Abstract
Soil salinity stress inhibits the growth of most beneficial soil fungi, thereby adversely affecting crop growth, though the underlying mechanisms remain poorly understood. Our study revealed that the beneficial fungus Trichoderma guizhouense NJAU4742 exhibited limited salt tolerance, with its growth being significantly suppressed under elevated salinity. To investigate the physiological, biochemical, and molecular responses of NJAU4742 to salt stress and its subsequent effects on tomato growth, we subjected NJAU4742 to X-ray irradiation, aiming to obtain mutants with altered salt tolerance. A forward mutant strain (designated M15) displaying near-complete loss of salt tolerance was successfully isolated. Comparative transcriptomic analysis between the wild type (wt) and M15 identified gene Tgmfs, a salt stress-responsive gene belonging to the major facilitator superfamily. By constructing Tgmfs knockout (Tgmfs-KO) and overexpression (Tgmfs-OE) strains, we observed that Tgmfs deletion caused intracellular Na+ accumulation in NJAU4742, prompting compensatory upregulation of Na+/K+-ATPase activity to maintain ion homeostasis. Concurrently, salt stress induced reactive oxygen species accumulation and oxidative stress in fungal cells, which was counteracted by enhanced superoxide dismutase activity and an elevated NAD+/NADH ratio, collectively boosting antioxidant defenses. Pot experiments demonstrated that the application of Tgmfs-OE or wt spore suspensions markedly improved tomato salt tolerance, with Tgmfs-OE treatment showing superior efficacy. This study advances our understanding of filamentous fungal salt adaptation mechanisms and their synergistic effects on plant resilience.
1. Introduction
Saline–alkali land refers to the land where the concentrations of soluble salts (such as Na+, Cl−, SO42−) and alkaline substances (such as Na2CO3) are excessively high, resulting in the deterioration of soil structure and the decline of soil fertility [1]. The area of saline–alkali land is extensive. At present, the total global area of saline–alkali land is approximately 1 billion hectares, and it is still confronted with severe challenges [2]. Saline–alkali land adversely affects the growth and yield of crops. According to statistics, approximately 20% of the world’s arable land is impacted by salinization, losing its agricultural production potential. High salt content induces salt stress in organisms [3,4,5]. Salt stress endangers plant growth through multiple pathways, such as osmotic imbalance, ion toxicity, metabolic disorder, and structural damage, and it causes the degradation of soil ecological functions [6,7,8]. Salt stress also causes similar harm to microorganisms by disrupting the physiological and biochemical processes of microorganisms, causing ion imbalance within and outside the cells, membrane damage, and decreased stability of proteins and nucleic acids [9,10]. To survive under these conditions, microorganisms have evolved a series of adaptive mechanisms to alleviate the impacts of salt stress. These mechanisms regulate gene expression, accumulate stress-resistant substances, and adjust the internal and external environment to maintain cell integrity [11]. The responses of microorganisms to salt stress are mainly driven by two mechanisms: osmotic stress and ionic stress. Osmotic stress stems from a high-salt environment, which elevates the extracellular osmotic pressure, leading to water loss within the cells and disrupting the osmotic balance [12,13]. On the other hand, ionic stress occurs when the ion concentration gradient between the intracellular and extracellular environments changes due to high salt levels, thereby disrupting ionic homeostasis [14]. The multi-faceted responses of microorganisms to salt stress encompass osmotic regulation, ionic homeostasis, accumulation of stress-resistant substances, and regulation of gene expression. Regarding osmotic regulation, microorganisms accumulate solutes such as proline, mannitol, and soluble sugars to maintain cellular osmotic balance. For instance, the HOG (High-Osmolarity Glycerol) pathway enhances the synthesis and transport of intracellular glycerol, maintaining the intracellular glycerol concentration to uphold osmotic balance within the cell, reduce water loss within the cell, and stabilize the cell membrane structure [15]. In terms of ionic homeostasis, they regulate the activity and expression of ion channels to control ion absorption and excretion. In Escherichia coli and Vibrio cholerae, Na+/H+ antiporters (NhaA, NhaB) regulate transmembrane transport to expel excessive intracellular Na+ while pumping H+ into the cell, alleviating ionic toxicity. Additionally, microorganisms accumulate stress-resistant substances such as antioxidants and heat shock proteins to counteract the oxidative stress triggered by salt stress [16,17,18,19]. Through random mutagenesis of the genome of Trichoderma guizhouense NJAU4742, we hope to discover new genes related to salt stress.
Trichoderma guizhouense NJAU4742 is a saprophytic filamentous fungus isolated from mature compost [20]. It can significantly promote plant root development by secreting auxin-like substances and ACC (1-Aminocyclopropane-1-Carboxylate) deaminase, and it can inhibit soil-borne pathogenic fungi such as Botrytis cinerea and Fusarium wilt [21,22]. The inhibition rate of Trichoderma guizhouense NJAU4742 on cucumber Botrytis cinerea and Fusarium spp. wilt can reach 100%. It can also enhance the resistance of crops to adverse conditions such as salinity and drought [23,24,25]. By analyzing the transcriptome data of NJAU4742, we discovered that a gene related to sodium ions (Na+) efflux was significantly upregulated under salt stress conditions, and this gene belongs to the MFS family. The Major Facilitator Superfamily (MFS) constitutes one of the largest membrane transporter families, being widely distributed among bacteria, fungi, plants, and animals [26]. The MFS family encompasses over 82 subfamilies and can be further categorized into sugar transporters, drug-resistance proteins, and lipid and signal molecule transporters based on their functionalities. MFS transporters primarily maintain ionic homeostasis by transporting small molecules (such as sugars, amino acids, ions, and drugs) across the membrane [27]. They play a crucial role in cellular metabolism, signal transduction, and environmental adaptation. There are two main transport modalities. One is symport, which utilizes the proton gradient as an energy source to co-transport substrates along with H+. The other is through conformational alterations. During the transport process, the protein switches between an extracellularly open state (binding the substrate) and an intracellularly open state (releasing the substrate) via an “alternating access” mechanism to achieve substrate transmembrane translocation [28,29]. Under salt stress conditions, MFS proteins transport Na+ or Cl− to the extracellular space or sequester them in vacuoles to mitigate ionic toxicity. For instance, MFS members in plants (such as SOS1 homologs) are implicated in Na+ efflux to maintain intracellular K+/Na+ balance [30]. Some MFS proteins are accountable for transporting osmoprotective substances such as proline and betaine to alleviate osmotic stress [31]. MFS also mediates the transmembrane transport of antioxidants such as glutathione (GSH) and ascorbic acid, enhancing the cell’s ability to scavenge reactive oxygen species (ROS) [32]. For example, the MFS member Qdr2 in yeast is involved in the intracellular accumulation of GSH to inhibit oxidative damage [33]. In microorganisms, MFS is more frequently investigated for its role in drug and toxin efflux. For instance, bacterial MFS proteins (such as LacY, EmrD, and QacA) act as efflux pumps to expel antibiotics (such as tetracycline) and environmental toxins, reducing intracellular drug concentrations and enhancing microbial drug resistance [34]. There are relatively scarce reports on maintaining intracellular ion balance in microorganisms under salt stress conditions.
Tomato is an annual herb of the Solanum genus in the Solanaceae family, with plants capable of reaching up to two meters in height. It originated in South America and is extensively cultivated in the United States, Russia, Italy, and China, being one of the most prevalently grown fruits and vegetables worldwide. It demonstrates strong adaptability and has a preference for warm and illuminated conditions. Tomatoes are rich in carotene, vitamin C, and B vitamins and possess high nutritional value, thereby being widely favored by people [35]. Tomato is a moderately salt-tolerant plant, which can survive in a certain degree of salt environment [36]. However, high salinity has adverse effects on its growth, development, and yield. Excessive soil salinity inhibits water uptake in tomato roots, primarily through osmotic stress and ion toxicity, and problems such as slow growth, yellowing leaves, and even death may occur. Salt stress can delay and inhibit the germination of tomato seeds and has a stronger inhibitory effect on the establishment of seedlings after germination. Inoculation with Piriformospora indica can increase tomato yield by 65% under salt stress conditions and reduce leaf Na+ content by 40% [37]. Inoculation with salt-tolerant bacteria or the application of Arbuscular Mycorrhiza (AM) fungi can alleviate salt stress, increasing the plant height and fresh weight of tomato seedlings by 27.11% and 58.65%, respectively [38,39]. In this experiment, Trichoderma was directly applied to a simulated coastal saline soil system to investigate a coastal saline soil planting system to investigate the influence of NJAU4742 on tomato growth under salt stress. This study employed forward mutagenesis of Trichoderma strains, coupled with transcriptomic and molecular biological analyses, to preliminarily elucidate the biological mechanisms underlying Trichoderma’s response to salt stress. Additionally, further research confirmed the link between Trichoderma’s salt tolerance and its plant growth-promoting effects on tomatoes.
