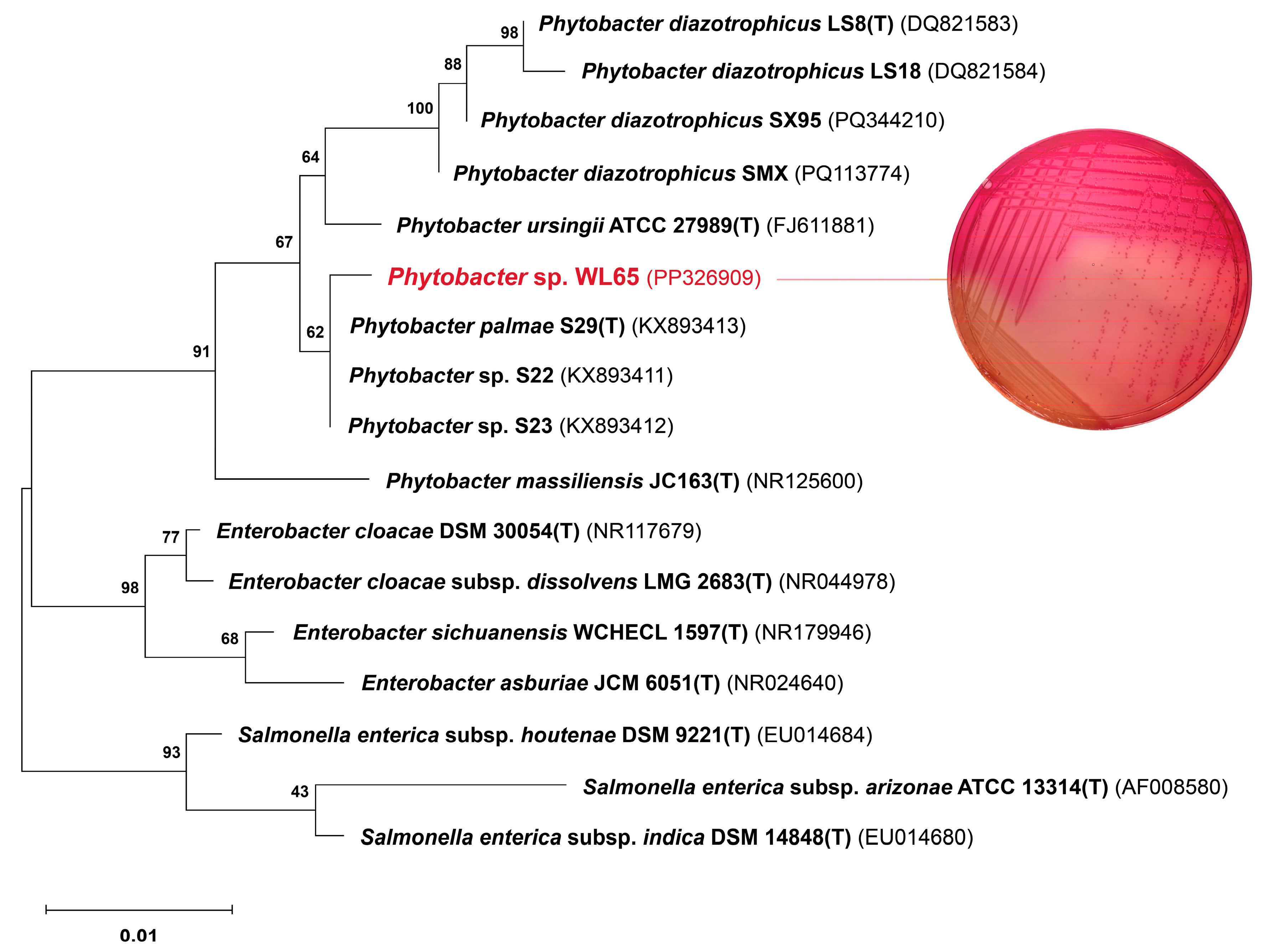
A phylogenetic tree based on the 16S rRNA gene sequences of four Phytobacter species and related genera was constructed, as shown in Figure 1. The analysis identified three major clades corresponding to the genera Enterobacter, Salmonella, and Phytobacter. Phytobacter sp. WL65 clustered within the Phytobacter clade, showing a high sequence similarity of 99.70% to P. palmae S29T. Comparatively, WL65 exhibited sequence similarities of 98.51%, 99.17%, and 98.44% to P. diazotrophicus DSM 17806T, P. ursingii ATCC 27989T, and P. massiliensis JC163T, respectively. Although WL65 demonstrated more than 99% similarity to both P. palmae and P. ursingii, its biochemical characteristics were more closely aligned with those of P. palmae, except that WL65 was negative for tryptophan deaminase.
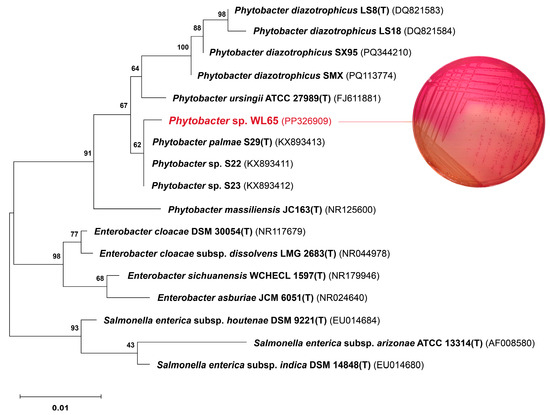
Figure 1.
Phylogenetic relationship of Phytobacter sp. WL65 and related genera based on 16S rRNA gene sequences using the maximum likelihood method. The numbers on the branches were the percentages of the bootstrap support derived from 1000 replications. Bar was the nucleotide substitutions per nucleotide. The MacConkey agar exhibited the pink colonies of Phytobacter sp. WL65.