2. Materials and Methods
2.1. Growth Conditions of Trichoderma guizhouense NJAU4742
Trichoderma guizhouense NJAU4742 was preserved by the Key Lab of Organic-Based Fertilizers of China and the Jiangsu Provincial Key Lab for Solid Organic Waste Utilization; its NCBI GenBank accession number is LVVK00000000.1. The spore suspension of Trichoderma guizhouense NJAU4742, preserved in glycerol at −80 °C, was thawed and inoculated onto Potato Dextrose Agar (PDA) plates; the contents of this medium are potato extract, glucose, and agar powder. These plates were incubated at 28 °C until the mycelium completely covered the surface (Figure S1). To facilitate the full development of green spores, the plates were further incubated under light conditions at 28 °C for 72 h. Sterile deionized water (3 mL) was then added to each plate, and the spores were carefully dislodged using a sterile spreader. The resulting mixture was filtered through four layers of sterile gauze (0.1–0.2 mm) to obtain Trichoderma spore suspension. The concentration of the spores was determined using a hemocytometer under a microscope. This spore suspension was subsequently stored at 4 °C for subsequent use.
2.2. Random Mutation of Trichoderma guizhouense NJAU4742
A suspension of fresh spores at a concentration of 1 × 107 spores·mL−1 was prepared and mixed with acetate buffer solution (pH 5.0) at a 1:1 ratio. This mixture was then aliquoted into sterilized 96-well plates, with 200 μL per well, under a laminar flow hood. The 96-well plates were sealed with sterilized breathable film (parafilm) and placed into an X-ray machine (RS 2000, RadSource, Brentwood, TN, USA). The spore solution was exposed to X-rays at a total dose of 250 Gy, which damaged the DNA and induced random mutations. Due to the high energy of the radiation, the exposure was conducted in three sessions (84 Gy, 84 Gy, and 82 Gy), with a 5 min interval between each session [40]. After irradiation, the spore solution was collected and mixed in the laminar flow hood. The irradiated mutant spore solution was diluted to a concentration of 105 spores·mL−1 and spread evenly (approximately 500 μL) on a PDA medium containing 1.5 M NaCl. A growth control of wt spores at 103 spores·mL−1 was also included. After 7 days, mutants showing significant differences in growth characteristics relative to wt were selected, and their mycelium was transferred to PDA plates for further growth. Following approximately 48–72 h of light exposure to induce sporulation, the spore solution was prepared with sterile water and filtered through four layers of gauze. Finally, spore separation was carried out on PDA medium with 1.5 M NaCl to isolate mutants for further experimental analysis.
2.3. Transcriptome Sequencing of the Wild Type and Mutant Strains Under Salt Stress
Total RNA was extracted using Trizol reagent (Thermofisher, Waltham, MA, USA, 15596018) according to the manufacturer’s procedure. Their quantity and purity were evaluated by the Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA, 5067-1511), for which high-quality RNA samples with RNA integrity number (RIN) > 7.0 were used to construct the sequencing library. The mRNA was purified from total RNA (5 μg) using Dynabeads Oligo (dT) (Thermofisher, Waltham, MA, USA) with two rounds of purification and was fragmented into short fragments using divalent cations under elevated temperature (Magnesium RNA Fragmentation Module (NEB, cat. e6150, Ipswich, MA, USA) at 94 °C for 5–7 min). The cleaved RNA fragments were reverse-transcribed to create the cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, cat. 1896649, Waltham, MA, USA), for the synthesis of U-labeled second-stranded DNAs with Escherichia coli DNA polymerase I (NEB, cat. m0209, Ipswich, MA, USA), RNase H (NEB, cat. m0297, Ipswich, MA, USA), and dUTP Solution (Thermofisher, cat. R0133, Waltham, MA, USA). The addition of an A-base to the blunt ends of each strand was prepared for ligation to the indexed adapters containing a T-base overhang. Dual-index adapters were ligated to the fragments, and their sizes were selected using AMPureXP beads. After treatment of the U-labeled second-stranded DNAs using the heat-labile UDG enzyme (NEB, cat. m0280, Ipswich, MA, USA), the ligated products were amplified with polymerase chain reaction under the following conditions: initial denaturation at 95 °C for 3 min; 8 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s; final extension at 72 °C for 5 min. The average insert size for the final cDNA libraries was 300 ± 50 bp. Finally, the 2 × 150 bp paired-end sequencing (PE150) was performed on an Illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) following the vendor’s recommended protocol. Hisat software (V.2 2.0.4) was used to align the sequencing data with the reference genome (https://bioinfo.njau.edu.cn/tgn4742/, accessed on 20 November 2021). The alignment results were assembled into transcripts to predict gene expression using StringTie. The differential expression of various functional genes was analyzed using edgeR and visualized using R language (V.4.2.0). Based on differentially expressed genes, GO (gene ontology) enrichment analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis were carried out for functional analysis and presented graphically.
2.4. Construct Functional Fragments for Gene Knockout and Overexpression
Gene knockout was performed using the high-fidelity PrimeSTAR® HS DNA Polymerase (TaKaRa; cat. R010A, Dalian, China) to amplify the upstream (1200 bp) and downstream (1200 bp) genomic fragments flanking the target gene. Hygromycin B resistance was employed as a selectable marker in this study. The functional fragments were then fused using an overlapping PCR strategy, resulting in a construct with the sequence arrangement of upstream-hygromycin B-downstream. Additionally, overexpression was accomplished by employing hygromycin B as the resistance gene and using a strong promoter the overexpression mutant strain [41]. The primer sequences are presented in Table S1.
2.5. Protoplast Transformation of Functional Fragments in Trichoderma guizhouense NJAU4742
Sterilized cellophane paper, autoclaved at 115 °C for 30 min, was carefully placed on the PDA medium within a laminar flow hood. Fresh NJAU4742 spores (1 × 108 spores·mL−1) were evenly distributed on the cellophane paper and incubated at 28 °C for 16 h. Concurrently, protoplasts were prepared using a lytic solution containing Solution A (6 M sorbitol, 0.5 M KH2PO4, pH 5.6) supplemented with Trichoderma lytic enzyme at a concentration of 0.75% (w/v). The cellophane with the 16-h-old Trichoderma mycelium was then immersed in the lytic solution and shaken at 28 °C at 100 rpm for 2 h. Following shaking, the Trichoderma mycelium was left in the solution and broken into fine fragments using a 5 mL pipette tip. The lytic solution containing mycelium fragments was filtered through sterile four-layer gauze into a sterile 50 mL centrifuge tube placed on ice. After centrifugation at 4 °C at 2000 rpm for 10 min, approximately 3–5 mL of the supernatant was collected and transferred to a sterile 15 mL centrifuge tube. The sample was centrifuged again under the same conditions for 10 min, the supernatant was discarded, and Solution B (10 M sorbitol, 0.5 M CaCl2·2H2O, 1 M Tris-HCl, pH 7.5) was added to prepare the protoplast solution. The protoplasts, target gene fragments, and PEG (Polyethylene Glycol) solution (250 g of 25% PEG 6000, 0.05 M CaCl2·2H2O, 1 M Tris-HCl, pH 7.5) were mixed in the following proportions: 77% protoplast solution, 19% PEG solution, and 4% fragments (v/v). After thorough mixing, the mixture was incubated on ice for 20 min. Then, 4 mL of PEG solution was added, mixed well, and incubated at room temperature for 5 min. Finally, 6 mL of Solution B was added and mixed thoroughly, and the mixture was evenly spread onto sucrose PDA containing 1 M sucrose (500 μL of the mixed solution per 90 mm diameter plate). After 16–24 h, when no obvious liquid droplets remained on the plates and numerous small, microcolonies appeared, the plates were overlaid with PDA medium containing hygromycin B (0.2 mg·mL−1). After 3–4 days, transformants showing resistance and growth on the medium surface were transferred to fresh hygromycin B PDA medium. Transformants with growth phenotypes similar to wt were selected for further studies [41].
2.6. Evaluate Intracellular Substrates Related to Salt Stress and Redox Status
The fresh spore suspension of the NJAU4742 knockout strain (Tgmfs-KO), and the overexpression strain (Tgmfs-OE) were cultivated on the PDA medium at 28 °C. PDA media with three different NaCl concentrations (0 M, 0.5 M, and 1.0 M) were prepared for inoculation after the sterilization at 115 °C for 30 min. About 2.5 μL of spore suspension (107 spores·mL−1) was inoculated on the PDA medium, which was covered with cool and sterilized cellophane paper to collect mycelia. After 7 days of growth, the mycelia were scraped, and their intracellular substance content was analyzed using the glycerol extraction kit (Applygen E1013, Beijing, China), SOD (superoxide dismutase) enzyme activity assay kit (Solarbio BC0170, Beijing, China), NAD+ and NADH content assay kit (Solarbio BC0310, Beijing, China), Na+/K+-ATPase activity assay kit (Solarbio BC0065, Beijing, China), and Sodium (Na) Colorimetric Assay Kit (Elabscience E-BC-K207-M, Wuhan, China). At least three biological replicates were carried out for each experiment, and the bars indicated the standard error of the mean (SEM).
2.7. Pot Experiments
The pot experiment was carried out in the Intelligent Climate Chamber of Nanjing Agricultural University, where the temperature was controlled within the range of 20–25 °C and the lighting duration was 12 h. A total of three major treatments were set up in the experiment, and 15 treatments are presented in Table S2, as follows: normal soil (EC = 27.93 mS/m) was paddy soil, salt soil (EC = 455.75 mS/m) was coastal saline soil, and half-salt soil (m/m, EC = 264.25 mS/m) was 50% normal soil and 50% salt soil. Each pot was filled with 1 kg of soil with a pore size ranging from 0.5 to 1 mm, and the diameter of the pot was 12 cm. One dwarf tomato seedling was planted in each pot, and there were 6 replicates for each treatment. Due to the excessively high salt concentration in the salt soil, the seedlings were prone to mortality. A total of 5 batches of seedlings were replaced from the start to the harvest of seedlings. All subsequent data were the data of the last batch of seedlings. On the day of transplanting the seedlings, 50 mL of 1/2 MS (Murashige and Skoog) nutrient solution was applied per pot, and the nutrient solution was replenished every 7 days thereafter. On the third day after transplanting the seedlings, 10 mL of Trichoderma spore solution (1% of the dry weight of the soil, v/m) was added to each pot, and 50 mL of water was applied per pot every 2–3 days.
Measurement of plant aboveground parts: when significant differences in biological traits occurred among treatments, plant height (measured with a ruler, cm), stem diameter (measured with a vernier caliper, cm), aboveground dry/fresh weight (measured with an electronic balance, g), and plant survival rate for different treatments were determined.
2.8. Data Process and Statistical Analysis
The data were expressed as the means of three replicates and analyzed using one-way ANOVA with the software SPSS16.0, and the LSD method was used to study the separation of soil properties of the difference between the significance of treatment (p < 0.05). Correlation analysis was performed using R software (V.3.3.3), and RDA was analyzed by Canoco (V.5.0).
3. Results
3.1. Screening of Irradiated Mutant Strains of Trichoderma guizhouense NJAU4742
On PDA plates without NaCl (0 M), all strains exhibited normal growth. However, with increasing NaCl concentrations, salt stress adversely affected the growth of all strains. While the growth of wt was inhibited under salt stress, it was still capable of some mycelial expansion. In stark contrast, the irradiated mutant strains were markedly more sensitive to salt stress than wt, showing severely impeded growth. Their mycelia failed to spread outward and instead contracted into compacted clumps. Despite this, a small portion of the mycelia managed to grow upward, resulting in the formation of denser colonies (Figure 1). By measuring the colony diameter differences of these colonies, it was discovered that the M15 mutant strain exhibited growth consistent with wt under normal circumstances. However, under salt stress conditions, its growth was the poorest among all strains (Figure S2). Despite their impaired salt tolerance, these mutants became valuable subjects for further investigation.
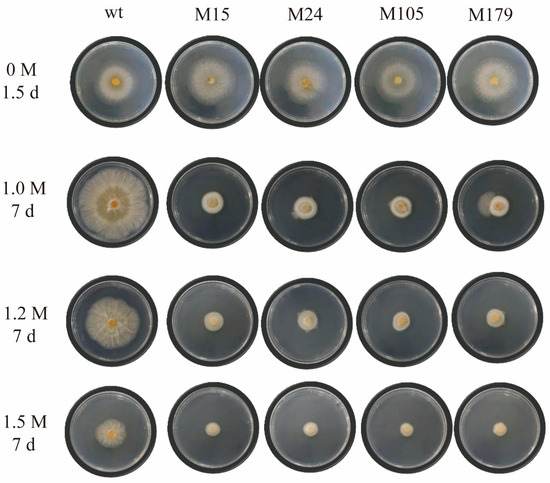
Figure 1.
The sensitivity of Trichoderma wild type and its mutant strains to salt stress. A total of four salt concentrations were established, with the strains cultured for 1.5 days under the 0 M condition and the other concentrations cultured for 7 days. Both the wt and four mutants were inoculated and tested.
3.2. Transcriptome Sequencing Result of the Wild-Type and Mutant Strains Under Salt Stress
Mutants with impaired salt tolerance exhibited marked growth differences compared to wt under salt stress conditions. At a 1.0 M NaCl concentration, these mutants’ growth was severely inhibited; their mycelia failed to spread and retracted into compacted clumps, indicating a disruption in the mutants’ primary intracellular salt tolerance mechanisms. To further investigate this phenotype, the mycelia of both wt and salt-tolerance-deficient mutants, cultured for 7 days under 1.0 M NaCl conditions, were collected for transcriptome sequencing with three biological replicates each. After sequencing, raw data were preprocessed, followed by a comprehensive statistical analysis, which included the raw read count, valid read count, the percentage of bases with a quality score of at least 20 (Q20%), the percentage of bases with a quality score of at least 30 (Q30%), and the GC content. The detailed statistical outcomes are presented in Table S3.
The statistical analysis revealed that the efficiency of our sample data exceeded 90.00%, and the quality, as measured by Q20%, was above 99.50%, indicating that the dataset met our quality standards. We then assessed the coverage of expressed genes, which represented the proportion of expressed genes out of the total gene pool. Preliminary analysis showed that, under 1.0 M NaCl conditions, the mutant strain M15 had 1448 upregulated genes and 1635 downregulated genes compared to wt under the same conditions (Figure 2A). Differentially expressed genes (DEGs) are the genes that are either upregulated or downregulated between samples or within the same sample after different treatments. Gene selection was based on fold change (FC) and significance level, with criteria set at FC ≥ 2 or FC ≤ 0.5 (absolute log2 FC ≥ 1) and q-value < 0.05 [42]. This threshold helped identify genes with significant statistical differences across multiple groups, leading to the identification of many salt-stress-specific DEGs. Under 1.0 M NaCl conditions, there were numerous highly reliable DEGs between the wt and the mutant strain across all transcripts. Specifically, 21 genes were significantly upregulated and 77 genes were significantly downregulated under salt stress in the mutant strain. The expression of gene A1A107451 in wt is upregulated by more than 1000 times compared to M15, so this gene will be studied further in the future (Figure 2B). Based on these screening results, we employed GO enrichment analysis and KEGG enrichment analysis to identify gene annotations and biosynthetic pathways highly associated with salt stress for further investigation.