3.2. Genome Sequencing and Phylogenomics Analysis
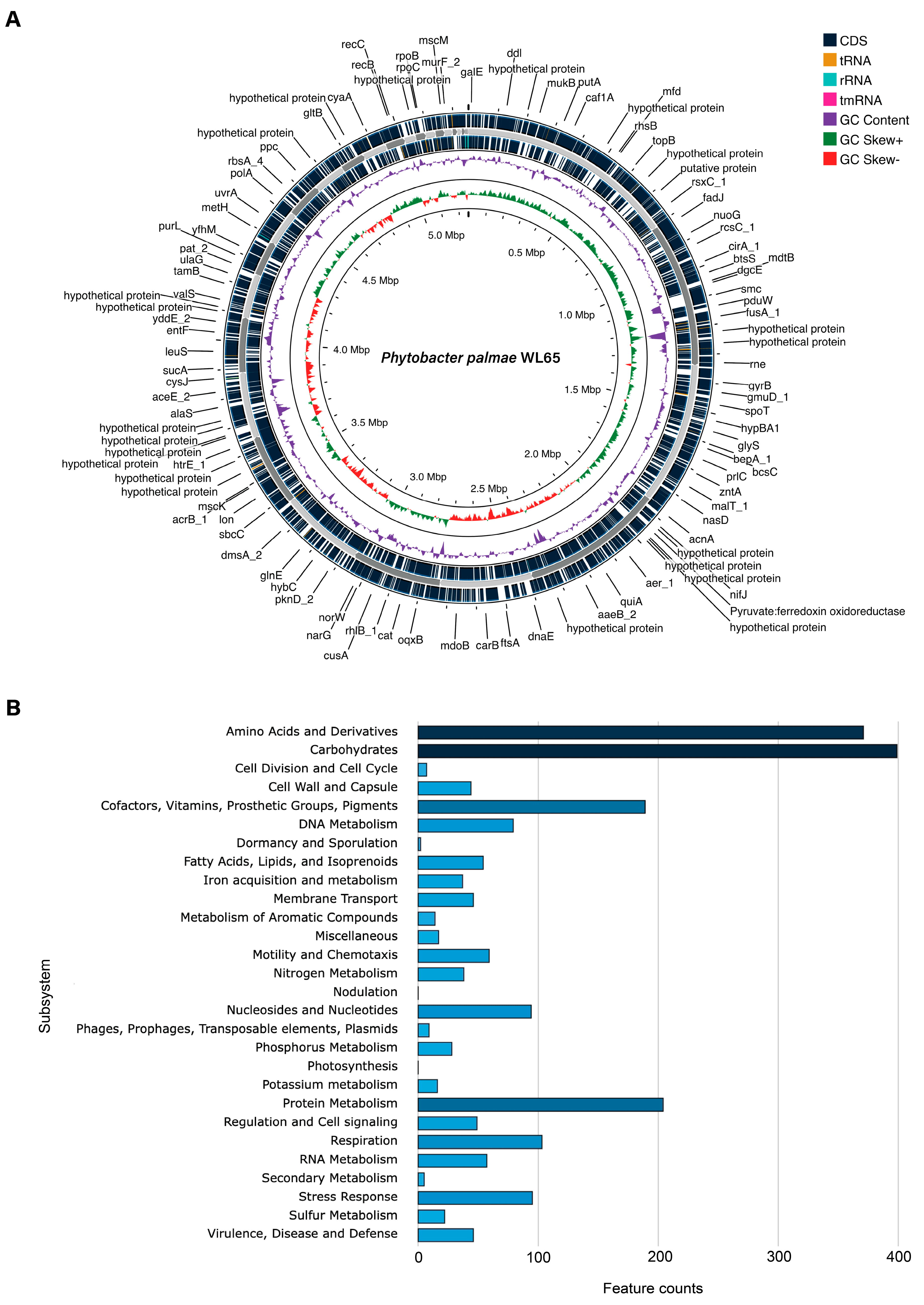
The draft genome sequence of isolate WL65 was analyzed, and its basic genome indices are summarized in Table 2, in comparison with the type strains P. palmae S29T. The complete genome has been deposited at DDBJ/ENA/GenBank under the accession JBLQZN000000000, and a circular chromosome map was generated using PATRIC, as shown in Figure 2A. The genome size was 5,253,439 bp (5.2 Mb), with a G + C content of 52.51% and consisted of 33 contigs. The genome contained 5210 coding sequences (CDS), 58 tRNAs, 4 rRNAs, 347 subsystems, an N50 value of 340,721, and an L50 value of 5. No antibiotic resistance genes, transporters, drug target genes, or plasmids were identified in the WL65 genome based on the analyses using the respective databases. A genome annotation of isolate WL65, indicating subsystem features, is presented in Figure 2B. These features include key biological processes, including carbohydrates, amino acids and derivatives, protein metabolism, cofactors, vitamins, prosthetic groups, stress response, nitrogen metabolism, phosphorus metabolism, etc. (Table S1). Functional annotation was performed using the FaCoP system and identified the genes associated with essential cellular processes, including transcription (227 genes, 11.8%), inorganic ion transport and metabolism (216 genes, 11.3%), carbohydrate transport and metabolism (211 genes, 11%), amino acid transport and metabolism (173 genes, 9%), and various gene distributions (Table S2 and Figure S1).

Table 2.
Overall statistical genome features of P. palmae WL65 and the type strain.
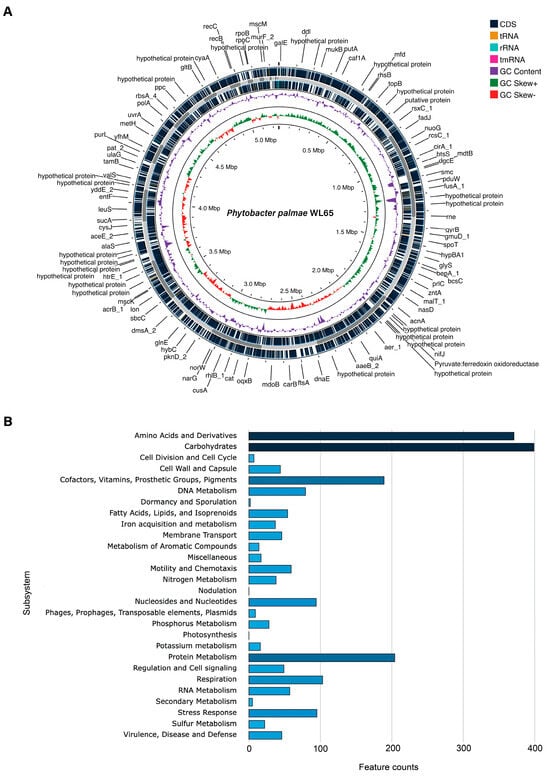
Figure 2.
The draft genome of P. palmae WL65: (A) A circular genome map was generated using the Proksee server, showing coding sequences (CDS), transfer RNA (tRNA), ribosomal RNA (rRNA), transfer messenger RNA (tmRNA), guanine-cytosine (GC) content, and GC skew; (B) a bar chart illustrating the subsystem category distribution analyzed using the SEED and RAST servers.