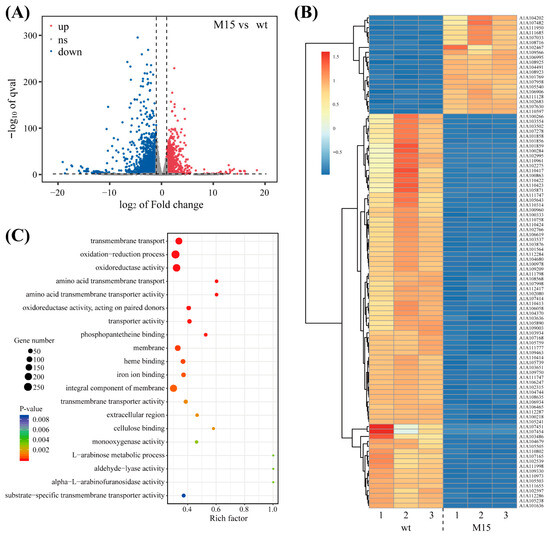
Figure 2.
Transcriptome analysis of wild type and mutant (M15) of Trichoderma guizhouense NJAU4742 under salt stress conditions. (A) Fold change and number summary of genes under 1.0 M NaCl treatment. (B) Cluster heatmap of differentially expressed genes. (C) At 1.0 M NaCl, the bubble map of the top 20 GO annotations was made, and it contained the number of differentially expressed genes and the annotation’s credibility and Rich factor.
Approximately 100 differentially expressed genes were identified, presenting a challenge in pinpointing the salt stress-specific response genes in NJAU4742. Gene enrichment analysis, which categorizes genes on the basis of genomic or database annotations, facilitated the identification of commonalities among these genes, such as shared functions or pathways. This approach helped uncover key biological processes involved in the molecular mechanisms of the salt stress response. The GO enrichment analysis, which encompasses molecular function (MF), cellular component (CC), and biological process (BP), was applied to the DEGs. Each GO term corresponds to a specific gene property, and by enriching the DEGs at the MF, CC, and BP levels, we gained insights into the subcellular localization and functional classification of genes involved in the salt stress response. The majority of DEGs were associated with biological processes such as redox reactions and transmembrane transport, with a significant localization to the cell membrane and nucleus. Their functional annotations were primarily related to redox reactions and catalytic activities (Figure 2C). This suggests that NJAU4742 modulates its response to salt stress by regulating membrane transport proteins and proteins involved in oxidative stress management.
Although the functions of the majority of DEGs were ascertained, further enrichment analysis was essential to pinpoint the key genes and assess the activation strength of specific pathways. The enrichment analysis encompassed not only the types of differential pathways but also the number of genes involved and their relative proportions, as indicated by the enrichment factor (Rich factor) (Figure S3). The analysis revealed that four pathways, involving over 150 DEGs, were highly significant, with the transmembrane transport pathway showing the highest enrichment factor, followed by redox and catalytic pathways. This underscored the critical role of transmembrane transport in NJAU4742’s response to stress. Genes associated with transmembrane transport constituted more than 30% of the total pathways (Rich factor > 0.3), indicating that this pathway is likely specific to salt stress responses.
3.3. Na+ Efflux Pumps in NJAU4742 Alleviate Salt Stress
A comparative analysis of DEGs between the irradiated mutant strain M15 and wt revealed a pronounced downregulation of a Na+ efflux pump gene, classified within the MFS. To further investigate whether Tgmfs was involved in NJAU4742’s salt stress response, we created Tgmfs knockout (Tgmfs-KO) and overexpression (Tgmfs-OE) strains. Comparative growth assays on 1.0 M NaCl-PDA media revealed that Tgmfs-KO strains exhibited significantly impaired salt tolerance after 7 days (Figure 3A). These results suggest that Tgmfs may mediate Na+ efflux, thereby maintaining ionic balance and allowing the strain to withstand high salt stress.
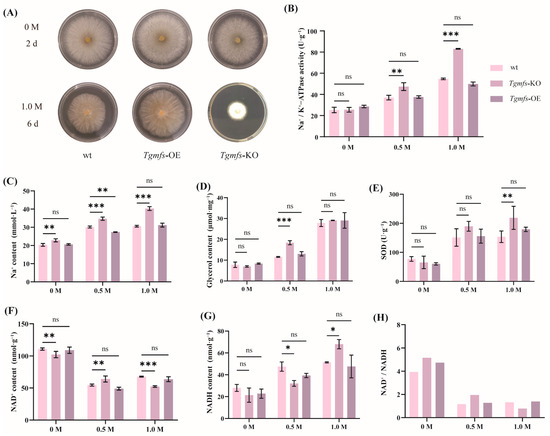
Figure 3.
The impact of Na+ efflux pump on the growth of Trichoderma guizhouense NJAU4742 under salt stress. (A) The biomass disparity between the wt and mutant strains under the condition of 1.0M NaCl. (B–H) Under three NaCl concentration conditions (0 M, 0.5 M, 1.0 M), the intracellular substances associated with salt stress in wt and mutant cells were measured, encompassing Na+/K+-ATPase activity (B), Na+ content (C), glycerol content (D), SOD activity (E), NAD+ content (F), NADH content (G), and NAD+/NADH ratio (H). ns: p > 0.05, * p < 0.05, ** p < 0.01, and *** p < 0.01.
3.3.1. Na+/K+-ATPase Regulates the Intracellular Ion Balance of NJAU4742
During salt stress, the disruption of the ionic balance between intracellular and extracellular compartments leads to excessive Na+ accumulation, which affects normal metabolism and physiological functions. Na+/K+-ATPase helps the cell cope with high salt environments by regulating the flow of Na+ and potassium ions to maintain the Na+/K+ balance. Both the wt and the Tgmfs-OE strains exhibited increased Na+/K+-ATPase activity with rising NaCl concentrations. However, this increase was significantly attenuated in wt and Tgmfs-OE strains compared to Tgmfs-KO. Specifically, in comparison to the 0 M NaCl condition, the Na+/K+-ATPase activity in the Tgmfs-KO strain increased by 225.50% under 1.0 M NaCl conditions (Figure 3B). The Tgmfs gene was a major Na+ extrusion pump in NJAU4742, helping the cell expel excess Na+ accumulation under salt stress. The absence of this gene in the knockout strain resulted in substantial Na+ accumulation inside the cell, which relied on other Na+ pumps or ion channels to expel the excess Na+.
3.3.2. The Variations of Intracellular Sodium Ions Within NJAU4742 Cells
The concentration of intracellular Na+ is a crucial indicator in cell physiology, which is mainly regulated by the Na+ pump (Na+/K+-ATPase) of the cell membrane and the s Na+ channels of the cell membrane. The intracellular Na+ contents of the three strains differed when exposed to varying NaCl concentrations. At 0 M, the intracellular Na+ contents of wt, Tgmfs-OE, and Tgmfs-KO were 20.36, 22.84, and 20.59 mmol/L, respectively. Under the condition of 0.5 M, the intracellular Na+ contents of all three strains increased. Specifically, they increased by 48.11%, 51.97%, and 32.83%, respectively, reaching 30.15, 34.71, and 27.35 mmol/L. At a concentration of 1.0 M, the three strains were subjected to higher salt stress, with intracellular Na+ contents being 30.61, 40.29, and 31.20 mmol/L, respectively. The changes in intracellular Na+ contents of wt and Tgmfs-OE were not significant compared to 0 M, while the change in Tgmfs-KO was considerable, increasing by 76.40% compared to 0 M and 16.08% compared to 0.5 M (Figure 3C).
3.4. Measurement of Intracellular Substances in Mutant Strains
3.4.1. Under Salt Stress, NJAU4742 Accumulates a Large Amount of Glycerol Intracellularly
When Trichoderma is exposed to salt stress, cells respond almost instantaneously within seconds by rapidly synthesizing and accumulating intracellular glycerol to alleviate the osmotic pressure difference between the inside and outside of the cell. Under no-salt-stress conditions, wt, Tgmfs-KO, and Tgmfs-OE all produced small amounts of glycerol, with values of 7.78 μmol·mg−1, 6.95 μmol·mg−1, and 8.36 μmol·mg−1, respectively, but with no significant (p > 0.05) differences. Under salt stress conditions, as the salt concentration increased, the glycerol content within the cells of both wt and mutant strains rose. At 0.5 M NaCl, the glycerol content in Tgmfs-KO was significantly (p < 0.05) higher than that in wt and Tgmfs-OE, increasing by 164.00% compared to 0 M NaCl. The glycerol contents in the wt and Tgmfs-OE were increased by 49.00% and 56.00%, respectively. At higher salt concentrations (1.0 M), the glycerol content in all three strains was significantly increased, but with no significant differences between them, suggesting that the intracellular glycerol content might have reached its maximum value. Under conditions with equivalent levels of intracellular glycerol, the poorer growth of Tgmfs-KO indicated that while Tgmfs deletion did not affect osmotic regulation, other stress factors caused by salt stress cannot be mitigated, thereby affecting the strain’s growth (Figure 3D).
3.4.2. Coenzyme I and SOD Relieve Oxidative Stress in NJAU4742 Cells
With increasing NaCl concentration, the SOD activity in both wt and mutant strains increased. Compared to 0 M NaCl, at 1.0 M NaCl, the SOD activity increased by 98.95% in wt, 234.27% in Tgmfs-KO, and 196.72% in Tgmfs-OE. These data indicated that Tgmfs-KO experienced severe oxidative stress under salt stress conditions. Meanwhile, the elevated SOD activity helped to maintain a higher level of antioxidant defense, aiding the cells in combating oxidative stress (Figure 3E).
In this study, at 0 M NaCl, the NAD+ levels in wt and Tgmfs-OE were 110.80 nmol·g−1 and 109 nmol·g−1, respectively. As the NaCl concentration increased to 0.5 M, NAD+ levels decreased to 54.66 nmol·g−1 and 49.04 nmol·g−1, respectively. However, at 1.0 M NaCl, the NAD+ content increased to 67.84 nmol·g−1 and 63.74 nmol·g−1, respectively. For Tgmfs-KO, the NAD+ levels were 102.20 nmol·g−1, 64.04 nmol·g−1, and 52.29 nmol·g−1 at 0 M, 0.5 M, and 1.0 M NaCl, respectively (Figure 3F). As salt concentration increased, salt stress significantly reduced intracellular NAD+, but at 1.0 M NaCl, NAD+ levels increased in wt and Tgmfs-OE, while they continued to decrease in Tgmfs-KO. NADH levels under different salt concentrations were also determined. The results indicated that salt stress significantly affected NADH levels, with changes opposite to those of NAD+. At different salt concentrations, intracellular NADH levels in the wt and mutant strains showed an increasing trend. Notably, the NADH content in Tgmfs-KO was 21.38 nmol·g−1 at 0 M NaCl and increased to 68 nmol·g−1 at 1.0 M NaCl with a rate of 218.00% (Figure 3G). The NAD+/NADH ratio reflects cellular energy metabolism and redox status. As NaCl concentration increased, this ratio decreased in both wt and mutant strains. Under salt stress conditions, the NAD+/NADH ratio was maintained at a lower level, with significant decreases in NAD+ levels (Figure 3H).
3.5. Effects of Salt-Tolerant Mutant Strain of Trichoderma on Tomato Biomass and Quality
The exogenous addition of different spore suspensions of Trichoderma strains can reflect their influences on the growth of tomato seedlings under salt-stress conditions. In this study, the results indicated that all tomato seedlings in all treatment conditions grew normally under normal soil conditions. However, as the salt concentration increased, it can be clearly seen in the figures that in the pots containing half salt soil, tomato seedlings began to show yellowing, wilting, and desiccation of leaves. In the untreated condition, the yellowing of tomato seedlings was the most obvious. Under pure salt soil conditions, the above situations were more obvious for tomato seedlings. Some seedlings showed severe wilting and needed external support to grow upward. In the untreated condition, the damage degree of tomato seedlings was the most severe (Figure 4 and Figure S4). The addition of Trichoderma alleviated the salt stress suffered by tomato seedlings in all treatment conditions. All tomato seedlings were able to survive and grow normally under normal soil conditions. With the increase in salt concentration, all the treated tomato seedlings began to die. Under the condition of half salt soil and salt soil, the highest survival rate was achieved in the Tgmfs-OE treatment, reaching 90% and 67.5%. Under these two conditions, the mortality rate of tomato seedlings was the highest in the untreated condition. Especially under pure salt soil conditions, the survival rate was merely 22.5% under the condition without Trichoderma treatment (Figure 5).

Figure 4.
Growth results of tomato seedlings under different treatment conditions. The growth discrepancies of tomato seedlings under various treatment conditions in three types of soil with distinct salinities.
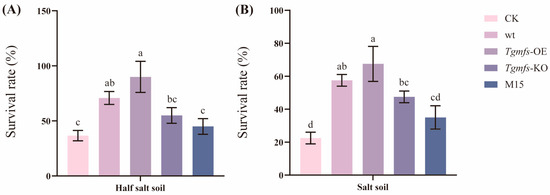
Figure 5.
Survival rate of tomato seedling. The variance of the survival rate of tomato seedlings in each treatment within the two types of soil with distinct salt concentrations was observed. (A) The survival rate of tomato seedlings in half salt soil. (B) The survival rate of tomato seedlings in salt soil. Different lowercase letters indicate statistically significant differences between treatments (one-way ANOVA, p < 0.05).
Different soil salt concentrations and the addition of different Trichoderma strains exert distinct influences on the aboveground physiological conditions of tomato plants; particularly, the effects on fresh weight and dry weight are conspicuously obvious. Since the tomato plants utilized in the experiment were dwarf tomato seedlings and tomatoes are moderately salt-tolerant plants, there were no significant disparities in plant height and stem diameter among the various treatments. All the indicators of the treatments with added Trichoderma strains are higher than those of the untreated control; especially, the indicators of the treatments with wt and Tgmfs-OE were significantly higher than those of the untreated control. Under normal soil conditions, the height, stem diameter, fresh weight, and dry weight of the treatment with Tgmfs-OE were the highest, at 20.47 cm, 4.73 mm, 5.78 g, and 0.55 g, respectively. Under the half-saline soil condition, the growth of tomato seedlings in all treatments was inhibited to a certain extent. The various indices of the Tgmfs-OE treatment were the highest, with values as follows: 16.85 cm, 4.44 mm, 5.06 g, and 0.465 g. Under pure saline conditions, the growth of tomato seedlings in all treatments is severely inhibited, and the overall growth was poorer than that in the first two salt concentration conditions. Among the four indicators of plant height, stem diameter, fresh weight, and dry weight, the treatment with Tgmfs-OE had the highest values for all indicators, at 16.17 cm, 4.33 mm, 3.82 g, and 0.41 g, respectively. In the treatments with Tgmfs-KO and M15, as these two mutant strains are significantly inhibited in their growth under salt stress conditions, the physiological indicators of tomato seedlings in these two treatments basically did not exhibit significant differences from those of the untreated control under salt stress conditions (Figure 6 and Figure S5).
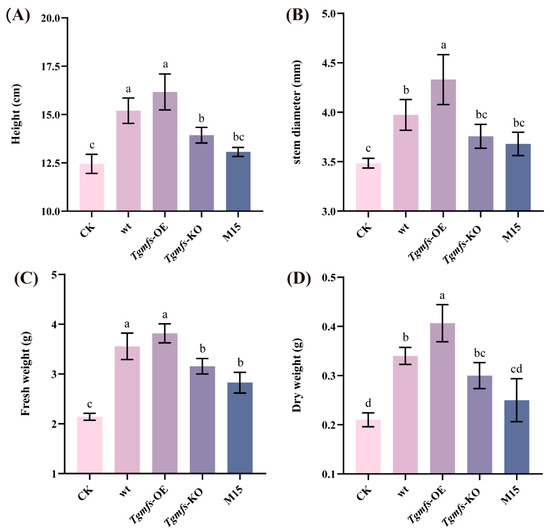
Figure 6.
The variation in biomass of tomato seedlings under salt soil conditions. (A–D) In the salt soil concentrations, the physiological indices of tomato seedlings in each treatment were measured, namely plant height (A), stem diameter (B), fresh weight (C), and dry weight (D). Different lowercase letters indicate statistically significant differences between treatments (one-way ANOVA, p < 0.05).