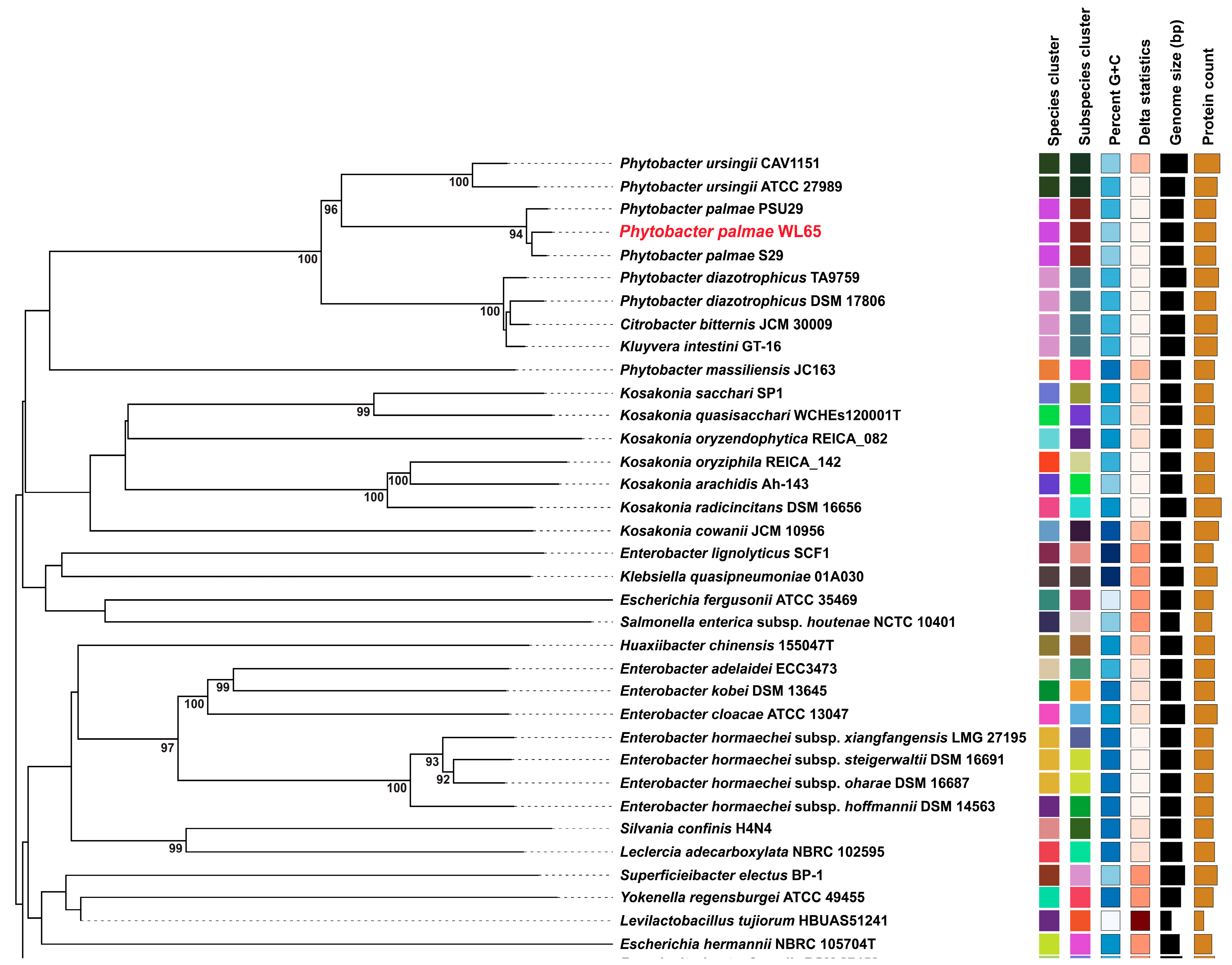
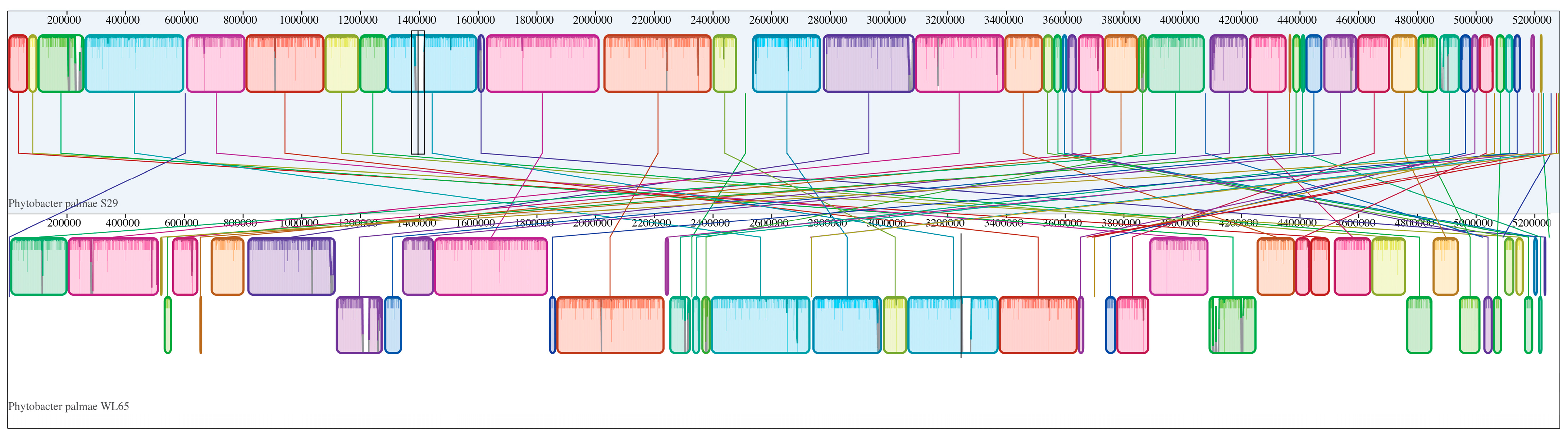
A phylogenomic tree was constructed to include the isolate WL65 and related species, as shown in Figure 3. The results clearly demonstrated that isolate WL65 clustered with P. palmae S29T and PSU29, forming a distinct group separate from other species. Pairwise genome comparisons were conducted (Table S3), and the digital DNA-DNA hybridization (dDDH) values between the isolate WL65 and P. palmae S29T and PSU29 were 94.1% and 92.9%, respectively (Figure 4 and Figure S2). Isolate WL65 showed a 47.9% dDDH similarity to P. ursingii ATCC 27989T and 45.0% to P. diazotrophicus DSM 17806T. Therefore, isolate WL65 was identified as P. palmae WL65.
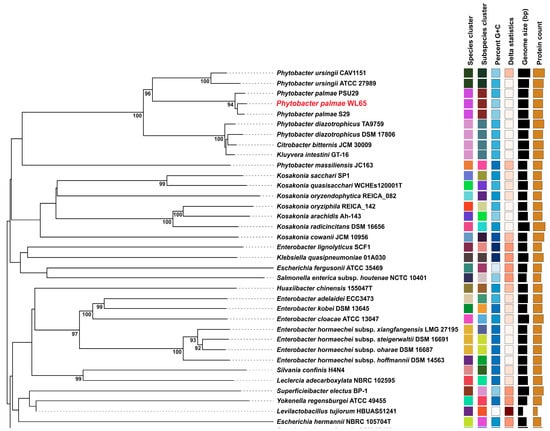
Figure 3.
Genome BLAST distance phylogeny (GBDP) tree. The tree, inferred with FastME 2.1.6.1 from GBDP, provides the distances calculated from the genome sequences. The branch lengths are scaled in terms of the GBDP distance formula d5. The numbers above the branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 61.7%. The tree was rooted at the midpoint.
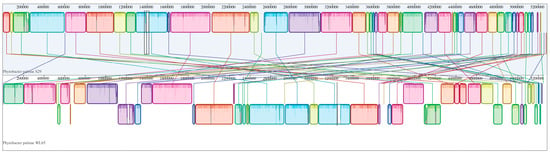
Figure 4.
Pairwise comparison of the P. palmae WL65 (bottom) genome with the type strain P. palmae S29T (top) using the progressive MAUVE algorithm via Geneious Prime software v.2025.0.3. The same colors represent homologous locally collinear blocks between genomes, connected by lines.