4. Discussion
Saline–alkali land refers to soil with excessively high levels of salt and alkaline substances. It is typically caused by various factors, such as improper irrigation, water pollution, poor land management, and climate change. As of 2021, saline–alkali land remains a prevalent global issue and poses a serious challenge. This type of soil adversely affects crop growth and yield. It is estimated that approximately 20% of the world’s arable land is affected by soil salinization, which leads to reduced agricultural productivity, threatening food security and rural economic development [43]. Additionally, it disrupts soil microbial activity and metabolism. Salt stress has been reported to exert multiple toxic effects on microorganisms, primarily through osmotic stress and ion toxicity [44].
Under salt stress conditions, Na+ is a key factor causing cellular toxicity. In high-salt environments, the extracellular Na+ concentration surrounding microbial cells increases sharply. This creates a significant osmotic pressure gradient between the extracellular and intracellular compartments, subjecting the cells to osmotic stress [45]. Consequently, water molecules rapidly exit the cells through membrane water channel proteins (e.g., AqpZ), leading to cell dehydration and plasmolysis [46]. To counteract this, microbial cells synthesize and accumulate compatible solutes (e.g., trehalose and glycerol in Saccharomyces cerevisiae) to maintain intracellular osmotic equilibrium. However, this process diverts energy from other physiological functions, slowing cell growth and reducing reproductive capacity [47,48]. Our study revealed that Trichoderma guizhouense NJAU4742 undergoes a similar response. Under salt stress, intracellular glycerol levels increased in the wt, Tgmfs-KO, and Tgmfs-OE strains, with all strains exhibiting slower growth compared to normal conditions, particularly Tgmfs-KO.
In high-salt environments, Na+ enters cells through non-selective cation channels along the concentration gradient, causing intracellular Na+ overload and toxicity [49]. Microorganisms maintain ion homeostasis by dynamically regulating Na+ influx and efflux. For example, Escherichia coli expels Na+ via Na+/H+ antiporters (e.g., NhaA and NhaB), while Bacillus subtilis enhances K+ uptake through the K+ transport system (KtrAB) to competitively inhibit Na+ influx [50,51]. In Trichoderma guizhouense NJAU4742, the TgMFS protein primarily mediates Na+ efflux to maintain intracellular Na+ homeostasis. Under salt stress, intracellular Na+ levels increased in wt, Tgmfs-KO, and Tgmfs-OE. The absence of TgMFS in Tgmfs-KO impaired Na+ efflux, resulting in higher intracellular Na+ levels compared to wt and Tgmfs-OE. However, residual Na+/K+-ATPase activity in Tgmfs-KO partially compensated for this defect by extruding Na+.
Excessive intracellular Na+ disrupts the ionic balance and metabolic processes, further impairing the electron transport chain in cellular respiration. This leads to electron leakage and reactive oxygen species (ROS) generation, inducing oxidative stress. For instance, in Escherichia coli, Na+ accumulation under salt stress inhibits cytochrome oxidase activity, causing electron leakage and superoxide anion (O2−) formation [52,53]. Nicotinamide adenine dinucleotide (NAD+/NADH) plays a critical role in energy metabolism and redox balance. Under salt stress, the NAD+/NADH ratio decreases due to inhibited NAD+ regeneration and increased ATP consumption for ion homeostasis [54]. Our findings aligned with this: in NJAU4742 strains (wt, Tgmfs-KO, and Tgmfs-OE), salt stress reduced NAD+ levels, increased NADH, and lowered the NAD+/NADH ratio.
Microorganisms combat oxidative stress through antioxidant systems [55,56,57]. Key enzymes include SOD, catalase (CAT), and peroxidase (POD). SOD catalyzes the conversion of superoxide anions to hydrogen peroxide and oxygen. In Bacillus subtilis, SOD effectively mitigates oxidative damage [58]. In our study, SOD levels significantly increased in all NJAU4742 strains under salt stress, with Tgmfs-KO requiring higher SOD activity to counteract severe Na+-induced oxidative stress. Studies demonstrate that Trichoderma enhances plant growth and salt tolerance. For example, Trichoderma volatiles improve Arabidopsis thaliana salt tolerance, while promoting wheat photosynthesis and water use efficiency under stress [59,60]. In this study, we evaluated the salt-tolerance and growth-promoting effects of NJAU4742 and its mutants on potted tomatoes in simulated coastal saline soil. Tomato seedlings were grown in soils of three salinity levels, with Trichoderma conidia suspensions applied via root irrigation. The results showed that low-salinity soil permitted normal growth, whereas high salinity caused high mortality. Trichoderma inoculation alleviated salt damage, enabling plant survival in saline soil [61].
By recruiting beneficial microorganisms under biotic and abiotic stresses, plants can enhance their stress resistance and promote growth. Abiotic stress induces shifts in root-associated microbial communities and facilitates the recruitment of stress-alleviating microorganisms. In response to abiotic stresses, plant roots secrete diverse metabolites that drive the regulation of the rhizosphere microbiome to mitigate these stresses. Studies have shown that salt-stressed plants secrete higher levels of metabolites compared to unstressed plants, including phenolic compounds, amino acids, organic acids, and sugars [62]. These metabolites facilitate the enrichment of beneficial microorganisms in the rhizosphere, aiding plants in resisting salt stress. For instance, wild soybeans under salt stress secrete purines, particularly xanthine, which induce motility in Pseudomonas (primarily through chemotaxis and flagellar assembly), leading to the enrichment of Pseudomonas and altering the root microbial community [63]. Plants host a diverse microbial community on and within their roots, leaves, and other organs, collectively known as the plant microbiome. Accumulating evidence indicates that the plant microbiota plays a critical role in enhancing plant adaptation and resistance to saline soils. Microorganisms associated with plant roots, such as plant growth-promoting rhizobacteria (PGPR) and endophytic bacteria, are essential for improving plant tolerance to high salinity. Microorganisms enhance salt tolerance via multiple pathways, including promoting nutrient acquisition, maintaining ion homeostasis, increasing osmolyte concentrations, scavenging excess ROS, and regulating plant hormones. PGPR can produce ACC deaminase, reducing ethylene accumulation in plants under salt stress. Plants with lower ethylene levels can better cope with salt-induced growth inhibition by associating with ACC deaminase-producing microorganisms [64,65]. Arbuscular mycorrhizal fungi (AMF) significantly enhance K+ uptake while reducing Na+ absorption, thereby lowering the Na+/K+ ratio and improving plant salt tolerance. Additionally, AMF-mediated K+ uptake increases with rising salinity levels [66].
5. Conclusions
This study demonstrates that the Tgmfs gene in NJAU4742 enables the organism to better respond to salt stress by eliminating the excess intracellular Na+ and alleviating Na+ toxicity. The overexpression strain Tgmfs-OE of NJAU4742 can more effectively aid tomatoes in their survival under salt stress conditions. These results provide crucial evidence that enhances our understanding of the relationship between the salt tolerance mechanisms of filamentous fungi and plant growth.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture15060610/s1, Figure S1: The growth condition of Trichoderma guizhouense NJAU4742 on PDA medium; Figure S2: The sensitivity of Trichoderma wild type (wt) and its mutant strains to salt stress (colony diameter); Figure S3: Differentially expressed genes belong to three GO annotation categories: biological process, cellular component, and molecular function; Figure S4: Scanning results of tomato roots under different treatment conditions; Figure S5: The impact of different treatments on tomato biomass; Table S1: PCR primers used in this study; Table S2: Soil setting scheme for pot experiments; Table S3: Sequencing sequence statistics and quality control.
Author Contributions
H.M.: methodology, software, supervision, writing—original draft, writing—review, and editing. H.W.: methodology and formal analysis. Y.X.: investigation and software. Q.H.: investigation and validation. T.L.: writing—review, editing, supervision, and data curation. D.L.: editing, supervision, and data curation. Q.S.: editing, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (32172680 and 32302679), the Natural Science Foundation of Jiangsu Province (BK20230991), and the China Postdoctoral Foundation under Grant Number 2024M751440.