3.3. Genome Analysis of P. palmae WL65 as Plant Growth-Promoting Rhizobacterium
P. palmae WL65 exhibited key plant growth-promoting bacterium traits, including nitrogen fixation, phosphate solubilization, indole-3-acetic acid (IAA) production, siderophore production, and ACC deaminase activity, as previously reported by Thamvithayakorn et al. [6]. The genomic analysis revealed the presence of genes associated with these beneficial characteristics (Table 3 and Table S4). The nitrogen fixation genes nifA, nifB, nifD, nifE, nifF, nifH, nifJ, nifK, nifL, nifS, nifU, nifW, and nifX indicate the strain’s capacity to convert atmospheric nitrogen (N2) into ammonia (NH3) by nitrogenase enzymes, thereby enhancing nitrogen availability for plants. Notably, nifH, nifD, and nifK encode the nitrogenase enzyme complex, which catalyzes nitrogen reduction, while nifA regulates nif gene expression. The presence of these genes suggests that P. palmae WL65 contributes significantly to nitrogen enrichment in the rhizosphere. The phosphate solubilization genes (pstA, pstB, pstC, pstS, phnC, phnD, phnE, and phnV) facilitate the uptake and mobilization of insoluble phosphate compounds, increasing phosphorus bioavailability. The presence of siderophore biosynthesis genes (fhuA, fhuB, fhuC, fhuD, fhuF, feoA, feoB, feoC, acrA, acrB, acrE, acrR, and acrZ) enables iron acquisition from the soil, improving plant iron uptake and alleviating iron deficiency stress. The potassium solubilization genes (gcd1 and gcd2) were characterized in the genome of P. palmae WL65. These genes encode dehydrogenase, producing gluconic acid to solubilize potassium, enhancing plant nutrient uptake and growth. The genes involved in the tryptophan-dependent IAA biosynthetic pathway (trpA, trpB, trpC, trpE, trpGD, trpR, and trpS) contribute to plant growth promotion by stimulating root elongation and lateral root formation.

Table 3.
Genes involved in plant growth-promoting traits based on RAST and Prokka.
For plant defense, detoxification, and stress tolerance genes, several key genes were identified. Genes encoding ACC deaminase (rimI, rimK, rimL, rimM, rimO, rimP, and rimJ) facilitate the degradation of 1-aminocyclopropane-1-carboxylate (ACC), an ethylene precursor, helping to mitigate plant stress under drought and salinity conditions. Additionally, P. palmae WL65 possesses hydrogen sulfide (H2S) biosynthesis genes (cysA–cysZ), which contribute to sulfur assimilation and plant defense. The sodA, sodB, and sodC genes encode the superoxide dismutase enzymes essential for oxidative stress protection, enhancing bacterial survival and plant vitality. Furthermore, the osmB, osmC, osmE, osmV, osmW, osmX, osmY, and oxyR genes encode peroxidases and oxidative stress-related proteins, supporting stress tolerance, detoxification, and plant growth promotion. Additionally, the phzF gene, responsible for phenazine production, enhances biocontrol and stress tolerance, making P. palmae WL65 more effective in sustainable agriculture.
The colonization and quorum sensing-related genes were also identified. Genes involved in biofilm formation (pgaA, pgaB, pgaC) contribute to bacterial adhesion, enhancing rhizosphere colonization and plant-microbe interactions. Additionally, the strain possesses genes associated with acetoin and butanediol biosynthesis (budC, alsT, and poxB), GABA production (gabR), glycine-betaine synthesis (proA, proB, proC, proP, proS, proX, proQ, proV, proW, proY, soxR, and soxS), and quorum sensing (luxR and luxS), which further support plant resilience and microbial communication. Notably, a significant proportion of hypothetical proteins were identified, suggesting the presence of novel functional elements that warrant further investigation. Importantly, no plasmids carrying antibiotic-resistant genes were detected, highlighting the genomic safety of P. palmae WL65 for agricultural applications.
3.4. Genome Analysis of Secondary Metabolism of P. palmae WL65 Using AntiSMASH
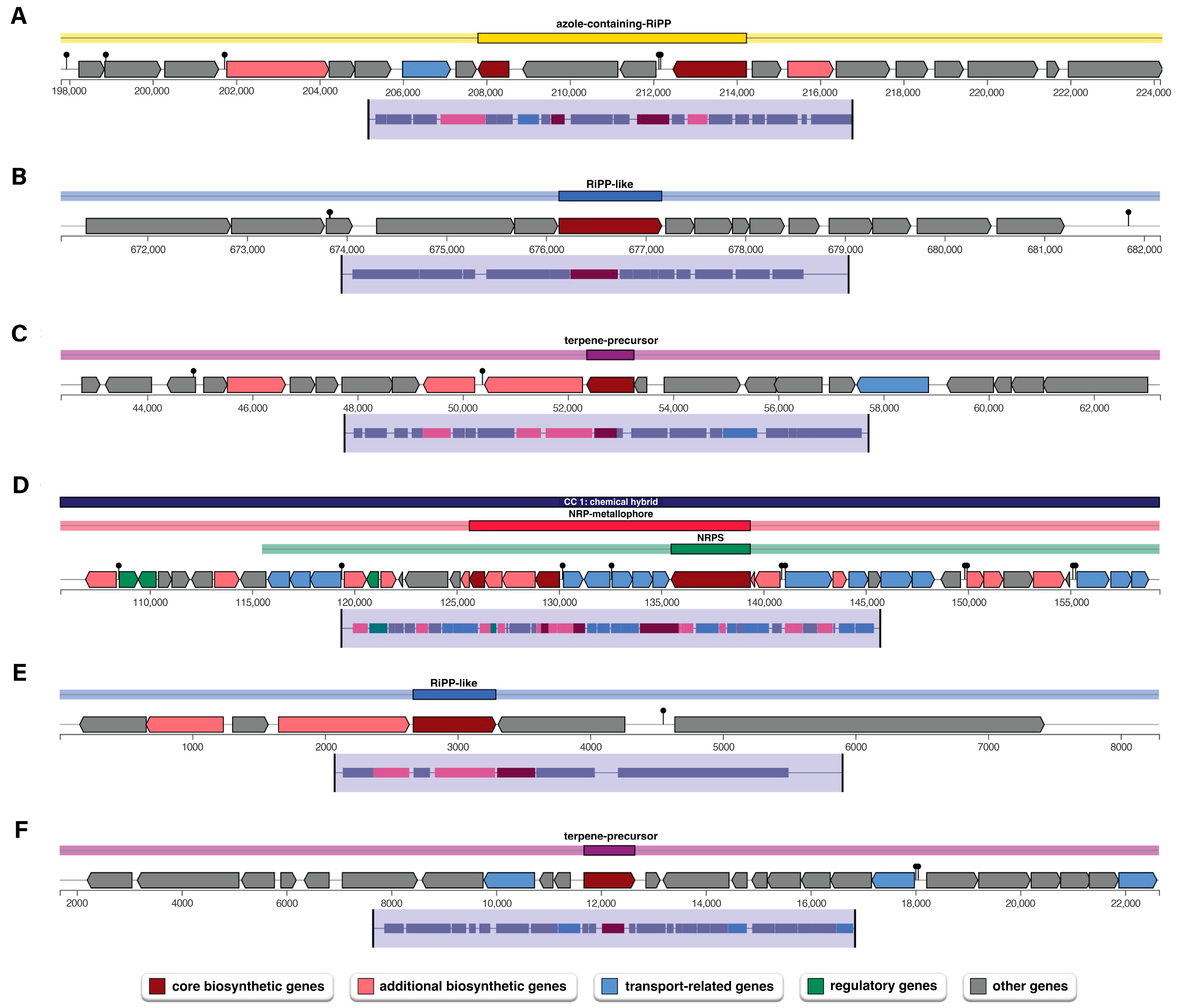
Antimicrobial activity is one of the key traits of plant growth-promoting bacterium for plant growth, particularly in suppressing phytopathogens. The genome analysis of P. palmae WL65 using AntiSMASH identified six gene clusters that are involved in the biosynthesis of secondary metabolites (Table 4 and Figure 5). Among these, the azole-containing RiPP cluster encodes ribosomal synthesized and post-translationally modified peptides (RiPPs) that are characterized by azole heterocycles. These compounds often exhibit antimicrobial, antifungal, or anticancer properties. The genome also contains RiPP-like gene clusters, which have been identified through genome mining and untargeted metabolomics, potentially encoding novel bioactive compounds with pharmaceutical applications, including antimicrobial and anticancer activities. Additionally, a terpene-precursor biosynthesis cluster was identified. Terpene precursors are metabolic intermediates produced from primary metabolism that form the basis for terpene production. These molecules undergo enzymatic modifications, such as cyclization, oxidation, and glycosylation, leading to the production of a diverse array of bioactive terpenoids commonly found in plants, fungi, and microorganisms. Notably, the genome of P. palmae WL65 contains a non-ribosomal peptide (NRP) metallophore biosynthetic cluster, which exhibits high similarity to the enterobactin biosynthetic pathway. Enterobactin, also known as enterochelin, is a high-affinity siderophore essential for iron acquisition. It facilitates ferric iron (Fe3⁺) scavenging from the environment and its transport into bacterial cells, enhancing bacterial survival and competitiveness under iron-limiting conditions.

Table 4.
Secondary metabolite biosynthetic gene clusters (BGCs) of P. palmae WL65 identified using AntiSMASH.
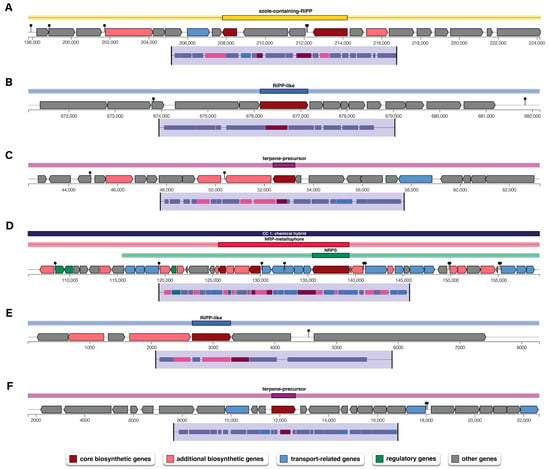
Figure 5.
Biosynthetic gene clusters (BGCs) encoding secondary metabolites in the P. palmae WL65 genome: (A) azole-containing-RiPP; (B) RiPP-like; (C) terpene-precursor; (D) NRP-metallophore, NRPS; (E) RiPP-like; and (F) terpene-precursor.
3.5. P. palmae WL65 as a Plant Growth-Promoting Rhizobacterium (PGPR) for Rice Cultivation in Pot Experiments
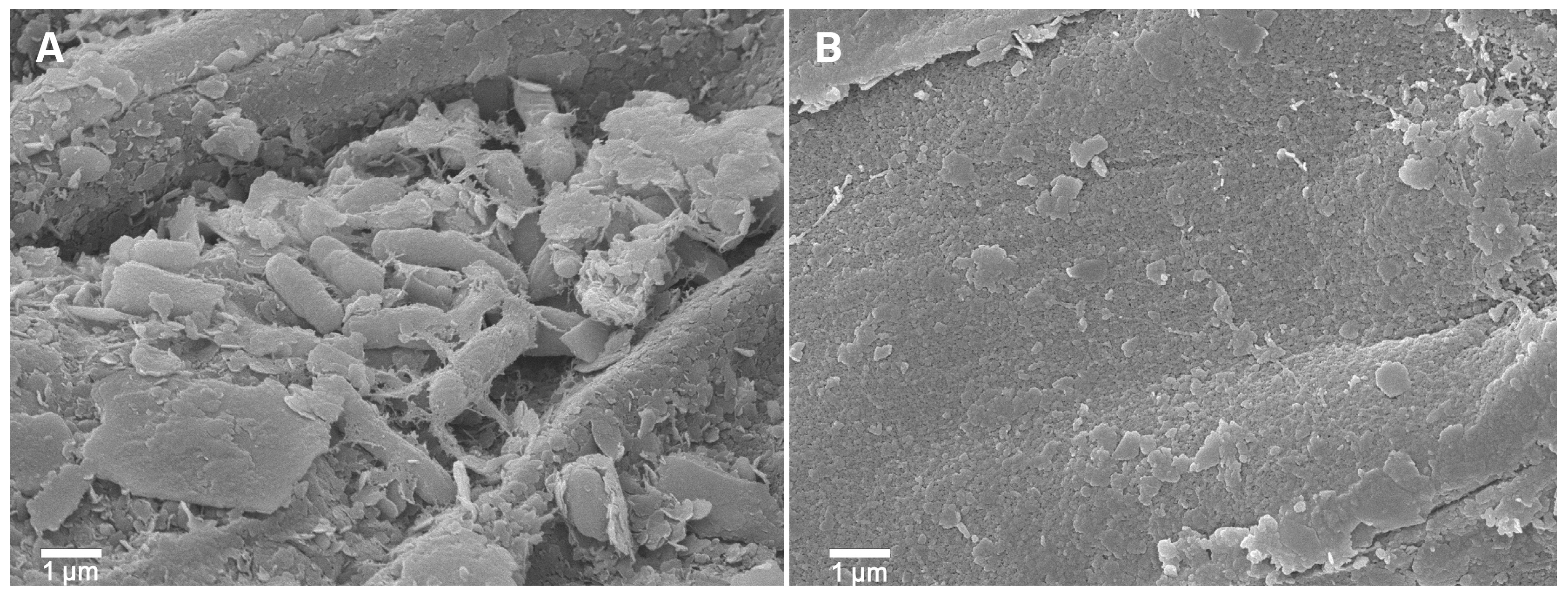
P. palmae WL65 exhibited key PGPR properties, as supported by the genome analysis. This strain has the potential as a PGPR for rice cultivation, given its origin in the rice rhizosphere. The ability of P. palmae WL65 to colonize rice roots was assessed using scanning electron microscopy (SEM), which revealed a high density of bacterial cells of similar size and shape surrounding the roots after four weeks of seedling cultivation, compared to the uninoculated control (Figure 6). This finding confirms the strain’s ability to effectively colonize plant roots, a critical trait for PGPR function capability. The enhanced root colonization observed may contribute to improved nutrient uptake and plant growth.
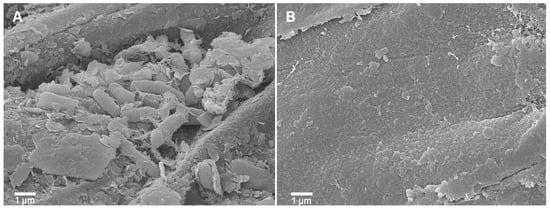
Figure 6.
SEM micrographs of PGPR colonization on rice roots (A) P. palmae WL65 and (B) uninoculated control.