Data Availability Statement
Source data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, Physiology, and Molecular Breeding Approaches for Improving Salt Tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Bouzouina, M.; Kouadria, R.; Lotmani, B. Fungal endophytes alleviate salt stress in wheat in terms of growth, ion homeostasis and osmoregulation. J. Appl. Microbiol. 2021, 130, 913–925. [Google Scholar] [CrossRef]
- Gonzalez, J.A.B.; Carrillo-Gonzalez, R.; Gonzalez-Chavez, M.d.C.A.; Sanchez, E.C.; Maruri, D.T. Selection of Salinity-Adapted Endorhizal Fungal Consortia from Two Inoculum Sources and Six Halophyte Plants. J. Fungi 2023, 9, 893. [Google Scholar] [CrossRef]
- Perez-Llano, Y.; Caridad Rodriguez-Pupo, E.; Druzhinina, I.S.; Chenthamara, K.; Cai, F.; Gunde-Cimerman, N.; Zalar, P.; Gostinčar, C.; Kostanjšek, R.; Folch-Mallol, J.L.; et al. Stress Reshapes the Physiological Response of Halophile Fungi to Salinity. Cells 2020, 9, 525. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, A.; Schillaci, M.; Balestrini, R. Mitigating the impact of soil salinity: Recent developments and future strategies. Ital. J. Agron. 2023, 18, 2173. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitas, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Saldana, C.; Villava, C.; Ramirez-Villarreal, J.; Morales-Tlalpan, V.; Campos-Guillen, J.; Chavez-Servin, J.; García-Gasca, T. Rapid and reversible cell volume changes in response to osmotic stress in yeast. Braz. J. Microbiol. 2021, 52, 895–903. [Google Scholar] [CrossRef]
- Proft, M.; Struhl, K. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell 2004, 118, 351–361. [Google Scholar] [CrossRef]
- Shirvanyan, A.; Mirzoyan, S.; Trchounian, K. Relationship between proton/potassium fluxes and central carbon catabolic pathways in different Saccharomyces cerevisiae strains under osmotic stress conditions. Process Biochem. 2023, 133, 309–318. [Google Scholar] [CrossRef]
- Wang, C.; Xiang, Y.; Qian, D. Current progress in plant V-ATPase: From biochemical properties to physiological functions. J. Plant Physiol. 2021, 266, 153525. [Google Scholar] [CrossRef]
- Anschutz, U.; Becker, D.; Shabala, S. Going beyond nutrition: Regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J. Plant Physiol. 2014, 171, 670–687. [Google Scholar] [CrossRef]
- Wang, Q.; Guan, C.; Wang, S.-M. Coordination of AtHKT1;1 and AtSOS1 facilitates Na+ and K+ homeostasis in Arabidopsis thaliana under salt stress. J. Plant Biol. 2014, 57, 282–290. [Google Scholar] [CrossRef]
- Li, T.; Liu, J.; Wang, Q.; Liu, Y.; Li, T.; Liu, D.; Shen, Q. Tr-milRNA1 Contributes to Lignocellulase Secretion under Heat Stress by Regulating the Lectin-Type Cargo Receptor Gene Trvip36 in Trichoderma guizhouence NJAU 4742. J. Fungi 2021, 7, 997. [Google Scholar] [CrossRef]
- Li, Y.; Shao, J.; Fu, Y.; Chen, Y.; Wang, H.; Xu, Z.; Feng, H.; Xun, W.; Liu, Y.; Zhang, N.; et al. The volatile cedrene from Trichoderma guizhouense modulates Arabidopsis root development through auxin transport and signalling. Plant Cell Environ. 2022, 45, 969–984. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Singh, P.C.; Mishra, A.; Chauhan, P.S.; Dwivedi, S.; Bais, R.T.; Tripathi, R.D. Trichoderma: A potential bioremediator for environmental clean up. Clean Technol. Environ. Policy 2013, 15, 541–550. [Google Scholar] [CrossRef]
- Huang, N.; Wang, W.; Yao, Y.; Zhu, F.; Wang, W.; Chang, X. The influence of different concentrations of bio-organic fertilizer on cucumber Fusarium wilt and soil microflora alterations. PLoS ONE 2017, 12, e0171490. [Google Scholar] [CrossRef]
- Cai, F.; Chen, W.; Wei, Z.; Pang, G.; Li, R.; Ran, W.; Shen, Q. Colonization of Trichoderma harzianum strain SQR-T037 on tomato roots and its relationship to plant growth, nutrient availability and soil microflora. Plant Soil 2015, 388, 337–350. [Google Scholar] [CrossRef]
- Pang, G.; Cai, F.; Li, R.; Zhao, Z.; Li, R.; Gu, X.; Shen, Q.; Chen, W. Trichoderma-enriched organic fertilizer can mitigate microbiome degeneration of monocropped soil to maintain better plant growth. Plant Soil 2017, 416, 181–192. [Google Scholar] [CrossRef]
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H., Jr. The major facilitator superfamily (MFS) revisited. FEBS J. 2012, 279, 2022–2035. [Google Scholar] [CrossRef]
- Hinchliffe, P.; Greene, N.P.; Paterson, N.G.; Crow, A.; Hughes, C.; Koronakis, V. Structure of the periplasmic adaptor protein from a major facilitator superfamily (MFS) multidrug efflux pump. FEBS Lett. 2014, 588, 3147–3153. [Google Scholar] [CrossRef]
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS). Chem. Rev. 2021, 121, 5289–5335. [Google Scholar] [CrossRef]
- Li, P.; Gu, Y.; Li, J.; Xie, L.; Li, X.; Xie, J. Mycobacterium tuberculosis Major Facilitator Superfamily Transporters. J. Membr. Biol. 2017, 250, 573–585. [Google Scholar] [CrossRef]
- Nino-Gonzalez, M.; Novo-Uzal, E.; Richardson, D.N.; Barros, P.M.; Duque, P. More Transporters, More Substrates: The Arabidopsis Major Facilitator Superfamily Revisited. Mol. Plant 2019, 12, 1182–1202. [Google Scholar] [CrossRef]
- Sand, M.; de Berardinis, V.; Mingote, A.; Santos, H.; Goettig, S.; Mueller, V.; Averhoff, B. Salt adaptation in Acinetobacter baylyi: Identification and characterization of a secondary glycine betaine transporter. Arch. Microbiol. 2011, 193, 723–730. [Google Scholar] [CrossRef]
- Frawley, E.R.; Crouch, M.-L.V.; Bingham-Ramos, L.K.; Robbins, H.F.; Wang, W.; Wright, G.D.; Fang, F.C. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc. Natl. Acad. Sci. USA 2013, 110, 12054–12059. [Google Scholar] [CrossRef] [PubMed]
- Rios, G.; Cabedo, M.; Rull, B.; Yenush, L.; Serrano, R.; Mulet, J.M. Role of the yeast multidrug transporter Qdr2 in cation homeostasis and the oxidative stress response. FEMS Yeast Res. 2013, 13, 97–106. [Google Scholar] [CrossRef]
- Paul, S.; Alegre, K.O.; Holdsworth, S.R.; Rice, M.; Brown, J.A.; McVeigh, P.; Kelly, S.M.; Law, C.J. A single-component multidrug transporter of the major facilitator superfamily is part of a network that protects Escherichia coli from bile salt stress. Mol. Microbiol. 2014, 92, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Z.; Li, X.; Guo, L.; Li, C.; Lai, J.; Han, Y.; Ye, W.; Miao, Y.; Deng, M.; et al. A gene cluster for polyamine transport and modification improves salt tolerance in tomato. Plant J. 2024, 120, 1706–1723. [Google Scholar] [CrossRef]
- Ming, X.; Xiang, L. (Eds.) Genetic Analysis of Several Traits Related to Salt Tolerance in Tomato. In Proceedings of the 3rd Conference on Key Technology of Horticulture, Shenyang, China, 17–18 December 2011. [Google Scholar]
- Abdelaziz, M.E.; Abdelsattar, M.; Abdeldaym, E.A.; Atia, M.A.M.; Mahmoud, A.W.M.; Saad, M.M.; Hirt, H. Piriformospora indica alters Na+/K+ homeostasis, antioxidant enzymes and LeNHX1 expression of greenhouse tomato grown under salt stress. Sci. Hortic. 2019, 256, 108532. [Google Scholar] [CrossRef]