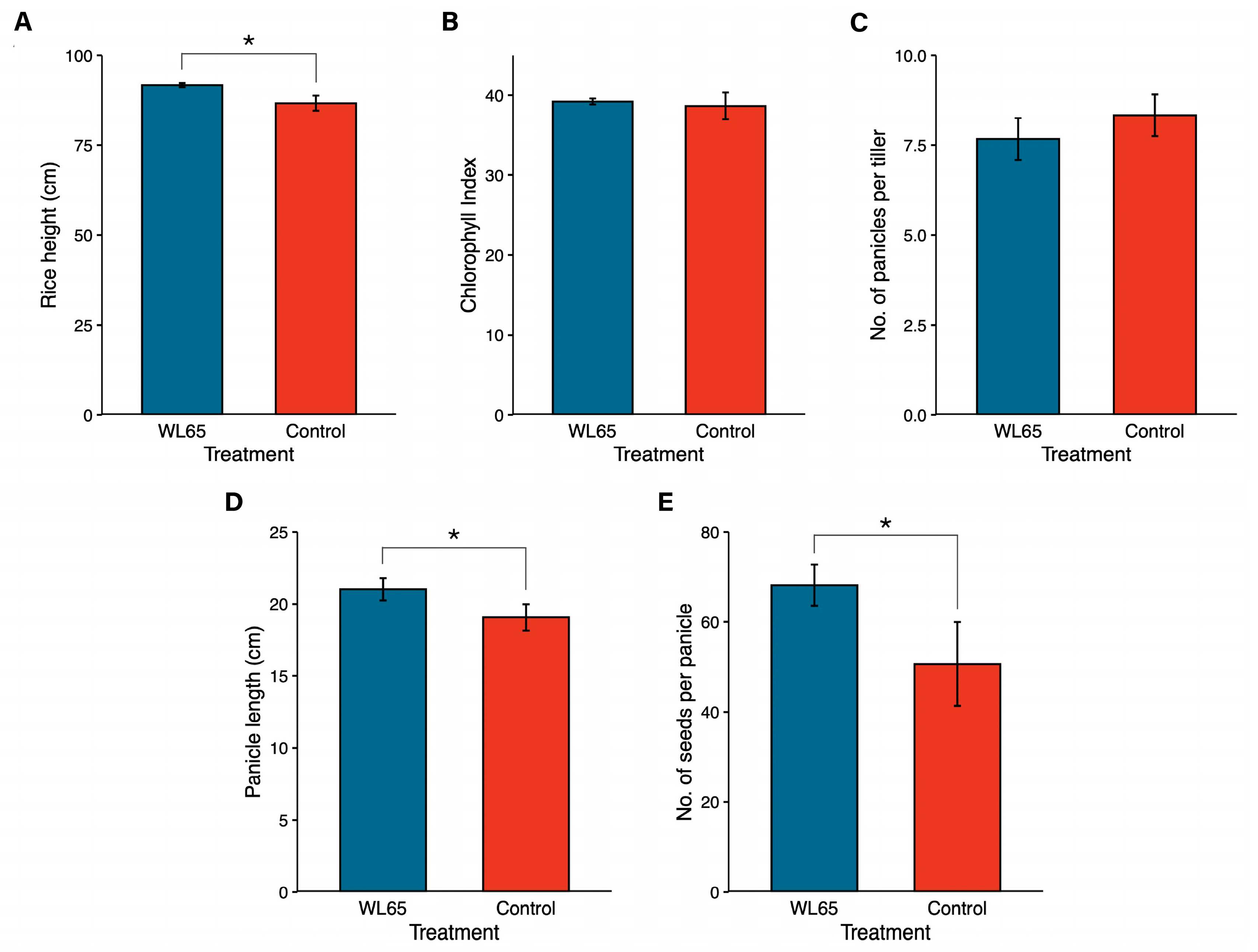
Further, P. palmae WL65 was evaluated as a PGPR in a pot experiment, where its impact on rice growth and soil physicochemical properties was assessed (Figure 7 and Figure S3 and Table S5). Rice plants inoculated with WL65 exhibited a significantly greater plant height (91.67 ± 0.58 cm) compared to the control (86.67 ± 2.08 cm), representing a 5.8% increase. Additionally, the panicle length (21.02 ± 0.77 cm) and the number of seeds per panicle (68.17 ± 4.54 seeds) were significantly higher than those in the control (19.07 ± 0.92 cm and 50.67 ± 9.29 seeds, respectively), corresponding to increases of 10.2% and 34.5%. However, the chlorophyll content index and the number of panicles per tiller did not differ significantly between the WL65 treatment and the control. The chlorophyll content indices were 39.22 ± 0.36 for WL65 and 38.65 ± 1.68 for the control. Similarly, the number of panicles per tiller was 7.67 ± 0.58 panicles in the WL65 treatment and 8.33 ± 0.58 panicles in the control. The physicochemical analysis of the soil samples post-harvest (Table 5) indicated that the organic matter, available phosphorus, potassium, calcium, and magnesium levels were higher in the WL65 treatment compared to the control, suggesting an improvement in soil fertility that was associated with bacterial inoculation.
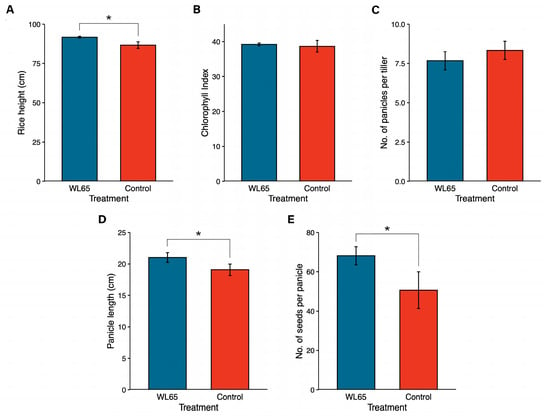
Figure 7.
Effects of P. palmae WL65 on rice growth parameters: (A) rice height; (B) chlorophyll index; (C) number of panicles per tiller; (D) panicle length compared to (E) the uninoculated control. * Indicates a statistically significant difference at p ≤ 0.05 (independent samples t-test).

Table 5.
Physicochemical properties of soil samples from the pot experiment after harvest.
4. Discussion
Phytobacter is a nitrogen-fixing bacterial genus consisting of only four species. In a recent study, we isolated rhizobacterial strains from the rhizosphere soils of rice (Oryza sativa L.) in northeast Thailand [6]. One isolate, Phytobacter sp. WL65, exhibiting outstanding plant growth-promoting properties, was selected for species identification. The biochemical and genomic characterization of WL65 confirmed its classification as P. palmae. This classification was supported by pairwise genome analysis and dDDH, which revealed a 94.1% genome similarity to the type strain P. palmae S29T and 92.9% similarity to P. palmae PSU29 [3,26]. Notably, these are the only two available genomic references for P. palmae in public databases. Interestingly, the biochemical profile of WL65 differed from that of P. palmae S29T, particularly in tryptophan metabolism. While S29 exhibited tryptophan deaminase activity, WL65 tested negative for this enzyme. Tryptophan deaminase, encoded by the tdcB gene, catalyzes the deamination of tryptophan, leading to the production of indole-3-pyruvate and other metabolites, which also contribute to indole production. Despite the absence of tdcB, WL65 exhibited indole production, suggesting the occurrence of an alternative metabolic pathway for indole production. The genome analysis revealed that WL65 possesses tnaA, encoding tryptophanase, an enzyme that directly converts tryptophan into indole, ammonia, and pyruvate. This finding suggests that WL65 relies on TnaA-mediated tryptophan degradation rather than the TdcB-dependent pathway observed in S29 [27]. The variation in tryptophan metabolism between these strains may be influenced by environmental conditions and horizontal gene transfer. Notably, tnaA is more commonly expressed under aerobic conditions, whereas tdcB is typically induced under anaerobic or fermentative conditions [27].