- Wu, F.; Chen, Z.; Xu, X.; Xue, X.; Zhang, Y.; Sui, N. Halotolerant Bacillus sp. strain RA coordinates myo-inositol metabolism to confer salt tolerance to tomato. J. Integr. Plant Biol. 2024, 66, 1871–1885. [Google Scholar] [CrossRef]
- Schmitz, L.; Yan, Z.; Schneijderberg, M.; de Roij, M.; Pijnenburg, R.; Zheng, Q.; Franken, C.; Dechesne, A.; Trindade, L.M.; van Velzen, R.; et al. Synthetic bacterial community derived from a desert rhizosphere confers salt stress resilience to tomato in the presence of a soil microbiome. ISME J. 2022, 16, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Tan, M.-Z.; Wang, R.-X.; Ye, F.-T.; Chen, Y.-P.; Luo, X.-M.; Feng, J.-X. Combination of genetic engineering and random mutagenesis for improving production of raw-starch-degrading enzymes in Penicillium oxalicum. Microb. Cell Factories 2022, 21, 272. [Google Scholar] [CrossRef]
- Miao, Y.; Xia, Y.; Kong, Y.; Zhu, H.; Mei, H.; Li, P.; Feng, H.; Xun, W.; Xu, Z.; Zhang, N.; et al. Overcoming diverse homologous recombinations and single chimeric guide RNA competitive inhibition enhances Cas9-based cyclical multiple genes coediting in filamentous fungi. Environ. Microbiol. 2021, 23, 2937–2954. [Google Scholar] [CrossRef]
- Choudhary, K.; Shih, N.P.; Deng, F.; Ledda, M.; Li, B.; Aviran, S. Metrics for rapid quality control in RNA structure probing experiments. Bioinformatics 2016, 32, 3575–3583. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhou, L.; Wang, X.; Wang, Z.; Li, P.; Dai, J.; Zhang, H.; Xie, Y. Nitratireductor luteus sp. nov. isolated from saline-alkali land. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2023, 116, 221–229. [Google Scholar] [CrossRef]
- Oren, A. Life in Hypersaline Environments. In Their World: A Diversity of Microbial Environments; Hurst, C.J., Ed.; Advances in Environtal Microbiology; Springer: Cham, Switzerland, 2016; pp. 301–339. [Google Scholar]
- Garg, N.; Manchanda, G. Role of Arbuscular Mycorrhizae in the Alleviation of Ionic, Osmotic and Oxidative Stresses Induced by Salinity in Cajanus cajan (L.) Millsp (pigeonpea). J. Agron. Crop Sci. 2009, 195, 110–123. [Google Scholar] [CrossRef]
- Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar] [CrossRef]
- Vindelov, J.; Arneborg, N. Saccharomyces cerevisiae and Zygosaccharomyces mellis exhibit different hyperosmotic shock responses. Yeast 2002, 19, 429–439. [Google Scholar] [CrossRef]
- Tanigawa, M.; Kihara, A.; Terashima, M.; Takahara, T.; Maeda, T. Sphingolipids Regulate the Yeast High-Osmolarity Glycerol Response Pathway. Mol. Cell. Biol. 2012, 32, 2861–2870. [Google Scholar] [CrossRef] [PubMed]
- Kumari, J.; Rathore, M.S. Na+/K+-ATPase a Primary Membrane Transporter: An Overview and Recent Advances with Special Reference to Algae. J. Membr. Biol. 2020, 253, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Padan, E.; Dwivedi, M. Overexpression, Isolation, Purification, and Crystallization of NhaA. In Methods Enzymology; Shukla, A.K., Ed.; Membrane Proteins—Engineering, Purification and Crystallization; Elsevier: Amsterdam, The Netherlands, 2015; Volume 557, pp. 135–148. [Google Scholar]
- Holtmann, G.; Bakker, E.P.; Uozumi, N.; Bremer, E. KtrAB and KtrCD: Two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 2003, 185, 1289–1298. [Google Scholar] [CrossRef]
- Rashid, A.N.; Janda, M.; Mahmud, M.Z.; Valentova, O.; Lenka, B.; Alekber, Q.A. Influence of Salt Stress on the FLG22-Induced ROS Production in Arabidopsis thaliana Leaves. Pak. J. Bot. 2021, 53, 1605–1610. [Google Scholar] [CrossRef]
- Swapnil, P.; Yadav, A.K.; Srivastav, S.; Sharma, N.K.; Srikrishna, S.; Rai, A.K. Biphasic ROS accumulation and programmed cell death in a cyanobacterium exposed to salinity (NaCl and Na2SO4). Algal Res.-Biomass Biofuels Bioprod. 2017, 23, 88–95. [Google Scholar]
- Wei, M.; Zhuang, Y.; Li, H.; Li, P.; Huo, H.; Shu, D.; Huang, W.; Wang, S. The cloning and characterization of hypersensitive to salt stress mutant, affected in quinolinate synthase, highlights the involvement of NAD in stress-induced accumulation of ABA and proline. Plant J. 2020, 102, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Srivastava, S.; Lokhande, V.H.; D’Souza, S.F.; Suprasanna, P. Salt stress reveals differential antioxidant and energetics responses in glycophyte (Brassica juncea L.) and halophyte (Sesuvium portulacastrum L.). Front. Environ. Sci. 2015, 3, 19. [Google Scholar] [CrossRef]
- Zhu, H.; Li, T.; Li, C.; Liu, Y.; Miao, Y.; Liu, D.; Shen, Q. Intracellular kynurenine promotes acetaldehyde accumulation, further inducing the apoptosis in soil beneficial fungi Trichoderma guizhouense NJAU4742 under acid stress. Environ. Microbiol. 2023, 25, 331–351. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Kato, T.; Watakabe, S.; Song, W.; Aikawa, S.; Furukawa, K. The respiratory chain provides salt stress tolerance by maintaining a low NADH/NAD+ ratio in Zymomonas mobilis. Microbiology 2015, 161, 2384–2394. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, D.; Wang, Z.; Guo, H.; Wang, Y.; Wang, X.; Dong, X. Oxidative stress response in atrazine-degrading bacteria exposed to atrazine. J. Hazard. Mater. 2012, 229, 434–438. [Google Scholar] [CrossRef]
- Gandhi, A.; Reichelt, M.; Goyal, D.; Vadassery, J.; Oelmueller, R. Trichoderma harzianum Protects the Arabidopsis Salt Overly Sensitive 1 Mutant Against Salt Stress. J. Plant Growth Regul. 2024. [Google Scholar] [CrossRef]
- Jalali, F.; Zafari, D.; Salari, H. Volatile organic compounds of some Trichoderma spp. increase growth and induce salt tolerance in Arabidopsis thaliana. Fungal Ecol. 2017, 29, 67–75. [Google Scholar] [CrossRef]
- Jiang, S.-Q.; Yu, Y.-N.; Gao, R.-W.; Wang, H.; Zhang, J.; Li, R.; Long, X.-H.; Shen, Q.-R.; Chen, W.; Cai, F. High-throughput absolute quantification sequencing reveals the effect of different fertilizer applications on bacterial community in a tomato cultivated coastal saline soil. Sci. Total Environ. 2019, 687, 601–609. [Google Scholar] [CrossRef]
- Li, H.; La, S.; Zhang, X.; Gao, L.; Tian, Y. Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J. 2021, 15, 2865–2882. [Google Scholar] [CrossRef]
- Zheng, Y.; Cao, X.; Zhou, Y.; Ma, S.; Wang, Y.; Li, Z.; Zhao, D.; Yang, Y.; Zhang, H.; Meng, C.; et al. Purines enrich root-associated Pseudomonas and improve wild soybean growth under salt stress. Nat. Commun. 2024, 15, 3520. [Google Scholar] [CrossRef]
- Margarita, S.; Sviatoslav, M.; Yelena, O.; Makhpal, Y. Can salt-adapted microorganisms alleviate salt stress in plants and enhance their non-specific resilience? Front. Agron. 2023, 5, 1287108. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, H. Plant salt response: Perception, signaling, and tolerance. Front. Plant Sci. 2023, 13, 1053699. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Guo, Z.; Zhou, L.; Xu, H.; Liu, C.; Yan, X. Advances in Identifying the Mechanisms by Which Microorganisms Improve Barley Salt Tolerance. Life 2024, 14, 6. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).