Although P. palmae S29T was originally isolated as an endophyte from oil palm tissues in Singapore, exhibiting strong nitrogen-fixing ability [3], our study identified WL65 from rice rhizosphere soil in Thailand, where it demonstrated nitrogen fixation along with additional plant growth-promoting properties [6]. This suggests that P. palmae is not plant-specific but rather a versatile species capable of colonizing different plant hosts. Few P. palmae isolates have been reported, including two nitrogen-fixing strains identified in the present study and the type strain. However, more recently, P. palmae PSU29 was isolated from a clinical sample in Southern Thailand [26]. The genome analysis of PSU29 revealed the presence of the oqxA and oqxB genes, which encode the OqxAB efflux pump. The oqxAB operon is commonly found in Enterobacteriaceae, particularly in Escherichia coli, Salmonella enterica, and Enterobacter spp. [28,29]. Interestingly, the genome analysis of P. palmae WL65 revealed the presence of only the oqxB gene, while oqxA was absent. This suggests a possible genomic variation or rearrangement in this strain. Unlike PSU29, WL65 does not have any mobile plasmids associated with antibiotic resistance, indicating that it lacks horizontally acquired resistance elements. Additionally, while PSU29 was identified in a clinical sample, WL65 was isolated from the rice rhizosphere, highlighting the environmental diversity of P. palmae and its potential adaptation to different ecological niches. However, the genome analysis of WL65 revealed the presence of key genes associated with plant growth-promoting (PGP) traits, including nitrogen fixation, phosphate solubilization, IAA biosynthesis, and siderophore production. These findings confirm the PGPR potential of WL65, as previously described by Thamvithayakorn et al. [6]. In addition to these well-established PGP traits, WL65 has several genes essential for bacterial survival, plant colonization, stress tolerance, and pathogen suppression, further supporting its role in sustainable agriculture. Notably, this study represents the first genomic analysis of PGPR-associated traits in P. palmae, providing insights into its secondary metabolite biosynthetic potential. One of the key findings was the presence of an enterobactin biosynthetic gene cluster. Enterobactin is a well-characterized siderophore that plays a crucial role in iron acquisition, a vital function for both bacterial competitiveness in the rhizosphere and plant health [30,31]. The ability of WL65 to produce siderophores further reinforces its potential as a beneficial PGPR strain capable of enhancing plant growth and resilience under nutrient-limited conditions. These findings support the potential of P. palmae WL65 as a promising candidate for biofertilizer development, contributing to sustainable and eco-friendly agricultural practices.
Thailand is one of the largest rice producers, having a rice diversity of over 100 rice varieties, including the hybrid rice RD79, developed by the Rice Department (RD). RD79 is among the most popular rice varieties in Thailand due to its high yield and resistance to rice blast disease [32]. Therefore, WL65 was selected as a potential PGPR for RD79 cultivation. The results demonstrated successful bacterial colonization on rice roots compared to the control. Root colonization represents one of the key characteristics of PGPR, as it allows beneficial bacteria to form a stable association with plants and thus exerts their beneficial growth-promoting effects. The colonization ability of WL65 might be attributed to biofilm-associated genes (pgaA, pgaB, and pgaC), which are essential for bacterial adhesion to plant roots. The pgaABC operon encodes proteins that are involved in the biosynthesis of the extracellular polymeric substance poly-β-1,6-N-acetyl-D-glucosamine (PNAG), which is important for surface attachment, biofilm formation, and structural integrity. More specifically, pgaC codes a glycosyltransferase-linking PNAG, pgaA codes for an outer membrane transporter of PNAG, and pgaB deacetylates PNAG to increase its adhesive properties [33,34]. Overall, the synergistic action of these functions helps to promote bacterial clustering, persistence in the rhizosphere, and sustain colonization of plant roots, which are all meant to enable effective PGPR activity. Additionally, biofilm formation enhances bacterial survival by increasing resistance to environmental stressors, promoting root surface attachment, stabilizing bacterial populations in the rhizosphere, and improving nutrient exchange [35]. The root colonization of WL65 in the rice rhizosphere was confirmed by SEM analysis. Notably, biofilm-forming PGPR such as Brucella sp. and Lysinibacillus macrolides have been shown to enhance wheat (Triticum aestivum L.) growth and productivity, leading to increases in plant height and grain yields by 16.7% and 17.5%, respectively, compared to the control [36]. The application of WL65 significantly improved the rice growth parameters, including the plant height, panicle length, and the number of seeds per panicle, compared to the control. These growth enhancements are likely associated with the production of phytohormones such as IAA, a key auxin involved in root and shoot development [37]. Additionally, the soil physicochemical properties after harvest indicated improved soil health in the WL65 treatment compared to the control. The higher organic matter content observed in the treated soil may result from the enhanced nutrient cycling driven by beneficial bacterial activity in the rhizosphere. Furthermore, the increased availability of phosphorus and potassium in the soil confirms the phosphate and potassium solubilization capabilities of WL65, supporting its role in enhancing soil fertility and plant nutrient uptake. Notably, calcium levels in the WL65 treatment increased by 27% compared to the control, contributing to soil organic carbon sequestration and reducing CO2 release [38]. Calcium also plays a crucial role in bacterial adhesion and modulating PGPR-induced defense responses against environmental stress [39]. These results indicate that P. palmae WL65 is a novel PGPR candidate with strong potential for rice farming due to its plant growth-promoting activity. Given that most PGPRs are not plant-specific, future research should explore their application in other crops. Moreover, WL65 can serve as an eco-friendly alternative to conventional biofertilizers, supporting plant growth and biomodification while contributing to sustainable agriculture.
5. Conclusions
This study provides the first genomic characterization of P. palmae WL65 as a PGPR. Whole-genome analysis confirmed its identity, revealing genes for nitrogen fixation, phosphate solubilization, IAA biosynthesis, siderophore production, and biofilm formation. Despite its genomic similarity to P. palmae S29T, WL65 exhibited metabolic differences in tryptophan metabolism. Its application in rice cultivation enhanced root colonization, plant growth, and yield while improving soil fertility. These findings highlight the potential of WL65 as a biofertilizer, contributing to sustainable agriculture and expanding our understanding of PGPR mechanisms for eco-friendly crop production.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture15070707/s1, Table S1: Subsystem of P. palmae WL65 distribution from SEED and Rapid Annotation using Subsystem Technology (RAST) servers; Table S2: Clusters of orthologous genes of P. palmae WL65 distribution using Functional Annotation and Classification of Proteins of Prokaryotes (FACoP); Figure S1: Clusters of orthologous genes distribution of P. palmae WL65; Table S3: Pairwise digital DNA-DNA hybridization (dDDH) comparisons of P. palmae WL65 genome with type-strain genomes; Figure S2: Pairwise comparisons of P. palmae WL65 genome with P. palmae PSU29 using the progressive mauve algorithm; Table S4: Genes involved in plant growth-promoting traits using RAST and Rapid Prokaryotic Genome Annotation (Prokka); Table S5: Effects of P. palmae WL65 on rice growth parameters in a pot experiment under greenhouse conditions; Figure S3: Rice pot experiment under greenhouse conditions showing WL65 treatment; T1 (left) and uninoculated control; T2 (right).
Author Contributions
Conceptualization, P.T., C.P. and N.S.; methodology, P.T. and N.S.; software, P.T.; validation, C.P., V.V. and N.S.; formal analysis, P.T. and N.S.; investigation, P.T.; resources, C.P.; data curation, P.T., C.P. and N.S.; writing—original draft preparation, P.T. and N.S.; writing—review and editing, C.P. and V.V.; supervision, N.S.; funding acquisition, P.T. and N.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Research Council of Thailand (NRCT) and ADAMA (THAILAND) LIMITED: grant no. N41A650169.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We would like to thank Pravech Ajawatanawong, Mahidol University, Thailand, for his help with whole-genome sequencing.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhang, G.X.; Peng, G.X.; Wang, E.T.; Yan, H.; Yuan, Q.H.; Zhang, W.; Lou, X.; Wu, H.; Tan, Z.Y. Diverse endophytic nitrogen-fixing bacteria isolated from wild rice Oryza rufipogon and description of Phytobacter diazotrophicus gen. nov. sp. nov. Arch. Microbiol. 2008, 189, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Madhaiyan, M.; Saravanan, V.S.; Blom, J.; Smits, T.H.; Rezzonico, F.; Kim, S.-J.; Weon, H.-Y.; Kwon, S.-W.; Whitman, W.B.; Ji, L. Phytobacter palmae sp. nov., a novel endophytic, N2 fixing, plant growth promoting Gamma proteobacterium isolated from oil palm (Elaeis guineensis Jacq.). Int. J. Syst. Evol. Microbiol. 2020, 70, 841–848. [Google Scholar] [CrossRef]
- Pillonetto, M.; Arend, L.N.; Faoro, H.; D’Espindula, H.R.; Blom, J.; Smits, T.H.; Mira, M.T.; Rezzonico, F. Emended description of the genus Phytobacter, its type species Phytobacter diazotrophicus (Zhang 2008) and description of Phytobacter ursingii sp. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yao, R.; Li, Y.; Wu, X.; Li, S.; An, Q. Proposal for unification of the genus Metakosakonia and the genus Phytobacter to a single genus Phytobacter and reclassification of Metakosakonia massiliensis as Phytobacter massiliensis comb. nov. Curr. Microbiol. 2020, 77, 1945–1954. [Google Scholar] [CrossRef]
- Almuzara, M.; Cittadini, R.; Traglia, G.; Haim, M.S.; De Belder, D.; Alvarez, C.; O’Connor, Z.; Ocampo, C.V.; Barberis, C.; Prieto, M. Phytobacter spp: The emergence of a new genus of healthcare-associated Enterobacterales encoding carbapenemases in Argentina: A case series. Infect. Prev. Pract. 2024, 6, 100379. [Google Scholar] [CrossRef]
- Thamvithayakorn, P.; Phosri, C.; Robinson-Boyer, L.; Limnonthakul, P.; Doonan, J.H.; Suwannasai, N. The synergistic impact of a novel plant growth-promoting rhizobacterial consortium and Ascophyllum nodosum seaweed extract on rhizosphere microbiome dynamics and growth enhancement in Oryza sativa L. RD79. Agronomy 2024, 14, 2698. [Google Scholar] [CrossRef]
- Aslanzadeh, J. Biochemical profile-based microbial identification systems. In Advanced Techniques in Diagnostic Microbiology; Springer: Boston, MA, USA, 2006; pp. 84–116. [Google Scholar]
- Brady, C.; Cleenwerck, I.; Venter, S.; Vancanneyt, M.; Swings, J.; Coutinho, T. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 2008, 31, 447–460. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bio Inform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Yaikhan, T.; Suwannasin, S.; Singkhamanan, K.; Chusri, S.; Pomwised, R.; Wonglapsuwan, M.; Surachat, K. Genomic characterization of multidrug-resistant Enterobacteriaceae clinical isolates from Southern Thailand hospitals: Unraveling antimicrobial resistance and virulence mechanisms. Antibiotics 2024, 13, 531. [Google Scholar] [CrossRef]
- Gong, F.; Yanofsky, C. Analysis of tryptophanase operon expression in vitro: Accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J. Biol. Chem. 2002, 277, 17095–17100. [Google Scholar] [CrossRef]
- Moosavian, M.; Khoshkholgh Sima, M.; Ahmad Khosravi, N.; Abbasi Montazeri, E. Detection of OqxAB efflux pumps, a multidrug-resistant agent in bacterial infection in patients referring to teaching hospitals in Ahvaz, Southwest of Iran. Int. J. Microbiol. 2021, 2021, 2145176. [Google Scholar] [CrossRef]
- Bharatham, N.; Bhowmik, P.; Aoki, M.; Okada, U.; Sharma, S.; Yamashita, E.; Shanbhag, A.P.; Rajagopal, S.; Thomas, T.; Sarma, M. Structure and function relationship of OqxB efflux pump from Klebsiella pneumoniae. Nat. Commun. 2021, 12, 5400. [Google Scholar] [CrossRef]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Bacterial siderophores: Classification, biosynthesis, perspectives of use in agriculture. Plants 2022, 11, 3065. [Google Scholar] [CrossRef]
- Chuchert, S.; Thongmak, J.; Daungnamkaew, B.; Sudpesrot, P.; Nakorn, W.N.; Marnmad, K.; Khomphet, T. Optimizing Temperature and Drying Conditions to Break Dormancy in Thai RD79 Rice Seeds. Indian J. Agric. Res. 2025, 1–9. [Google Scholar] [CrossRef]
- Itoh, Y.; Rice, J.D.; Goller, C.; Pannuri, A.; Meisner, J.J.; Beveridge, T.J.; Preston III, J.F.; Rom, T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamine. J. Bacteriol 2008, 190, 3670–3680. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.J.; Tu, I.F.; Tseng, T.S.; Tsai, Y.H.; Wu, S.H. The deficiency of poly-β-1,6-N-acetyl-glucosamine deacetylase trigger A. baumannii to convert to biofilm-independent colistin-tolerant cells. Sci. Rep. 2023, 13, 2800. [Google Scholar] [CrossRef]
- Ajijah, N.; Fiodor, A.; Pandey, A.K.; Rana, A.; Pranaw, K. Plant growth-promoting bacteria (PGPB) with biofilm-forming ability: A multifaceted agent for sustainable agriculture. Diversity 2023, 15, 112. [Google Scholar] [CrossRef]
- Rafique, M.; Naveed, M.; Mumtaz, M.Z.; Niaz, A.; Alamri, S.; Siddiqui, M.H.; Waheed, M.Q.; Ali, Z.; Naman, A.; Rehman, S.U. Unlocking the potential of biofilm-forming plant growth-promoting rhizobacteria for growth and yield enhancement in wheat (Triticum aestivum L.). Sci. Rep. 2024, 14, 15546. [Google Scholar] [CrossRef]
- Lata, D.L.; Abdie, O.; Rezene, Y. IAA-producing bacteria from the rhizosphere of chickpea (Cicer arietinum L.): Isolation, characterization, and their effects on plant growth performance. Heliyon 2024, 10, e39702. [Google Scholar] [CrossRef]
- Shabtai, I.A.; Wilhelm, R.C.; Schweizer, S.A.; Höschen, C.; Buckley, D.H.; Lehmann, J. Calcium promotes persistent soil organic matter by altering microbial transformation of plant litter. Nat. Commun. 2023, 14, 6609. [Google Scholar] [CrossRef]
- Wang, C.; Luan, S. Calcium homeostasis and signaling in plant immunity. Curr. Opin. Plant Biol. 2024, 77, 102485. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).